Abstract
Prior studies have demonstrated that inhibition of CHK1 can promote the activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) and phosphorylation of histone H2AX and that inhibition of poly(ADP-ribose) polymerase 1 (PARP1) can affect growth factor-induced ERK1/2 activation. The present studies were initiated to determine whether CHK1 inhibitors interacted with PARP1 inhibition to facilitate apoptosis. Transient expression of dominant-negative CHK1 raised basal ERK1/2 activity and prevented CHK1 inhibitors from activating ERK1/2. CHK1 inhibitors modestly increased the levels of PARP1 ADP ribosylation and molecular or small-molecule inhibition of PARP1 blocked CHK1 inhibitor-stimulated histone H2AX phosphorylation and activation of ERK1/2. Stimulated histone H2AX phosphorylation was ataxia telangiectasia-mutated protein-dependent. Multiple CHK1 inhibitors interacted in a greater than additive fashion with multiple PARP1 inhibitors to cause transformed cell-killing in short-term viability assays and synergistically killed tumor cells in colony-formation assays. Overexpression of BCL-xL or loss of BAX/BAK function, but not the function of BID, suppressed CHK1 inhibitor + PARP1 inhibitor lethality. Inhibition of BCL-2 family protein function enhanced CHK1 inhibitor + PARP1 inhibitor lethality and restored drug-induced cell-killing in cells overexpressing BCL-xL. Thus, PARP1 plays an important role in regulating the ability of CHK1 inhibitors to activate ERK1/2 and the DNA damage response. An inability of PARP1 to modulate this response results in transformed cell death mediated through the intrinsic apoptosis pathway.
Introduction
Multiple CHK1 inhibitors, including 7-hydroxystaurosporine (UCN-01) and 5-(3-fluoro-phenyl)-3-ureido-thiophene-2-carboxylic acid (S)-piperidin-3-ylamide hydrochloride (AZD7762) are currently being evaluated as antineoplastic agents in clinical trials, both alone and in combination with chemotherapeutic agents and ionizing radiation (Mow et al., 2001; Prudhomme, 2006). These agents are proposed to enhance the toxicity of chemotherapeutic drugs by inhibition of CHK1 with subsequent inappropriate cell cycle progression after DNA damage (Graves et al., 2000). Inhibition of CHK1 may directly promote the activation of the protein phosphatase CDC25C and can also interfere with CDC25C elimination by blocking its binding to 14-3-3 proteins and subsequent degradation (Peng et al., 1997; Graves et al., 2000). The CHK1 inhibitor UCN-01 is known to have many additional intracellular kinase targets, including the downstream effector of phosphatidylinositol 3-kinase, 3-phosphoinositide-dependent kinase 1, and “classic” protein kinase C isoforms (Komander et al., 2003).
Based on initial phase I studies, the maximal free achievable concentration of UCN-01 in human plasma was believed to be at or below ∼100 nM with a long plasma half-life because of UCN-01 binding to human α1 acidic glycoprotein (Fuse et al., 1998, 2005; Sausville et al., 1998; Hagenauer et al., 2004; Dees et al., 2005). Nonetheless, the combination of UCN-01 with topotecan or cisplatin has shown some preliminary evidence of patient activity (Hotte et al., 2006; Perez et al., 2006). We have noted in a wide variety of tumor cell types that UCN-01 activates the ERK1/2 pathway and that pharmacological or genetic inhibition of the ERK1/2 pathway dramatically potentiates apoptosis and suppresses tumor growth in vivo (Dai et al., 2001, 2002, 2008; McKinstry et al., 2002; Hawkins et al., 2005; Hamed et al., 2008). We have reported previously that the novel CHK1 inhibitor AZD7762 interacts with MEK1/2 inhibitors and farnesyltransferase inhibitors in a manner similar to that of UCN-01 to kill malignant hematopoietic cells in vitro (Pei et al., 2008). Thus, multiple CHK1 inhibitors can interact with multiple MEK1/2 inhibitors to promote tumor cell killing. It has been noted that CHK1 inhibition leads to the formation of single- and double-stranded DNA breaks, as judged by increased phosphorylation of the atypical histone H2AX, often referred to as γH2AX (Syljuåsen et al., 2005; Bucher and Britten, 2008). Thereafter, we also noted that UCN-01, in addition to activating ERK1/2, promotes increased phosphorylation of histone H2AX, indicative that DNA damage was occurring because of the inhibition of CHK1 function and that inhibition of ERK1/2 further enhanced histone H2AX phosphorylation before induction of apoptosis (Dai et al., 2008). Thus, CHK1-dependent regulation of ERK1/2 may play an important role in DNA damage-sensing and repair in transformed cells.
Cells contain multiple complexes of proteins that regulate DNA damage-sensing and repair responses. One central protein in the regulation of multiple forms of DNA repair processes is poly(ADP-ribose) polymerase 1 (PARP1), because of its central role in DNA repair, particularly nonhomologous end joining, and has been pharmacologically targeted for cancer therapeutics with inhibitors that block its ADP ribosylation and repair function (Schreiber et al., 2002, 2006; Rodon et al., 2009). Indeed, multiple PARP1 inhibitors have been developed with several in clinical use, including 10-(4-methyl-piperazin-1-ylmethyl)-2H-7-oxa-1,2-diaza-benzo[de]anthracen-3-one (GPI15427), 10-(aminomethyl)-4,5,6,7-tetrahydro-1H-cyclopenta[a]pyrrolo[3,4-c]carbazole-1,3(2H)-dione (CEP6800), 8-hydroxy-2-methyl-4(3H)-quinazolinone (NU1025), and olaparib (AZD2281) (Graziani and Szabó, 2005). Although initially noted for its role in the repair of DNA strand breaks, PARP1 has been shown to have a much wider range of biological actions and participates in the regulation of transcription, DNA replication, apoptosis, and modulating reactive oxygen species levels (Spina Purrello et al., 2002; McCabe et al., 2006; Quénet et al., 2009). We and others have noted that signaling from the epidermal growth factor receptor can regulate PARP1 activity, in part through regulation of the ERK1/2 pathway (Hagan et al., 2007).
Based on the fact that CHK1 inhibitors activate ERK1/2 and promote H2AX phosphorylation, and that PARP1 function has been linked to ERK1/2 signaling, we investigated whether the inhibition of PARP1 function modulated the activation of cell signaling pathways induced by CHK1 inhibitor treatment. Our data demonstrate that CHK1-induced phosphorylation of ERK1/2 and H2AX is blunted or abolished when PARP1 function or expression is reduced. A reduced ability of cells to increase ERK1/2 activation correlated with a synergistic induction of cell-killing that was mediated through the intrinsic apoptosis pathway.
Materials and Methods
Materials.
Phospho-/total-ERK1/2 antibodies, GAPDH, 10H ADP ribosylation, PARP1, phospho-/total-CHK1, ataxia telangiectasia-mutated (ATM), and phospho-/total-H2AX antibodies were all purchased from Cell Signaling Technology, Inc. (Danvers, MA). Terminal deoxynucleotidyl transferase dUTP nick-end labeling kits were purchased from PerkinElmer Life and Analytical Sciences (Waltham, MA) and Roche Applied Science (Mannheim, Germany), respectively. Trypsin-EDTA, RPMI 1640 medium, and penicillin-streptomycin were purchased from Invitrogen (Carlsbad, CA). MDA-MB-231, MCF7, SKBR3, BT474, and PANC1 cells were purchased from the American Type Culture Collection (Manassas, VA). The 4T1 line was kindly provided by Dr. A. Larner (Virginia Commonwealth University, Richmond, VA). Simian virus 40 Large T mouse embryonic fibroblasts lacking the expression of various proapoptotic BH3 domain proteins were kindly provided by Dr. S. Korsmeyer (Harvard University, Boston, MA). The plasmid to express dominant-negative CHK1 was kindly supplied by Dr. Steven Grant (Virginia Commonwealth University). 2-(2-Chloro-4-iodo-phenylamino)-N-cyclopropylmethoxy-3,4-difluoro- benzamide(PD184352), 2′-amino-3′-methoxyflavone (PD98059), NU1025, N-(5,6-dihydro-6-oxo-2-phenanthridinyl)-2-acetamide hydrochloride (PJ34), and 4-(3′-chloroanilino)-6,7-dimethoxy-quinazoline (AG1478) were purchased from Calbiochem (San Diego, CA). The validated siRNA molecules used knockdown ATM from QIAGEN (Valencia, CA). UCN-01 was purchased from Sigma-Aldrich (St. Louis, MO). AZD7762 and AZD2281 were purchased from Axon Medchem (Groningen, the Netherlands). UCN-01 was purchased from Sigma-Aldrich. Obatoclax (GX15-070) was supplied by GeminX Pharmaceuticals (Malvern, PA).
Culture and In Vitro Exposure of Cells to Drugs.
Tumor cells for the studies in this manuscript were cultured at 37°C [5% (v/v) CO2] in vitro using RPMI 1640 medium supplemented with 10% (v/v) fetal calf serum. In vitro vehicle/UCN-01/PD184352/AZD7762/PJ34 and so forth treatment was from a 100 mM stock solution of each drug, and the maximal concentration of vehicle (DMSO) in media was 0.02% (v/v).
Cell Treatments, SDS-PAGE, and Western Blot Analysis.
For in vitro analyses of short-term apoptosis effects, cells were treated with vehicle/drugs or their combination for the indicated times. Cells for colony formation assays were plated at 250 to 4000 cells/well in sextuplicate and for in vitro assays 14 h after plating were treated with the individual or the drug combination(s) at a fixed increasing dose ratio according to the method of Chou and Talalay (1984) for 48 h followed by drug removal. Then, 10 to 14 days after exposure or tumor isolation, plates were washed in phosphate-buffered saline, fixed with methanol, and stained with a filtered solution of crystal violet [5% (w/v)]. After washing with tap water, the colonies were counted both manually (by eye) and digitally using a ColCount plate reader (Oxford Optronics, Oxford, England). Data presented are the arithmetic mean (± S.E.M.) from both counting methods from multiple studies. Colony formation was defined as a colony of 50 cells or greater.
For SDS-PAGE and immunoblotting, cells were plated at 5 × 105 cells/cm2 and treated with therapeutic drugs at the indicated concentrations, and after the indicated time of treatment, they were lysed with whole-cell lysis buffer (0.5 M Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, and 0.02% bromphenol blue), and the samples were boiled for 30 min. The boiled samples were loaded onto 10 to 14% SDS-PAGE, and electrophoresis was run overnight. Proteins were electrophoretically transferred onto 0.22 μm of nitrocellulose and immunoblotted with various primary antibodies against different proteins. All immunoblots were visualized by use of an Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
Short-Term Cell Viability Assays after Drug Exposure.
Cells were isolated at the indicated times by trypsinization and either were subjected to trypan blue cell viability assay by counting in a light microscope or were fixed to slides and stained using a commercially available Diff-Quick (GEIMSA, Oviedo, Spain) assay kit.
Recombinant Adenoviral Vectors: Infection In Vitro.
We generated and purchased recombinant adenoviruses noted previously to express constitutively activated MEK1 or AKT proteins and mitochondrial protective protein BCL-xL (Vector Biolabs, Philadelphia, PA). Unless otherwise stated, cells were infected with these adenoviruses at an approximate multiplicity of infection (m.o.i.) of 50. As noted above, cells were further incubated for 24 h to ensure adequate expression of transduced gene products before drug exposures.
siRNA Transfection In Vitro.
Approximately a 10 nM concentration of a defined prevalidated siRNA (Ambion, Austin, TX) was diluted into 50 μl of growth media lacking FBS and penicillin/streptomycin. Based on the manufacturer's instructions, an appropriate amount of Lipofectamine 2000 reagent (usually 1 μl) (Invitrogen, Carlsbad, CA) was diluted into a separate vial containing media lacking FBS or penicillin/streptomycin. The two solutions were incubated separately at room temperature for 5 min and then mixed together (vortexed) and incubated at room temperature for 30 min. The mixture was added to each well (slide or 12-well plate) containing an appropriate amount (∼ 0.5 ml) of penicillin/streptomycin and FBS-free medium. Cells were incubated for 2 to 4 h at 37°C with gentle rocking. Media were then replaced with 1 ml of 1× penicillin/streptomycin and FBS-containing media.
Data Analysis.
Comparison of the effects between various in vitro drug treatments was performed after analysis of variance using the Student's t test. Differences with a p value of <0.05 were considered statistically significant. Experiments shown are the means of multiple individual points from multiple studies (± S.E.M.). Median dose-effect isobologram colony-formation analyses to determine synergism of drug interaction were performed according to the methods of Chou and Talalay (1984) using the CalcuSyn program for Windows (Biosoft, Cambridge, UK). Cells were treated with agents at an escalating fixed concentration drug dose. A combination index of <1.00 indicates synergy of interaction between the two drugs; a combination index of ∼1.00 indicates an additive interaction; a combination index (CI) value of >1.00 indicates antagonism of action between the agents.
Results
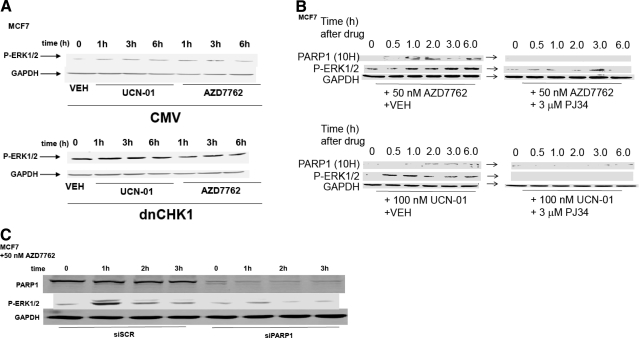
We have published previously that MEK1/2 inhibitors interact with UCN-01 in a synergistic manner to kill mammary tumor cells in vitro and in vivo (Hamed et al., 2008). To prove or refute whether UCN-01 and a chemically unrelated CHK1 inhibitor, AZD7762, were mediating their ERK1/2-activating effects via inhibition of CHK1, we made use of a plasmid to express dominant-negative CHK1. Expression of a dominant-negative CHK1 protein in MCF7 cells enhanced basal levels of ERK1/2 phosphorylation within 24 h and blunted the ability of UCN-01 or AZD7762 to stimulate ERK1/2 phosphorylation (Fig. 1A).
Fig. 1.
Inhibition of CHK1 enhances ERK1/2 activation in a PARP-1-dependent fashion. A, MCF7 cells were transfected with either an empty vector control plasmid or a plasmid to express dominant-negative CHK1 (dnCHK1). Twenty-four hours after transfection, cells were treated with vehicle (VEH, DMSO), UCN-01 (100 nM), or AZD7762 (50 nM). Cells were isolated at the indicated time points and subjected to SDS-PAGE followed by immunoblotting to determine the phosphorylation of ERK1/2 (P-ERK1/2) or the expression of GAPDH. Data are from a representative of two separate studies. B, MCF7 cells were treated with vehicle (VEH, DMSO) or the PARP-1 inhibitor PJ34 (3 μM) followed 30 min later by CHK1 inhibitors UCN-01 (100 nM) or AZD7762 (50 nM). Cells were isolated 0 to 6 h after CHK1 inhibitor addition, as indicated. Cell lysates were subjected to SDS-PAGE followed by immunoblotting to determine the phosphorylation of ERK1/2 (P-ERK1/2), the ADP ribosylation of PARP-1 (10H antibody), or the expression of GAPDH. Data are from a representative of three separate studies. C, MCF7 cells were transfected with either a scrambled nonspecific siRNA (siSCR, 20 nM) or an siRNA to knock down the expression of PARP-1. Twenty-four hours after transfection, cells were treated with AZD7762 (50 nM). Cells were isolated at the indicated time points and subjected to SDS-PAGE followed by immunoblotting to determine the phosphorylation of ERK1/2 (P-ERK1/2), the expression of PARP-1, or the expression of GAPDH. Data are from a representative of two separate studies.
UCN-01 was shown previously in malignant blood tumor cells to increase the phosphorylation of histone H2AX, indicative of DNA damage (Dai et al., 2008). Based on this observation, we determined whether another marker of the DNA damage response in tumor cells, PARP1 ADP ribosylation, could be visualized. Treatment of MCF7 breast cancer cells with either UCN-01 or AZD7762 increased PARP1 ADP ribosylation, as judged using the anti-poly(ADP ribose) 10H antibody (Fig. 1B). It is noteworthy that increased ERK1/2 phosphorylation correlated with elevated PARP1 (10H) reactivity. Coexposure of cells to the PARP inhibitor PJ34 blocked CHK1 inhibitor-induced PARP1 activation and PARP1 ADP ribosylation. To confirm our findings using a molecular approach, we knocked down the expression of PARP1. Knockdown of PARP1 expression in breast cancer cells significantly reduced AZD7762-induced activation of ERK1/2 (Fig. 1C). Thus, CHK1 inhibitor-induced ERK1/2 activation requires functional expression of PARP1.
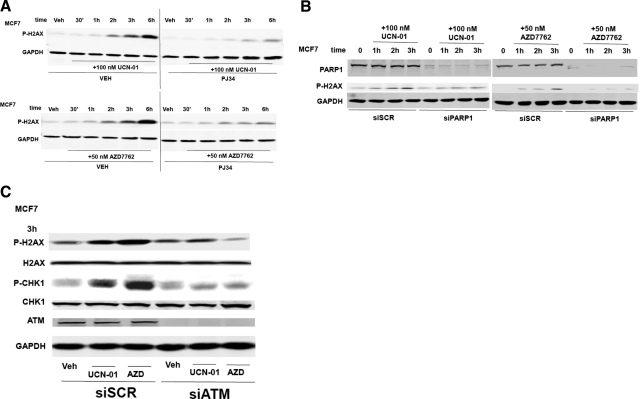
In breast cancer cells, UCN-01 and AZD7762 rapidly increased H2AX phosphorylation (Fig. 2, A and B). Inhibition of PARP1, either by use of PJ34 or by knockdown of PARP1 expression, significantly reduced the induction of H2AX phosphorylation by the CHK1 inhibitors. In other model systems, phosphorylation of H2AX has been shown to be mediated by the ATM protein, and PARP1 plays a key role in permitting ATM activation. Knockdown of ATM expression prevented UCN-01 or AZD7762 from increasing H2AX phosphorylation (Fig. 2C). It is noteworthy that both CHK1 inhibitors promoted a compensatory increase in CHK1 phosphorylation, which was also ATM-dependent. Together, the data in Figs. 1 and 2 demonstrate that CHK1 inhibitor-mediated phosphorylation of both ERK1/2 and H2AX requires PARP1 function and that phosphorylation of H2AX after CHK1 inhibitor exposure requires expression of ATM.
Fig. 2.
PARP-1 is essential for CHK1 inhibitor-induced phosphorylation of histone H2AX. A, MCF7 cells were treated with vehicle (VEH, DMSO) or the PARP-1 inhibitor PJ34 (3 μM) followed 30 min later by CHK1 inhibitors UCN-01 (100 nM) or AZD7762 (50 nM). Cells were isolated 0 to 6 h after CHK1 inhibitor addition, as indicated. Cell lysates were subjected to SDS-PAGE followed by immunoblotting to determine the phosphorylation of H2AX or the expression of GAPDH. Data are from a representative of three separate studies. B, MCF7 cells were transfected with either a scrambled nonspecific siRNA (siSCR, 20 nM) or an siRNA known to induce down-expression of PARP-1. Twenty-four hours after transfection, cells were treated with UCN-01 (100 nM) or AZD7762 (50 nM). Cells were isolated at the indicated time points and subjected to SDS-PAGE followed by immunoblotting to determine the phosphorylation of H2AX, the expression of PARP-1, or the expression of GAPDH. Data are from a representative of two separate studies. C, MCF7 cells were transfected with nonspecific siRNA control (siSCR) or an siRNA to knock down ATM (siATM). Twenty-four hours after transfection, cells were treated with vehicle (VEH, DMSO) or CHK1 inhibitors UCN-01 (100 nM) or AZD7762 (50 nM). Cells were isolated 3 h after CHK1 inhibitor addition, as indicated. Cell lysates were subjected to SDS-PAGE followed by immunoblotting to determine the phosphorylation of H2AX/CHK1 or the expression of GAPDH, ATM, CHK1, and H2AX. Data are from a representative of three separate studies.
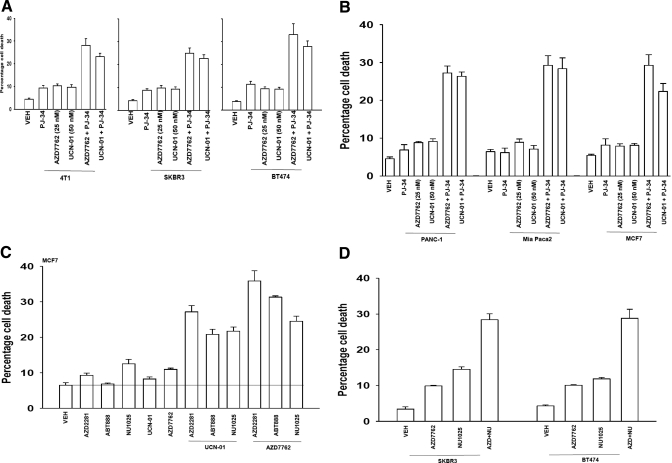
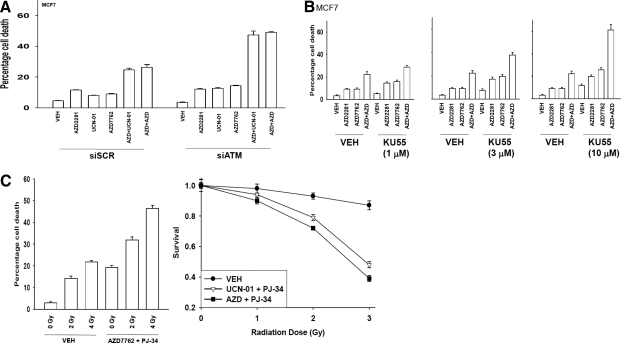
We next explored the survival of PARP1-inhibited cells after CHK1 inhibitor treatment. Inhibition of PARP1 promoted CHK1 inhibitor lethality in a range of breast cancer cells (Fig. 3A). Very similar data were obtained in pancreatic cancer cells (Fig. 3B). In agreement with data using short-term viability assays, median dose-effect colony formation assays, as judged by CI values of less than 1.00, demonstrated a synergy of drug interaction in killing tumor cells (Tables 1 and 2). PARP1 inhibitors are presently generating a significant level of clinical interest, and we determined whether other more clinically relevant PARP1 inhibitors recapitulated the lethal effects of PJ34 or siRNA knockdown of PARP1. The clinically relevant PARP1 inhibitors veliparib (ABT888), NU1025, and AZD2281 enhanced the lethality of UCN-01 and of AZD7762 in breast cancer cells (Fig. 3C). Similar data were obtained in other breast cancer cells (Fig. 3D). Because CHK1 inhibitor-induced ATM activation was PARP1-dependent, we determined the impact of inhibiting ATM function on drug combination lethality. Knockdown of ATM expression significantly enhanced the lethality of PARP1 inhibitor + CHK1 inhibitor lethality, suggesting that in the absence of PARP1 + CHK1 signaling, the compensatory activation of ATM is a protective signal (Fig. 3E). Similar data were obtained when a clinically relevant ATM inhibitor was used instead of siRNA knockdown (Fig. 3F). Because manipulation of PARP1/CHK1 function was leading to a DNA damage response in tumor cells, and inhibition of ATM further enhanced this effect, we next determined whether drug exposure enhanced tumor cell radiosensitivity. In both short-term and long-term colony assays, inhibition of PARP1 + CHK1 function enhanced the toxic effects of exposure to ionizing radiation (Fig. 3G).
Fig. 3.
PARP-1 inhibition enhances the toxicity of CHK1 inhibitors in transformed cells. A, breast cancer cells were plated in triplicate and treated with vehicle (VEH, DMSO), PJ34 (3 μM), UCN-01 (50 nM), or AZD7762 (25 nM). Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay is the mean of all data points from three studies ± S.E.M. B, MCF7 breast cancer and PANC-1 and MiaPaca2 pancreatic cancer cells were plated in triplicate and treated with vehicle (VEH, DMSO), PJ34 (3 μM), UCN-01 (50 nM), or AZD7762 (25 nM). Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay is the mean of all data points from three studies ± S.E.M. C, MCF7 cells were plated in triplicate and treated with vehicle (VEH, DMSO), NU1025 (10 μM), AZD2281 (0.5 μM), ABT888 (1.0 μM), and/or AZD7762 (25 nM), or UCN-01 (50 nM). Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay is the mean of all data points from three studies ± S.E.M. D, SKBR3 and BT474 cells were plated in triplicate and treated with vehicle (VEH, DMSO), NU1025 (10 μM), and/or AZD7762 (25 nM). Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay is the mean of all data points from three studies ± S.E.M. E, MCF7 cells were transfected with nonspecific siRNA control (siSCR) or an siRNA to knock down ATM (siATM). Twenty-four hours after transfection, cells were treated with vehicle (VEH, DMSO) and/or by AZD7762 (25 nM) or UCN-01 (50 nM). Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay is the mean of all data points from three studies ± S.E.M. F, MCF7 cells were plated in triplicate and treated with vehicle (VEH, DMSO), AZD2281 (0.5 μM), AZD7762 (25 nM), or AZD2281 + AZD7762 in combination. Thirty minutes after exposure, cells are treated with vehicle (DMSO) or with increasing concentrations of the ATM inhibitor 2-(4-morpholinyl)-6-(1-thianthrenyl)-4H-pyran-4-one (KU55933) (1–10 μM). Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay is the mean of all data points from three studies ± S.E.M. G, left, MCF7 cells were plated and treated with vehicle (VEH, DMSO) or the PARP-1 inhibitor PJ34 (3 μM) followed 30 min later by CHK1 inhibitor AZD7762 (25 nM). Cells were irradiated (4 Gy) and used for short-term viability assays 48 h after exposure and for viability determined using trypan blue exclusion. Right, MCF7 cells were plated in sextuplicate as single cells, and 12 h after plating, cells were treated with vehicle (VEH, DMSO) or the PARP-1 inhibitor PJ34 (3 μM) followed 30 min later by CHK1 inhibitors UCN-01 (50 nM) or AZD7762 (25 nM). Cells were irradiated 30 min after drug additions. Forty-eight hours after drug exposure, the media were changed, and cells were cultured in drug-free media for an additional 10 to 14 days (n = 2 ± S.E.M.).
TABLE 1.
CHK1 inhibitors synergize with PARP1 inhibitors to kill pancreatic carcinoma cells
PANC-1 (pancreatic) and MiaPaca2 (pancreatic) carcinoma cells were plated as single cells (250–2000 cells/well) in sextuplicate, and 12 h after this plating, the infected cells were treated with vehicle (DMSO), the PARP1 inhibitor PJ34 (0.75–3.0 μM), the CHK1 inhibitors UCN-01 (22.5–37.5 nM) or AZD7762 (6.25–25.0 nM), or the combinations of the PARP1 and CHK1 inhibitor drugs combined, as indicated at a fixed concentration ratio to perform median dose-effect analyses for the determination of synergy. Forty-eight hours after drug exposure, the media were changed, and cells were cultured in drug-free media for an additional 10 to 14 days. Cells were fixed, stained with crystal violet, and colonies of >50 cells/colony were counted. Colony formation data were entered into the CalcuSyn program, and CI values were determined. A CI value of less than 1.00 indicates synergy.
| AZD7762 | PJ34 | Fa | CI | UCN01 | PJ34 | Fa | CI | |
|---|---|---|---|---|---|---|---|---|
| nM | μM | nM | μM | |||||
| Panc1 | 6.25 | 0.75 | 0.26 | 0.40 | 12.5 | 0.75 | 0.27 | 0.41 |
| 12.5 | 1.50 | 0.39 | 0.48 | 25.0 | 1.50 | 0.42 | 0.44 | |
| 25.0 | 3.00 | 0.62 | 0.43 | 50.0 | 3.00 | 0.54 | 0.58 | |
| MiaPaca2 | 6.25 | 0.75 | 0.38 | 0.51 | 12.5 | 0.75 | 0.37 | 0.45 |
| 12.5 | 1.50 | 0.45 | 0.68 | 25.0 | 1.50 | 0.44 | 0.68 | |
| 25.0 | 3.00 | 0.68 | 0.62 | 50.0 | 3.00 | 0.74 | 0.40 |
Fa, fraction affected.
TABLE 2.
CHK1 inhibitors synergize with PARP1 inhibitors to kill mammary carcinoma cells
MCF7 carcinoma cells plated as single cells (250–2000 cells/well) in sextuplicate, and 12 h after this plating, the infected cells were treated with vehicle (VEH, DMSO), the PARP1 inhibitor PJ34 (0.75–3.0 μM), the CHK1 inhibitors UCN-01 (22.5–37.5 nM) or AZD7762 (6.25–25.0 nM), or the combinations of the PARP1 and CHK1 inhibitor drugs, as indicated at a fixed concentration ratio to perform median dose-effect analyses for the determination of synergy. Forty-eight hours after drug exposure, the media were changed, and cells were cultured in drug-free media for an additional 10 to 14 days. Cells were fixed, stained with crystal violet, and colonies of >50 cells/colony were counted. Colony formation data were entered into the CalcuSyn program, and CI values were determined. A CI value of less than 1.00 indicates synergy.
| AZD7762 | PJ34 | Fa | CI | UCN01 | PJ34 | Fa | CI |
|---|---|---|---|---|---|---|---|
| nM | μM | nM | μM | ||||
| 6.25 | 0.75 | 0.16 | 0.49 | 12.5 | 0.75 | 0.17 | 0.53 |
| 12.5 | 1.50 | 0.33 | 0.52 | 25.0 | 1.50 | 0.32 | 0.54 |
| 25.0 | 3.00 | 0.51 | 0.54 | 50.0 | 3.00 | 0.43 | 0.47 |
Fa, fraction affected.
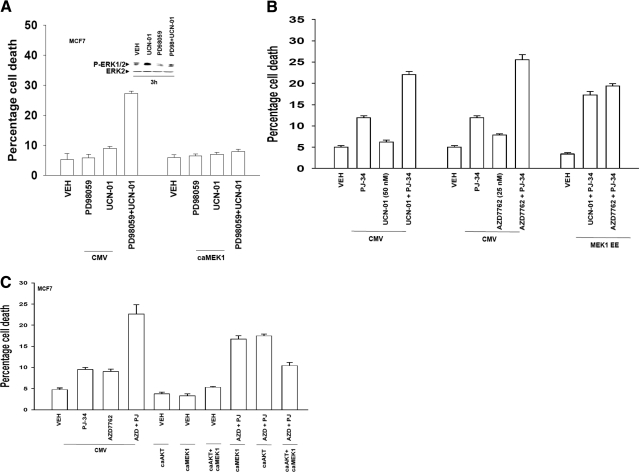
In Figs. 1 and 2, we noted that loss of PARP1 function suppressed CHK1 inhibitor-induced activation of ERK1/2. Inhibition of CHK1 inhibitor-induced ERK1/2 activation using an MEK1/2 inhibitor enhanced CHK1 inhibitor toxicity, an effect that was blocked by overexpressing an activated form of MEK1 (Fig. 4A). However, expression of a constitutively activated MEK1 protein only partially suppressed the toxicity of PARP1 inhibitor + CHK1 inhibitor treatment (Fig. 4B). Expression of an activated form of AKT significantly suppressed PARP1 inhibitor + CHK1 inhibitor lethality, and combined expression of activated MEK1 and AKT proteins abolished drug toxicity (Fig. 4C).
Fig. 4.
Inhibition of CHK1 inhibitor-induced ERK1/2 activation is not the sole molecular mechanism of drug interaction. A, MCF7 cells were infected in triplicate at an m.o.i. of 50 with either an empty vector adenovirus (CMV) or with an adenovirus to express constitutively activated MEK1 EE. Twenty-four hours after infection, cells were treated with vehicle (VEH, DMSO), PD98059 (25 μM), or UCN-01 (50 nM) as indicated. Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay are the means of all data points from three studies ± S.E.M. B, MCF7 cells were infected in triplicate at an m.o.i. of 50 with either an empty vector adenovirus (CMV) or with an adenovirus to express constitutively activated MEK1 EE. Twenty-four hours after infection, cells were treated as indicated with vehicle (VEH, DMSO), PJ34 (3 μM), UCN-01 (50 nM), or AZD7762 (25 nM). Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay are the means of all data points from three studies ± S.E.M. C, MCF7 cells were infected in triplicate at an m.o.i. of 50 with either an empty vector adenovirus (CMV) or with adenoviruses to express constitutively activated MEK1 EE and/or constitutively activated AKT. Twenty-four hours after infection, cells were treated as indicated with vehicle (VEH, DMSO), PJ34 (3 μM), and/or AZD7762 (25 nM) as indicated. Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay are the means of all data points from three studies ± S.E.M.
Based on the cell survival findings in previous figures, including evidence that ERK1/2 signaling promoted MCL-1 and BCL-xL expression, we determined the apoptosis pathway(s) being induced by the combination of CHK1 and PARP1 inhibitors. Transformed mouse embryonic fibroblasts genetically deleted for BAX/BAK were resistant to drug combination lethality (Fig. 5A). In contrast, cells that were deleted for the caspase 8 substrate BID or for BIM did not exhibit any reduction in drug lethality (Fig. 5A, data not shown). Overexpression of BCL-2 family proteins has been shown to block CHK1 inhibitor + MEK1/2 inhibitor lethality (Grant and Dent, 2007). Overexpression of BCL-xL suppressed CHK1 inhibitor + PARP1 inhibitor lethality that was reversed by the addition of a small-molecule inhibitor of BCL-2 family proteins, 2-amino-6-bromo-a-cyano-3-(ethoxycarbonyl)-4H-1-benzopy ran-4-acetic acid ethyl ester (HA14-1) (Fig. 5B). Data similar to that for HA14-1 were obtained when a clinically relevant BCL-2/BCL-xL/MCL-1 inhibitor, obatoclax (GX15-070), was used. Together, these findings demonstrate that CHK1 inhibitors synergize with PARP1 inhibition to kill multiple carcinoma cell types via the intrinsic apoptosis pathway.
Fig. 5.
Loss of BAX/BAK function abolishes the toxic interaction between CHK1 inhibitors and PARP-1 inhibitors; cell killing is potentiated by inhibitors of BCL-2/BCL-xL function. A, transformed mouse embryonic fibroblasts [MEF; wild type, WT; deleted for BAX and BAK, BAX/BAK(−/−); deleted for BID, BID(−/−)] were plated in triplicate and treated with vehicle (VEH, DMSO), PJ34 (3 μM), UCN-01 (50 nM), or AZD7762 (25 nM). Cells were isolated 48 h after exposure, and viability was determined using trypan blue exclusion. Data for each assay are the means of all data points from three studies ± S.E.M. B, PANC-1 and MCF7 cells were infected with either an empty vector adenovirus (CMV) or with an adenovirus to express BCL-XL. Twenty-four hours after infection, cells were pretreated for 30 min with vehicle (VEH, DMSO) or HA14-1 (10 μM) and then treated as indicated with vehicle (VEH, DMSO) or PJ34 (3 μM) and UCN-01 (50 nM). Cells were isolated 48 h after exposure, and viability was determined in triplicate using trypan blue exclusion. Data for each assay are the means of all data points from two studies ± S.E.M. C, MCF7 cells were infected with either an empty vector adenovirus (CMV) or with an adenovirus to express BCL-XL. Twenty-four hours after infection, cells were pretreated for 30 min with vehicle (VEH, DMSO) or obatoclax (GX15-070, 50 nM) and then treated as indicated with vehicle (VEH, DMSO) or PJ34 (3 μM) and UCN-01 (50 nM). Cells were isolated 48 h after exposure, and viability was determined in triplicate using trypan blue exclusion. Data for each assay is the mean of all data points from two studies ± S.E.M. *, p < 0.05 less than corresponding value in empty vector virus-infected cells; #, p < 0.05 greater than corresponding value in empty vector-infected cells not treated with obatoclax; $, greater than corresponding value in BCL-xL-infected cells treated with obatoclax.
Discussion
Previous studies by this group have argued that MEK1/2 inhibitors or farnesyltransferase inhibitors interact with the CHK1 inhibitor UCN-01 to promote tumor cell-specific killing in a wide variety of malignancies including breast, prostate, and multiple hematological cell types (Grant and Dent, 2007). The net output of the cytoprotective RAS-MEK1/2-ERK1/2 pathway has been shown previously to be a critical determinant of tumor cell survival (Riches et al., 2008). Furthermore, activation of this cascade has been observed as a compensatory response of tumor cells to various environmental stresses, including cytotoxic drugs. The present studies were initiated to determine whether CHK1 inhibitors, which cause ERK1/2 activation and a DNA damage response, interact with inhibitors of PARP1; PARP1 is a protein that plays a key role in DNA repair and regulation of ERK1/2 signaling. Based on the expression of a dominant-negative CHK1 protein, UCN-01 and AZD7762-induced activation of ERK1/2 was dependent on inhibition of CHK1; furthermore, expression of dominant-negative CHK1 enhanced basal levels of ERK1/2 phosphorylation arguing for a central regulatory role between CHK1 and the RAF-MEK-ERK1/2 pathway. Thus, our findings argue that inhibition of CHK1 is essential, in part, for the activation of ERK1/2 to occur by CHK1 inhibitors.
Suppression of CHK1 function has been shown to cause DNA damage in transformed cells as judged by increased H2AX phosphorylation. The damage-stimulated phosphorylation of H2AX has been associated with the actions of the ATM protein (Riches et al., 2008). An additional hallmark of the cellular DNA damage response is activation of PARP1 (Rodon et al., 2009). PARP1 activation results in ADP ribosylation of multiple DNA repair complex proteins, transcription factors, and PARP1 itself. As a result of this effect on multiple repair proteins, loss of PARP1 function promotes genomic instability and leads to hyperactivation of CHK1 with increased cell numbers in G2 phase (Lu et al., 2006). This is also of interest because other groups have postulated the chemotherapy-sensitizing effect of CHK1 inhibitors is due to abrogation of the G2 checkpoint (Prudhomme, 2006). In our studies, two chemically distinct CHK1 inhibitors rapidly promoted H2AX phosphorylation and increased PARP1 ADP ribosylation. Inhibition of PARP1 function blocked CHK1 inhibitor-induced H2AX phosphorylation and blocking CHK1 inhibitor-induced activation of ERK1/2. The inhibition of induced H2AX phosphorylation by PARP inhibition is probably explained by the requirement that ATM has for PARP1 function in being able to become activated after DNA damage and in our studies, knockdown of ATM blocked CHK1 inhibitor-induced H2AX phosphorylation (Haince et al., 2007). And of note, ATM/checkpoint pathway signaling has been linked previously in one of our prior studies to the regulation of the ERK1/2 pathway (Golding et al., 2007).
We presented evidence previously that inhibition of CHK1-induced ERK1/2 activation further enhanced H2AX phosphorylation, indicative that loss of ERK1/2 signaling increased the amount of DNA damage being induced by the CHK1 inhibitor (Dai et al., 2008). This correlated with a subsequent profound induction of apoptosis. The present work demonstrated that inhibition of PARP1 blocked not only ERK1/2 activation but also H2AX phosphorylation. However, despite blocking the apparent DNA damage-signaling response, we found that PARP1 inhibitors significantly enhanced the lethality of CHK1 inhibitors. Based on the use of BAX/BAK(−/−) cells and the expression of BCL-xL, the induction of mitochondrial dysfunction was shown to play a primary role in the synergistic induction of cell killing after treatment of cells with a PARP1 inhibitor and CHK1 inhibitors. It is noteworthy that mammary carcinoma cells with very low basal levels of ERK1/2 activity and that are relatively noninvasive such as MCF7 were apparently as susceptible to being killed by exposure to PARP1 inhibitor and CHK1 inhibitors as were mammary carcinoma cells and pancreatic cancer cells with very high basal levels of ERK1/2 activity and that are highly invasive, such as MDA-MB-231 and PANC-1. Simian virus 40 large T antigen-transformed fibroblasts that are not tumorigenic in mice were also sensitive to the drug schedule, although in agreement with prior findings, we have found that multiple nontransformed/nonestablished cell types such as primary mammary epithelial cells and CD34+ stem cells are insensitive to being killed by the CHK1 inhibitor + PARP1 pathway inhibitor combined drug exposure regimen (A. Yacoub and P. Dent, unpublished observations). Together, our data suggest that CHK1 function plays a key role in maintaining cell viability in transformed cells and does so, in part, by regulating ERK1/2 pathway signaling as part of a DNA damage response.
Overexpression of mitochondrial BCL-2 family members has been shown in many tumor cell systems to raise the apoptotic threshold of tumor cells (Cory and Adams, 2005; Lee and Gautschi, 2006; Konopleva et al., 2008). Because the potentiation of CHK1 inhibitor lethality by PARP1 inhibition occurs primarily by promoting mitochondrial dysfunction, it would be assumed that over time, one of the mechanisms by which cells could survive this treatment will be a viability selection based on increased expression of BCL-2 family members. With this general possibility in mind for multiple chemotherapeutic treatments, several drug companies have developed small-molecule inhibitors of BCL-2, BCL-xL, and MCL-1, including the drugs gossypol, ABT-737 (Oltersdorf et al., 2005), navitoclax (ABT-263), and GX15-070 (Nguyen et al., 2007; Tse et al., 2008). In the present studies, we noted that a commercially available inhibitor of BCL-2 and BCL-XL, HA14-1, significantly enhanced the lethality of the two drug (CHK1 inhibitor + PARP1 inhibitor) regimen. Prior studies have also shown that HA14-1 can overcome the protective effect of BCL-xL in cells treated with UCN-01 and PD184352 (Hamed et al., 2008). Furthermore, the clinically relevant BCL-2 inhibitor obatoclax also enhanced (CHK1 inhibitor + PARP1 inhibitor) toxicity and overcame the protective effect of BCL-xL overexpression. Together, these findings demonstrate that the potentiation of CHK1 inhibitor lethality by PARP1 inhibitors can be profoundly enhanced by additional destabilization of mitochondrial function via inhibition of BCL-2 family member activity(ies).
In conclusion, inhibition of PARP1 blocks CHK1 inhibitor-induced activation of both the DNA damage-response machinery and of ERK1/2. Studies beyond the scope of this article are required to determine whether this drug combination alters tumor cell survival in vivo.
Supplementary Material
This work was funded by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01-DK52825]; the National Institutes of Health National Cancer Institute [Grants P01-CA104177; R01-CA108325; R01-CA150214]; Department of Defense Awards [Grants DAMD17-03-1-0262; W81XWH-10-1-0009]; The Jim Valvano “Jimmy V” Foundation; The Goodwin Foundation; and the Universal Inc. Professorship in Signal Transduction Research (P.D.).
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.067199.
- UCN-01
- 7-hydroxystaurosporine
- ERK
- extracellular signal-regulated kinase
- MEK
- mitogen-activated protein kinase kinase
- m.o.i.
- multiplicity of infection
- DMSO
- dimethyl sulfoxide
- ATM
- ataxia telangiectasia-mutated
- PAGE
- polyacrylamide gel electrophoresis
- CI
- combination index
- FBS
- fetal bovine serum
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siRNA
- small interfering RNA
- PARP1
- poly(ADP-ribose) polymerase 1
- AZD7762
- 5-(3-fluoro-phenyl)-3-ureido-thiophene-2-carboxylic acid (S)-piperidin-3-ylamide hydrochloride
- GPI15427
- 10-(4-methyl-piperazin-1-ylmethyl)-2H-7-oxa-1,2-diaza-benzo[de]anthracen-3-one
- NU1025
- 8-hydroxy-2-methyl-4(3H)-quinazolinone
- AZD2281
- olaparib
- EE
- [Glu218,Glu222]Mek1
- PD184352
- 2-(2-chloro-4-iodo-phenylamino)-N-cyclopropylmethoxy-3,4-difluoro-benzamide
- PD98059
- 2′-amino-3′-methoxyflavone
- PJ34
- N-(5,6-dihydro-6-oxo-2-phenanthridinyl)-2-acetamide hydrochloride
- AG1478
- 4-(3′-chloroanilino)-6,7-dimethoxy-quinazoline
- GX15-070
- obatoclax
- ABT888
- veliparib
- HA14-1
- 2-amino-6-bromo-a-cyano-3-(ethoxycarbonyl)-4H-1-benzopy ran-4-acetic acid ethyl ester
- ABT-263
- navitoclax
- KU55933
- 2-(4-morpholinyl)-6-(1-thianthrenyl)-4H-pyran-4-one
- CEP6800
- 10-(aminomethyl)-4,5,6,7-tetrahydro-1H-cyclopenta[a]pyrrolo[3,4-c]carbazole-1,3(2H)-dione.
References
- Bucher N, Britten CD. (2008) G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer 98:523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S, Adams JM. (2005) Killing cancer cells by flipping the Bcl-2/Bax switch. Cancer Cell 8:5–6 [DOI] [PubMed] [Google Scholar]
- Chou TC, Talalay P. (1984) Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22:27–55 [DOI] [PubMed] [Google Scholar]
- Dai Y, Chen S, Pei XY, Almenara JA, Kramer LB, Venditti CA, Dent P, Grant S. (2008) Interruption of the Ras/MEK/ERK signaling cascade enhances Chk1 inhibitor-induced DNA damage in vitro and in vivo in human multiple myeloma cells. Blood 112:2439–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Landowski TH, Rosen ST, Dent P, Grant S. (2002) Combined treatment with the checkpoint abrogator UCN-01 and MEK1/2 inhibitors potently induces apoptosis in drug-sensitive and -resistant myeloma cells through an IL-6-independent mechanism. Blood 100:3333–3343 [DOI] [PubMed] [Google Scholar]
- Dai Y, Yu C, Singh V, Tang L, Wang Z, McInistry R, Dent P, Grant S. (2001) Pharmacological inhibitors of the mitogen-activated protein kinase (MAPK) kinase/MAPK cascade interact synergistically with UCN-01 to induce mitochondrial dysfunction and apoptosis in human leukemia cells. Cancer Res 61:5106–5115 [PubMed] [Google Scholar]
- Dees EC, Baker SD, O'Reilly S, Rudek MA, Davidson SB, Aylesworth C, Elza-Brown K, Carducci MA, Donehower RC. (2005) A phase I and pharmacokinetic study of short infusions of UCN-01 in patients with refractory solid tumors. Clin Cancer Res 11:664–671 [PubMed] [Google Scholar]
- Fuse E, Kuwabara T, Sparreboom A, Sausville EA, Figg WD. (2005) Review of UCN-01 development: a lesson in the importance of clinical pharmacology. J Clin Pharmacol 45:394–403 [DOI] [PubMed] [Google Scholar]
- Fuse E, Tanii H, Kurata N, Kobayashi H, Shimada Y, Tamura T, Sasaki Y, Tanigawara Y, Lush RD, Headlee D, et al. (1998) Unpredicted clinical pharmacology of UCN-01 caused by specific binding to human alpha1-acid glycoprotein. Cancer Res 58:3248–3253 [PubMed] [Google Scholar]
- Golding SE, Rosenberg E, Neill S, Dent P, Povirk LF, Valerie K. (2007) Extracellular signal-related kinase positively regulates ataxia telangiectasia mutated, homologous recombination repair, and the DNA damage response. Cancer Res 67:1046–1053 [DOI] [PubMed] [Google Scholar]
- Grant S, Dent P. (2007) Simultaneous interruption of signal transduction and cell cycle regulatory pathways: implications for new approaches to the treatment of childhood leukemias. Curr Drug Targets 8:751–759 [DOI] [PubMed] [Google Scholar]
- Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O'Connor PM, Piwnica-Worms H. (2000) The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem 275:5600–5605 [DOI] [PubMed] [Google Scholar]
- Graziani G, Szabó C. (2005) Clinical perspectives of PARP inhibitors. Pharmacol Res 52:109–118 [DOI] [PubMed] [Google Scholar]
- Hagan MP, Yacoub A, Dent P. (2007) Radiation-induced PARP activation is enhanced through EGFR-ERK signaling. J Cell Biochem 101:1384–1393 [DOI] [PubMed] [Google Scholar]
- Hagenauer B, Maier-Salamon A, Thalhammer T, Zöllner P, Senderowicz A, Jäger W. (2004) Metabolism of UCN-01 in isolated perfused rat liver: role of Mrp2 in the biliary excretion of glucuronides. Oncol Rep 11:1069–1075 [DOI] [PubMed] [Google Scholar]
- Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. (2007) Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem 282:16441–16453 [DOI] [PubMed] [Google Scholar]
- Hamed H, Hawkins W, Mitchell C, Gilfor D, Zhang G, Pei XY, Dai Y, Hagan MP, Roberts JD, Yacoub A, et al. (2008) Transient exposure of carcinoma cells to RAS/MEK inhibitors and UCN-01 causes cell death in vitro and in vivo. Mol Cancer Ther 7:616–629 [DOI] [PubMed] [Google Scholar]
- Hawkins W, Mitchell C, McKinstry R, Gilfor D, Starkey J, Dai Y, Dawson K, Ramakrishnan V, Roberts JD, Yacoub A, et al. (2005) Transient exposure of mammary tumors to PD184352 and UCN-01 causes tumor cell death in vivo and prolonged suppression of tumor regrowth. Cancer Biol Ther 4:1275–1284 [DOI] [PubMed] [Google Scholar]
- Hotte SJ, Oza A, Winquist EW, Moore M, Chen EX, Brown S, Pond GR, Dancey JE, Hirte HW. (2006) Phase I trial of UCN-01 in combination with topotecan in patients with advanced solid cancers: a Princess Margaret Hospital Phase II Consortium study. Ann Oncol 17:334–340 [DOI] [PubMed] [Google Scholar]
- Komander D, Kular GS, Bain J, Elliott M, Alessi DR, Van Aalten DM. (2003) Structural basis for UCN-01 (7-hydroxystaurosporine) specificity and PDK1 (3-phosphoinositide-dependent protein kinase-1) inhibition. Biochem J 375:255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M, Watt J, Contractor R, Tsao T, Harris D, Estrov Z, Bornmann W, Kantarjian H, Viallet J, Samudio I, et al. (2008) Mechanisms of antileukemic activity of the novel Bcl-2 homology domain-3 mimetic GX15-070 (obatoclax). Cancer Res 68:3413–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Gautschi O. (2006) Clinical development of SRC tyrosine kinase inhibitors in lung cancer. Clin Lung Cancer 7:381–384 [DOI] [PubMed] [Google Scholar]
- Lu HR, Wang X, Wang Y. (2006) A stronger DNA damage-induced G2 checkpoint due to over-activated CHK1 in the absence of PARP-1. Cell Cycle 5:2364–2370 [DOI] [PubMed] [Google Scholar]
- McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka MZ, et al. (2006) Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 66:8109–8115 [DOI] [PubMed] [Google Scholar]
- McKinstry R, Qiao L, Yacoub A, Dai Y, Decker R, Holt S, Hagan MP, Grant S, Dent P. (2002) Inhibitors of MEK1/2 interact with UCN-01 to induce apoptosis and reduce colony formation in mammary and prostate carcinoma cells. Cancer Biol Ther 1:243–253 [DOI] [PubMed] [Google Scholar]
- Mow BM, Blajeski AL, Chandra J, Kaufmann SH. (2001) Apoptosis and the response to anticancer therapy. Curr Opin Oncol 13:453–462 [DOI] [PubMed] [Google Scholar]
- Nguyen M, Marcellus RC, Roulston A, Watson M, Serfass L, Murthy Madiraju SR, Goulet D, Viallet J, Bélec L, Billot X, et al. (2007) Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci USA 104:19512–19517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435:677–681 [DOI] [PubMed] [Google Scholar]
- Pei XY, Dai Y, Chen S, Bodie WW, Kramer LB, Dent P, Grant S. (2008) The MEK1/2 inhibitor AZD6244 (ARRY-142886) interacts synergistically with the novel Chk1 inhibitor AZD7762 to induce apoptosis in human multiple myeloma cells, in Proceedings of the American Association of Cancer Research; 2008 Sep 22–25; Philadelphia, PA pp LB-103, American Association of Cancer Research, Philadelphia, PA [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. (1997) Mitotic and G2 checkpoint control: regulation of 14–3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277:1501–1505 [DOI] [PubMed] [Google Scholar]
- Perez RP, Lewis LD, Beelen AP, Olszanski AJ, Johnston N, Rhodes CH, Beaulieu B, Ernstoff MS, Eastman A. (2006) Modulation of cell cycle progression in human tumors: a pharmacokinetic and tumor molecular pharmacodynamic study of cisplatin plus the Chk1 inhibitor UCN-01 (NSC 638850). Clin Cancer Res 12:7079–7085 [DOI] [PubMed] [Google Scholar]
- Prudhomme M. (2006) Novel checkpoint 1 inhibitors. Recent Pat Anticancer Drug Discov 1:55–68 [DOI] [PubMed] [Google Scholar]
- Quénet D, El Ramy R, Schreiber V, Dantzer F. (2009) The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol 41:60–65 [DOI] [PubMed] [Google Scholar]
- Riches LC, Lynch AM, Gooderham NJ. (2008) Early events in the mammalian response to DNA double-strand breaks. Mutagenesis 23:331–339 [DOI] [PubMed] [Google Scholar]
- Rodon J, Iniesta MD, Papadopoulos K. (2009) Development of PARP inhibitors in oncology. Expert Opin Investig Drugs 18:31–43 [DOI] [PubMed] [Google Scholar]
- Sausville EA, Lush RD, Headlee D, Smith AC, Figg WD, Arbuck SG, Senderowicz AM, Fuse E, Tanii H, Kuwabara T, et al. (1998) Clinical pharmacology of UCN-01: initial observations and comparison to preclinical models. Cancer Chemother Pharmacol 42 (Suppl):S54–S59 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Amé JC, Dollé P, Schultz I, Rinaldi B, Fraulob V, Ménissier-de Murcia J, de Murcia G. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277:23028–23036 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7:517–528 [DOI] [PubMed] [Google Scholar]
- Spina Purrello V, Cormaci G, Denaro L, Reale S, Costa A, Lalicata C, Sabbatini M, Marchetti B, Avola R. (2002) Effect of growth factors on nuclear and mitochondrial ADP-ribosylation processes during astroglial cell development and aging in culture. Mech Ageing Dev 123:511–520 [DOI] [PubMed] [Google Scholar]
- Syljuåsen RG, Sørensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. (2005) Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol 25:3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, et al. (2008) ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res 68:3421–3428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.