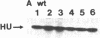Abstract
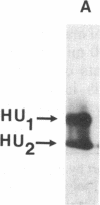
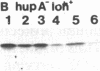
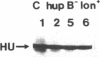
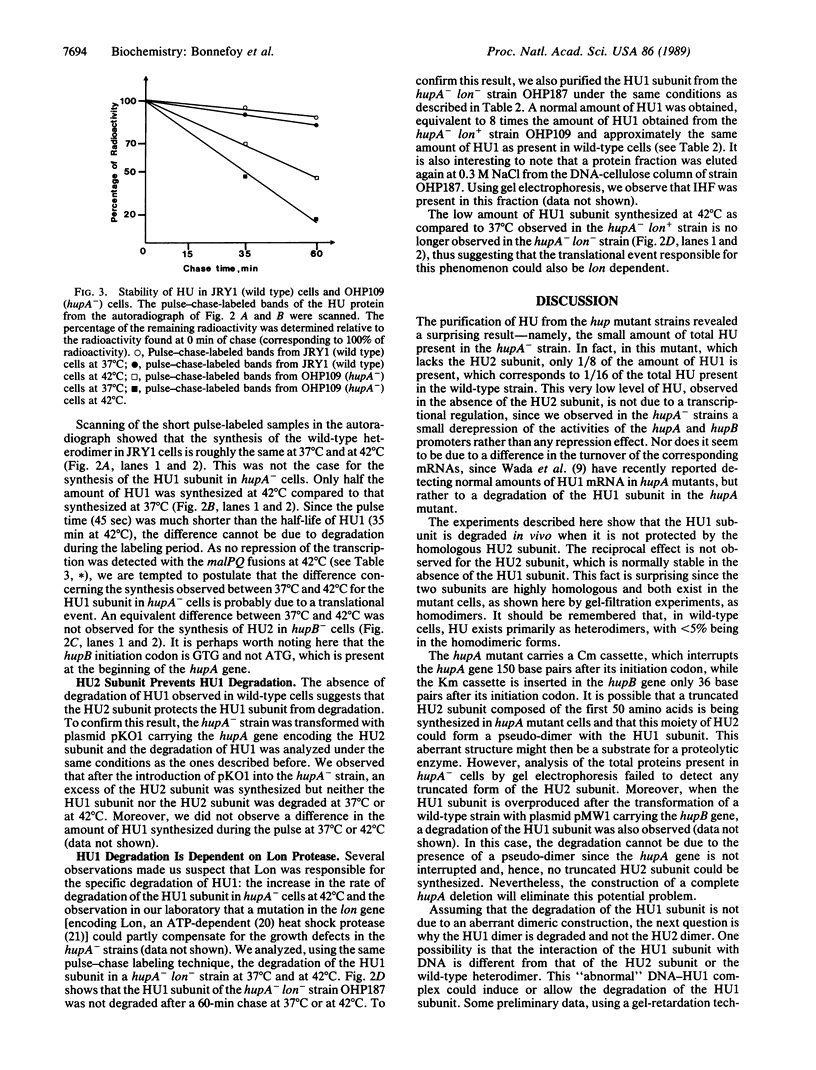
HU, the major DNA binding protein of Escherichia coli, exists in solution as a heterodimer composed of two highly homologous subunits: HU1, encoded by hupB; HU2, encoded by hupA. The purification of the HU protein from hupA- or hupB- bacteria showed that the hupB mutant strains synthesize normal amounts of the HU2 subunit (which corresponds to 60% of the total HU present in wild-type cells). On the contrary, the amount of HU1 present in hupA mutant strains corresponds to only 6% of the total HU present in wild-type cells. We showed by fusions of the hupB and hupA promoters to the malPQ operon that the absence of one subunit has no major effect on the transcription rate of the gene encoding the other subunit. Analysis of the stability of the HU1 and HU2 subunits, using pulse-chase labeling experiments, showed that the HU1 subunit is degraded specifically in the absence of the HU2 subunit and, moreover, that this degradation is dependent on the presence of the Lon protease.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Broyles S. S., Pettijohn D. E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol. 1986 Jan 5;187(1):47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- Chin D. T., Goff S. A., Webster T., Smith T., Goldberg A. L. Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La. J Biol Chem. 1988 Aug 25;263(24):11718–11728. [PubMed] [Google Scholar]
- Craigie R., Arndt-Jovin D. J., Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon N. E., Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(2):424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S. A., Casson L. P., Goldberg A. L. Heat shock regulatory gene htpR influences rates of protein degradation and expression of the lon gene in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6647–6651. doi: 10.1073/pnas.81.21.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S. A., Goldberg A. L. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985 Jun;41(2):587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- Holck A., Kleppe K. Affinity of protein HU for different nucleic acids. FEBS Lett. 1985 Jun 3;185(1):121–124. doi: 10.1016/0014-5793(85)80753-5. [DOI] [PubMed] [Google Scholar]
- Huisman O., Faelen M., Girard D., Jaffé A., Toussaint A., Rouvière-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J Bacteriol. 1989 Jul;171(7):3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y., Osato K., Wada M., Imamoto F. Cloning and sequencing of the HU-2 gene of Escherichia coli. Mol Gen Genet. 1987 Sep;209(2):408–410. doi: 10.1007/BF00329674. [DOI] [PubMed] [Google Scholar]
- Kano Y., Wada M., Nagase T., Imamoto F. Genetic characterization of the gene hupB encoding the HU-1 protein of Escherichia coli. Gene. 1986;45(1):37–44. doi: 10.1016/0378-1119(86)90129-0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Maurizi M. R. Degradation in vitro of bacteriophage lambda N protein by Lon protease from Escherichia coli. J Biol Chem. 1987 Feb 25;262(6):2696–2703. [PubMed] [Google Scholar]
- Maurizi M. R., Trisler P., Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J Bacteriol. 1985 Dec;164(3):1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Gottesman S. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc Natl Acad Sci U S A. 1983 Jan;80(2):358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987 Oct 9;51(1):101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A., Flamm E., Weisberg R. A., Miller H. I. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987 Sep;169(9):4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Kjeldgaard N. O. Native Escherichia coli HU protein is a heterotypic dimer. FEBS Lett. 1979 Oct 15;106(2):297–300. doi: 10.1016/0014-5793(79)80518-9. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Yaniv M., Germond J. E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979 Jun;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Schoemaker J. M., Gayda R. C., Markovitz A. Regulation of cell division in Escherichia coli: SOS induction and cellular location of the sulA protein, a key to lon-associated filamentation and death. J Bacteriol. 1984 May;158(2):551–561. doi: 10.1128/jb.158.2.551-561.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storts D. R., Markovitz A. Construction and characterization of mutations in hupB, the gene encoding HU-beta (HU-1) in Escherichia coli K-12. J Bacteriol. 1988 Apr;170(4):1541–1547. doi: 10.1128/jb.170.4.1541-1547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Ingigliardi D., Raibaud O. A convenient technique to compare the efficiency of promoters in Escherichia coli. Nucleic Acids Res. 1985 Aug 26;13(16):5919–5926. doi: 10.1093/nar/13.16.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Kano Y., Ogawa T., Okazaki T., Imamoto F. Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J Mol Biol. 1988 Dec 5;204(3):581–591. doi: 10.1016/0022-2836(88)90357-9. [DOI] [PubMed] [Google Scholar]