Abstract
Multiple HIV-1 subtypes and circulating recombinant forms (CRFs) are known to cocirculate in Africa. In West Africa, the high prevalence of CRF02_AG, and cocirculation of subtype A, CRF01_AE, CRF06_cpx, and other complex intersubtype recombinants has been well documented. Mali, situated in the heart of West Africa, is likely to be affected by the spread of recombinant subtypes. However, the dynamics of the spread of HIV-1 recombinant subtypes as well as nonrecombinant HIV-1 group M subtypes in this area have not been systematically assessed. Herein, we undertook genetic analyses on full-length env sequences derived from HIV-1-infected individuals living in the capital city of Mali, Bamako. Of 23 samples we examined, 16 were classified as CRF02_AG and three had a subsubtype A3. Among the remaining HIV-1 strains, CRF06_cpx and CRF09_cpx were each found in two patients. Comparison of phylogenies for six matched pol and full-length env sequences revealed that two strains had discordant subtype/CRF designations between the pol and env regions: one had A3polCRF02_AGenv and the other had CRF02_AGpolA3env. Taken together, our study demonstrated the high prevalence of CRF02_AG and complexity of circulating HIV-1 strains in Mali. It also provided evidence of ongoing virus evolution of CRF02_AG, as illustrated by the emergence of more complex CRF02_AG/A3 intersubtype recombinants in this area.
The remarkable viral diversity of HIV-1 has resulted in the classification of nine subtypes (A to D, F to H, J, and K), five subsubtypes (A1, A2, A3, F1, and F2), and a number of intersubtype recombinant strains within the major group M.1,2 Some recombinants have already attained considerable prevalence and geographic coverage worldwide and become important strains in the pandemic. Among the known 37 circulating recombinant forms (CRFs, CRF01 to CRF37), at least eight are believed to originate from West and Central Africa, making this region a hotspot for intersubtype recombination.3 In West and Central Africa, CRF02_AG is of great importance in the pandemic, accounting for the majority of new HIV-1 infections in this region.1,4–9 The CRF02_AG, like pure subtypes, is also involved in recombination events. Indeed, the emergence of more complex intersubtype recombinants containing fragments of CRF02_AG, such as CRF09_cpx, CRF30_0206, or the recently described CRF36_cpx and CRF37_cpx, has been reported in certain West and Central African countries.10–13
Mali, a nation with an estimated population of 12.3 million,14 is one of a few sub-Saharan African countries with a low prevalence of the HIV/AIDS epidemic in the general population. The adult (aged 15–49 years) HIV prevalence was estimated to be at 1.7% (1.3 - 2.1%) in 2005.15 Despite the low rate of HIV/AIDS infection in Mali today, a wider spread of the epidemic is currently anticipated. Poverty, poor health conditions, and certain cultural practices are among a variety of factors that could possibly contribute to the spread of HIV infection in Mali. Migration of seasonal workers to and from neighboring counties such as Côte d'Ivoire and Burkina Faso with higher rates of HIV infection (7.1% and 2.0%, respectively) is another potentially significant contributor to the spread of HIV infection in this country.15
Mali is a landlocked country and shares its borders with Senegal and Mauritania on the west; Algeria on the northeast; Niger on the east; and Burkina Faso, Côte d'Ivoire, and Guinea on the south. High rates of HIV infection and high genetic diversity of HIV strains have been reported in some of the neighboring West and Central African countries.11,15–22 Given that Mali has important trade and travel links with such countries, it is likely that Mali is affected by the spread of recombinant subtypes that are prevalent in the West and Central African region. However, with the exception of the single survey conducted in 1995, covering commercial sex workers in Bamako, Mali,23 the dynamics of the spread of HIV-1 recombinant subtypes as well as nonrecombinant HIV-1 group M subtypes in this area have not been systematically assessed.
Twenty-three patients were included in the present study. Blood specimens were randomly collected from HIV-1 sero-positive, epidemiologically unlinked individuals who were seen at the University Hospital of Bamako in Mali between 2003 and 2005 (Table 1). All individuals provided written informed consent for the use of patient samples that was approved by the Institutional Review Boards at the University of Bamako, Mali, and the National Institute of Allergy and Infectious Diseases. HIV was reported to have been sexually acquired between 1998 and 2004. HIV-1 subtype assignments were determined based on the full-length env sequences. Additional analysis on phylogenies of the pol gene was carried out for six patients out of 23. For the remaining 17 patients, the pol gene sequencing was not done due to lack of biological specimens.
Table 1.
Patient Characteristics and HIV-1 Subtype Informationa
| |
|
|
|
|
|
|
HIV-1 subtype |
|
|---|---|---|---|---|---|---|---|---|
| Patient ID | Gender | Profession | Year sampling | Probable year of infection | Probable location of HIV infection | Sample source | env | pol |
| MAL01 | F | Housewife | 2003 | 1998 | Bamako, Mali | PBMC provirus | CRF02 | |
| MAL04 | F | Housewife | 2003 | 1998 | Bamako, Mali | PBMC provirus | CRF02 | |
| MAL06 | M | Driver | 2003 | 1998 | Bamako, Mali | PBMC provirus | A3 | |
| MAL07 | F | Housewife | 2003 | 1998 | Bamako, Mali | PBMC provirus | CRF02 | |
| MAL12 | F | Housewife | 2003 | 1998 | Bamako, Mali | PBMC provirus | CRF02 | |
| MAL13 | F | Trader | 2003 | 1998 | Bamako, Mali | PBMC provirus | CRF02 | |
| MAL14 | F | Trader | 2003 | 1998 | Côte d'Ivoire | PBMC provirus | CRF09 | |
| MAL019 | M | Trader | 2003 | 1999 | Bamako, Mali | PBMC provirus | CRF06 | |
| MAL125 | F | Artist | 2003 | 1999 | Bamako, Mali | PBMC provirus | CRF02 | |
| MAL127 | F | Housewife | 2003 | 1999 | Bamako, Mali | Plasma virus | CRF02 | |
| MAL144 | F | Housewife | 2003 | 1999 | Bamako, Mali | PBMC provirus | CRF02 | |
| MAL152 | M | Manager | 2003 | 1999 | Bamako, Mali | Plasma virus | CRF02 | |
| MAL154 | F | Housewife | 2003 | 1999 | Bamako, Mali | PBMC provirus | A3 | |
| MAL155 | M | Carpenter | 2003 | 2000 | n.a. | Plasma virus | CRF02 | |
| MAL162 | n.a. | n.a. | 2003 | n.a. | n.a. | Plasma virus | CRF02 | |
| MAL164 | n.a. | n.a. | 2003 | n.a. | n.a. | Plasma virus | CRF02 | |
| MAL08 | F | Housewife | 2004 | 2004 | Bamako, Mali | Plasma virus | CRF02 | CRF02 |
| MAL27 | M | Military | 2004 | 2004 | Bamako, Mali | Plasma virus | CRF02 | A3 |
| MAL29 | M | Carpenter | 2004 | 2003 | Bamako, Mali | Plasma virus | CRF09 | CRF09 |
| MAL30 | F | Housewife | 2004 | 2004 | Bamako, Mali | Plasma virus | A3 | CRF02 |
| MAL40 | F | Nurse | 2004 | 2004 | Bamako, Mali | Plasma virus | CRF02 | CRF02 |
| MAL41 | F | Nurse | 2004 | 2004 | Bamako, Mali | Plasma virus | CRF02 | CRF02 |
| MAL56 | F | Housewife | 2005 | n.a. | Bamako, Mali | Plasma virus | CRF06 | n.a. |
The HIV-1 subtype/CRF designation of env and pol genes was determined based on the nearest reference strain found by the neighbor-joining tree method. n.a., not available.
Peripheral blood was separated by centrifugation at 400 × g for 5min, and plasma was aliquoted and stored at −80°C. Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples by Ficoll-Hypaque gradient density, pelletted, and stored at −80°C until use. Viral RNA was extracted from plasma samples using the QIAamp Viral RNA Mini kit (Qiagen, Inc., Valencia, CA). Due to limited plasma sample availability, proviral DNA derived from PBMCs instead of plasma RNA was used for amplification of the HIV-1 gene for the following 11 patients: MAL01, 04, 06, 07, 12, 13, 14, 019, 125, 144, and 154. Reverse transcription was performed with the Superscript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) as previously described with minor modifications24 by using −9146 5′-CTGCCAATCAGGGAAGTAGCCTTGTGT-3′ as the cDNA primer. DNA was extracted from PBMCs using the PureGene genomic DNA isolation kit (Gentra Systems, Minneapolis, MN). Nested polymerase chain reaction (PCR) amplification of a 3-kbp fragment, encompassing the entire env gene and 5′-portion of the nef gene, was performed with the Expand High Fidelity PCR System (Roche Applied Science, Indianapolis, IN) as previously described25 using the following primer sets: +5954 (sense) 5′-GGCTTAGGCATCTCCTATGGCAGGAAGAA-3′ and −9146 (antisense) 5′-CTGCCAATCAGGGAAGTAGCCTTGTGT-3′ in a first round reaction; +6202 (sense) 5′-AGAAAGAGCAGAAGACAGTGGCAATGA-3′ and −9068 (antisense) 5′-TAGCCCTTCCAGTCCCCCCTTTTCTTTTA-3′ in a second round reaction. Each round of PCR consisted of 30 cycles, and amplifications were performed as follows: the initial denaturation at 94°C for 2 min; 10 cycles of amplification (94°C for 15 s, 63°C for 30 s, 68°C for 4 min); and an additional 20 cycles of amplification (94°C for 15 s, 63°C for 30 s, 68°C for 4 min + 5 s per cycle); and the final extension at 68°C for 7 min. The PCR products were gel-purified on 0.8% agarose gels using the S.N.A.P. UV-Free Gel Purification Kit (Invitrogen, Carlsbad, CA) and then cloned into pCR2.1-TOPO vector (TOPO TA Cloning it, Invitrogen, Carlsbad, CA) for sequence analysis of individual molecular clones. The DNA was sequenced with the ABI BigDye Terminator v3.1 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and analyzed with the ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA). On average 8–10 molecular clones were generated, and one single clone for each patient has been sequenced and analyzed. Amplification and sequencing of the HIV-1 pol gene were performed with the TRUGENE HIV-1 Genotyping kit (Bayer). Sequence segments were assembled with the Sequencher (Gene Codes Corporation, Ann Arbor, MI).
The completed sequences were aligned with reference sequences of appropriate HIV-1 subtypes and circulating recombinant forms (CRFs) (obtained from the Los Alamos HIV Sequence Database, http://www.hiv.lanl.gov) by using the Se-Al (Sequence Alignment Editor, v2.0a11, Rambaut, A. Department of Zoology, University of Oxford, UK), taking into account the protein sequences. Phylogenetic relationships among different subtypes were estimated by use of the neighbor-joining method26 with the PAUP* program (Swofford, DL. 2003. PAUP*. Phylogenetic analysis using parsimony and other methods. Version 4.0b10. Sinauer Associates, Sunderland, MA). Statistical support for various nodes in the neighbor-joining tree was obtained by 1000 replications of the bootstrap procedure.27 Gaps were ignored for affected pairwise comparisons. The sequence data used in this study have been deposited in GenBank and are available under the accession numbers EU480455–EU480489.
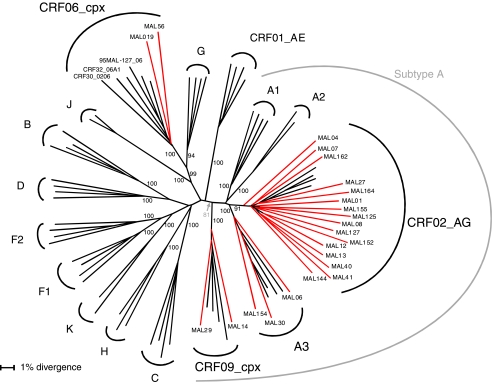
Phylogenetic analysis of 23 full-length env sequences revealed that all 23 clustered within the HIV-1 group M clade (Fig. 1). The majority of group M sequences (21 of 23) fell within the subtype A clade. Sixteen of the 21 subtype A sequences were further classified as CRF02_AG, three as sub-subtype A3, and two sequences clustered with recently described CRF09_cpx (Fig. 1 and Tables 1 and 2). The remaining two sequences clustered together within the CRF06_cpx radiation with a bootstrap value of 100%. An additional pruning analysis, which consists of removing respective reference sequences from an alignment and rerunning the phylogenetic analysis, revealed that our CRF02_AG, subsubtype A3, CRF09_cpx, and CRF06_cpx sequences all maintained their distinct clustering patterns even when their respective reference sequences were absent, further confirming the stability of the tree topology and subtype assignments (data not shown). In addition, the CRF02_AG, CRF09_cpx, and subsubtype A3 sequences clustered separately and distantly from the crown groups of A1 viruses. Similarly, CRF06_cpx sequences clustered among themselves being outside the crown group of the pure subtype G reference sequences. Taken together, these data imply that CRF02_AG, CRF09_cpx, subsubtype A3, and CRF06_cpx have been in circulation for some time.
FIG. 1.
Phylogenetic classification of 23 Malian HIV-1 isolates based on the full-length env region. A representative sequence from each Malian patient (highlighted in red) and 116 reference subtypes and available CRFs (at least two representative sequences of subtypes, subsubtypes, and of CRF01 to CRF15, CRF18, CRF19 as well as a prototype sequence from CRF16, CRF17, CRF20–CRF25, CRF28–CRF33, and CRF35–CRF37) were initially used to construct the tree; some references have been omitted here for clarity. For a complete list of reference sequences used in the analysis, see Table 2. A phylogenetic tree was constructed by the neighbor-joining method with the HKY85 model of evolution. Bootstrap percentile values from 1000 replications are shown at nodes defining major grouping of sequences. Clustering of subtype/subsubtype sequences are delineated in the figure. Previously generated sequences from Mali, 95ML127 (CRF06), were included in the analysis. Recently classified CRF30_020611 and CRF32_06A1,31 both containing CRF06_cpx segments in their env region, were also included in the analysis and found within the CRF06_cpx radiation with a high bootstrap value of 100%.
Table 2.
Classification by Subtype
| Subtype | Reference strain | Accession no. | Country code | Country |
|---|---|---|---|---|
| A1 | Q23 | AF004885 | KE | Kenya |
| A1 | SE7253 | AF069670 | SE | Sweden |
| A1 | 92UG037 | U51190 | UG | Uganda |
| A1 | 98UG57136 | AF484509 | UG | Uganda |
| A2 | 97CDKTB48 | AF286238 | CD | Congo |
| A2 | 94CY017 | AF286237 | CY | Cyprus |
| A3 | DDJ360 | AY521630 | SE | Sweden |
| A3 | DDI579 | AY521629 | SE | Sweden |
| A3/02 | DDJ362 | AY521632 | SE | Sweden |
| A3/02 | DDJ364 | AY521633 | SE | Sweden |
| B | HXB2 | K03455 | FR | France |
| B | 671_00T36 | AY423387 | NL | Netherlands |
| B | BK132 | AY173951 | TH | Thailand |
| B | 1058_11 | AY331295 | US | USA |
| C | BR025-d | U52953 | BR | Brazil |
| C | ETH2220 | U46016 | ET | Ethiopia |
| C | 95IN21068 | AF067155 | IN | India |
| C | SK164B1 | AY772699 | ZA | South Africa |
| D | ELI | K03454 | CD | Congo |
| D | 4412HAL | AY371157 | CM | Cameroon |
| D | A280 | AY253311 | TZ | Tanzania |
| D | 94UG114 | U88824 | UG | Uganda |
| F1 | VI850 | AF077336 | BE | Belgium |
| F1 | 93BR020_1 | AF005494 | BR | Brazil |
| F1 | FIN9363 | AF075703 | FI | Finland |
| F1 | MP411 | AJ249238 | FR | France |
| F2 | 0016BBY | AY371158 | CM | Cameroon |
| F2 | MP255 | AJ249236 | CM | Cameroon |
| F2 | MP257 | AJ249237 | CM | Cameroon |
| F2 | CM53657 | AF377956 | CM | Cameroon |
| G | DRCBL | AF084936 | BE | Belgium |
| G | HH8793 | AF061641 | KE | Kenya |
| G | 92NG083 | U88826 | NG | Nigeria |
| G | SE6165 | AF061642 | SE | Sweden |
| H | VI991 | AF190127 | BE | Belgium |
| H | VI997 | AF190128 | BE | Belgium |
| H | 90_056 | AF005496 | CF | Central African Republic |
| J | SE7887 | AF082394 | SE | Sweden |
| J | SE7022 | AF082395 | SE | Sweden |
| K | EQTB11C | AJ249235 | CD | Congo |
| K | MP535 | AJ249239 | CM | Cameroon |
| 01_AE | CM240 | U54771 | TH | Thailand |
| 01_AE | 90CF11697 | AF197340 | CF | Central African Republic |
| 01_AE | 90CF402 | U51188 | CF | Central African Republic |
| 01_AE | 90CF4071 | AF197341 | CF | Central African Republic |
| 02_AG | IBNG | L39106 | NG | Nigeria |
| 02_AG | DJ263 | AF063223 | FR (DJ) | Djibouti |
| 02_AG | DJ264 | AF063224 | FR (DJ) | Djibouti |
| 02_AG | SE7812 | AF107770 | SE | Sweden |
| 03_AB | KAL153_2 | AF193276 | RU | Russian Federation |
| 03_AB | 98BY10443 | AF414006 | BY | Belarus |
| 03_AB | RU98001 | AF193277 | RU | Russian Federation |
| 04_cpx | CY032 | AF049337 | CY | Cyprus |
| 04_cpx | 97PVCH | AF119820 | GR | Greece |
| 04_cpx | 97PVMY | AF119819 | GR | Greece |
| 05_DF | VI1310 | AF193253 | BE | Belgium |
| 05_DF | VI961 | AF076998 | BE | Belgium |
| 05_DF | X492 | AY227107 | ES | Spain |
| 06_cpx | BFP90 | AF064699 | AU | Australia |
| 06_cpx | 95ML84 | AJ245481 | ML | Mali |
| 06_cpx | 95ML127 | AJ288982 | ML | Mali |
| 06_cpx | 97SE1078 | AJ288981 | SN | Senegal |
| 07_BC | CN54 | AX149771 | CN | China |
| 07_BC | 97CN001 | AF286226 | CN | China |
| 07_BC | 98CN009 | AF286230 | CN | China |
| 07_BC | CNGL179 | AF503396 | CN | China |
| 08_BC | GX_6F | AY008715 | CN | China |
| 08_BC | 98CN006 | AF286229 | CN | China |
| 09_cpx | 96GH2911 | AY093605 | GH | Ghana |
| 09_cpx | 96SN1795 | AY093603 | SN | Senegal |
| 09_cpx | 99DE4057 | AY093607 | US | USA |
| 09_cpx | 00IC_10092 | AJ866553 | CI | Côte d'Ivoire |
| 10_CD | BF061 (TZBF061) | AF289548 | TZ | Tanzania |
| 10_CD | BF071 | AF289549 | TZ | Tanzania |
| 10_CD | BF110 | AF289550 | TZ | Tanzania |
| 11_cpx | GR17 | AF179368 | GR | Greece |
| 11_cpx | 0186ND | AY371149 | CM | Cameroon |
| 11_cpx | 1816 | AF492624 | CM | Cameroon |
| 11_cpx | MP1298 | AJ291719 | FR | France |
| 12_BF | ARMA159 | AF385936 | AR | Argentina |
| 12_BF | X1241 | AY536238 | ES | Spain |
| 12_BF | URTR17 | AY037272 | UY | Uruguay |
| 13_cpx | 1849 (96CM_1849) | AF460972 | CM | Cameroon |
| 13_cpx | 3226MN | AY371154 | CM | Cameroon |
| 13_cpx | 4164 | AF460974 | CM | Cameroon |
| 14_BG | X397 | AF423756 | ES | Spain |
| 14_BG | X475 | AF423758 | ES | Spain |
| 14_BG | X605 | AF450096 | ES | Spain |
| 15_01B | MU2079 | AF516184 | TH | Thailand |
| 15_01B | OUR1331 | AF529572 | TH | Thailand |
| 15_01B | R2399 | AF530576 | TH | Thailand |
| 16_A2D | 97KR004 | AF286239 | KR | Korea |
| 18_cpx | CM53379 | AF377959 | CM | Cameroon |
| 18_cpx | CU76 | AY586540 | CU | Cuba |
| 19_cpx | CU38 | AY588970 | CU | Cuba |
| 19_cpx | CU7 | AY894994 | CU | Cuba |
| O | ANT70 | L20587 | BE | Belgium |
| CRF17_BF | ARMA038 | AY037281 | AR | Argentina |
| CRF20_BG | CB228 | AY900577 (gag-pol) | CU | Cuba |
| CRF21_A2D | 99KE_KER2003 | AF457051 (KER2003) | KE | Kenya |
| CRF22_01A1 | CM53122 | AY037284S2 | CM | Cameroon |
| CRF23_BG | CB118 | AY900571 | CU | Cuba |
| CRF24_BG | CB378 | AY900574 | CU | Cuba |
| CRF25_cpx | 02CM_1918LE | AY371169 (1918LE) | CM | Cameroon |
| CRF26_AU | Pending | |||
| CRF27 | Pending | |||
| CRF28_BF | BREPM12609 | DQ085873 | BR | Brazil |
| CRF29_BF | BREPM16704 | DQ085876 | BR | Brazil |
| CRF30_0206 | 00NE36 | AJ508597 (NE36) | NE | Niger |
| CRF31_BC | 04BR142 | AY727527 | BR | Brazil |
| CRF32_06A1 | EE0369 | AY535660 | EE | Estonia |
| CRF33_01B | 05MYKL007 | DQ366659 | MY | Malaysia |
| CRF34_01B | Pending | TH | Thailand | |
| CRF35_AD | AF026 | EF158040 | AF | Afghanistan |
| CRF36_cpx | 00CMNYU830 | EF087994 | CM | Cameroon |
| CRF37_cpx | 00CMNYU926 | EF116594 | CM | Cameroon |
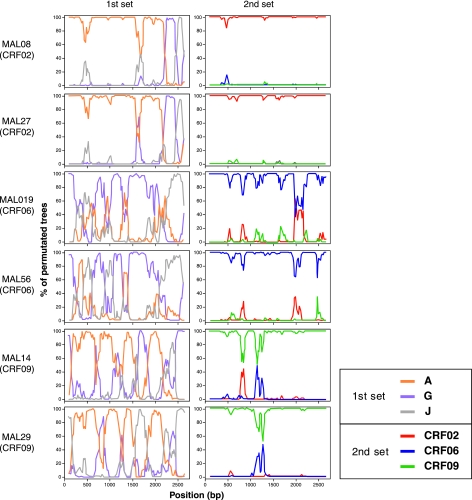
To confirm the clade assignments and the recombinant structure of newly generated Mali sequences, bootscanning analyses were performed with the use of SimPlot software.28 All Mali CRF sequences: CRF02 (16 sequences), CRF06 (two sequences), and CRF09 (two sequences) were subjected to bootscanning analysis by being plotted against references of nonrecombinant subtypes and prototype CRFs. Bootscanning plots of two representative sequences for CRF02 (MAL08 and 27), CRF06 (MAL019 and 56), and CRF09 (MAL14 and 29) are shown in Fig. 2. CRF02 sequences found in Mali (represented by MAL08 and 27 in Fig. 2) showed a similar breakpoint profile of the CRF02 prototype: IbNG, having a mosaic genome structure, involving subtypes A and G. CRF06 sequences derived from the Mali patients (MAL019 and 56) had multiple breakpoints, involving segments from subtypes G and J, which is in line with the mosaic pattern previously identified for the CRF06 prototype: BFP90. CRF09 sequences found in Mali patients (MAL14 and 29) had a complex genome structure, involving short stretch of segments derived from subtypes A and G with multiple breakpoints. The bootscanning analysis involving prototypic CRF sequences confirmed the similarity between the CRF09 sequences from Mali and the prototype CRF09: 96GH2911.
FIG. 2.
Bootscanning analyses of 2.6-kb env regions of HIV-1 sequences of CRF02 (MAL08 and 27), CRF06 (MAL019 and 56), and CRF09 (MAL14 and 29). Two sets of bootscanning were performed: The first set of bootscanning plots depicts relationship of the Mali sequences to the representative strains of HIV-1 nonrecombinant subtypes (A: 94SE_Q23, G: DRCBL, and J: 93SE_7887). The second set of bootscanning plots delineates relationship of the Mali sequences to the prototype strain of CRF02 (IbNG), CRF06 (BFP90), and CRF09 (96GH2911). For the bootscanning plots, the SimPlot software performed bootscanning on neighbor-joining trees by using SEQBOOT (100 replicates), DNADIST (with the Kimura's two-parameter method and a transition/transversion ratio of 2.0), and CONSENSE from the PHYLIP package,32 for a 200 bp window moving along the alignment with increments of 20 bp.
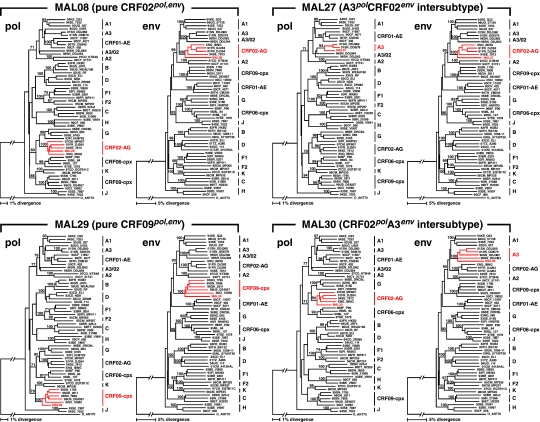
Next, we examined the pol and env gene fragments of the Mali specimens for concordance of subtypes, which supports the clade assignment. We observed that four of the six specimens studied (MAL08, MAL29, MAL40, and MAL41) had concordant subtypes in the pol and env (Fig. 3 and Table 3). On the other hand, the sequence of MAL30 was a mosaic of CRF02_AG and subsubtype A3. The analyses indicated that the sequence from subject MAL30 was grouped with high bootstrap values to CRF02_AG reference sequences in the pol gene, and the env portion of the genome clustered with high bootstrap values with subsubtype A3 reference sequences (Fig. 3). Similarly, the viral sequence from MAL27 was composed of a mosaic of A3 in the pol and CRF02_AG in the env regions (Fig. 3). Since amplification of the pol and env gene fragments was performed separately, it is not possible to determine whether these A3/CRF02_AG viruses truly represent intersubtype recombinants or dual infection in the individuals who carried discordant subtypes. Further analysis involving the full-length HIV-1 genome sequencing is warranted for the precise intersubtype assignments. In addition, no known major drug resistance-associated mutations were detected in these Mali isolates.
FIG. 3.
Subregion tree analysis of the pol and full-length env sequences. Four representative patterns are shown. Each gene segment was subjected to separate phylogenetic analyses based on the neighbor-joining method to identify the subtype or circulating recombinant form (CRF) origin of the segment. The stability of the nodes was assessed by the use of bootstrap resampling of 1000 replications. Bootstrap values above 70% are shown. The clusters for subtype/subsubtypes and CRF01_AE, CRF02_AG, CRF06_cpx, and CRF09_cpx are outlined with a vertical bar on the right of each tree. The genotype assignments of each Mali strain in the respective phylogenetic analysis are highlighted in red in each tree. All the HIV-1 reference sequences were retrieved from the HIV sequence database, Los Alamos National Library. For a complete list of reference sequences used in the analysis, see Table 3.
Table 3.
Pol and env Trees
| Pol tree |
env tree |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subtype | Sequence name | Accession no. | Country code | Country | Subtype | Sequence name | Accession no. | Country code | Country |
| A1 | Q23 | AF004885 | KE | Kenya | A1 | Q23 | AF004885 | KE | Kenya |
| A1 | SE7253 | AF069670 | SE | Sweden | A1 | SE7253 | AF069670 | SE | Sweden |
| A1 | U455 | M62320 | UG | Uganda | A1 | 98UG57136 | AF484509 | UG | Uganda |
| A1 | 92UG037 | U51190 | UG | Uganda | A1 | 92UG037 | U51190 | UG | Uganda |
| A2 | 97CDKTB48 | AF286238 | CD | Congo | A2 | 97CDKTB48 | AF286238 | CD | Congo |
| A2 | 94CY017 | AF286237 | CY | Cyprus | A2 | 94CY017 | AF286237 | CY | Cyprus |
| A3 | DDJ360 | AY521630 | SE | Sweden | A3 | DDJ360 | AY521630 | SE | Sweden |
| A3 | DD1579 | AY521629 | SE | Sweden | A3 | DD1579 | AY521629 | SE | Sweden |
| A3/02 | DDJ362 | AY521632 | SE | Sweden | A3/02 | DDJ362 | AY521632 | SE | Sweden |
| A3/02 | DDJ364 | AY521633 | SE | Sweden | A3/02 | DDJ364 | AY521633 | SE | Sweden |
| B | HXB2 | K03455 | FR | France | B | HXB2 | K03455 | FR | France |
| B | RF | M17451 | US | USA | B | 671_00T36 | AY423387 | NL | Netherlands |
| B | JFRL | U63632 | Us | USA | B | BK132 | AY173951 | TH | Thailand |
| B | WEAU160 | U21135 | US | USA | B | 1058_11 | AY331295 | US | USA |
| C | BR025-d | U52953 | BR | Brazil | C | BR025-d | U52953 | BR | Brazil |
| C | ETH2220 | U46016 | ET | Ethiopia | C | ETH2220 | U46016 | ET | Ethiopia |
| C | 96BW0502 | AF110967 | BW | Botswana | C | 95IN21068 | AF067155 | IN | India |
| C | 95IN21068 | AF067155 | IN | India | |||||
| D | ELI | K03454 | CD | Congo | D | ELI | K03454 | CD | Congo |
| D | NDK | M27323 | CD | Congo | D | 4412HAL | AY371157 | CM | Cameroon |
| D | 84ZR085 | U88822 | CD | Congo | D | A280 | AY253311 | TZ | Tanzania |
| D | 94UG114 | U88824 | UG | Uganda | D | 94UG114 | U88824 | UG | Uganda |
| F1 | VI850 | AF077336 | BE | Belgium | F1 | VI850 | AF077336 | BE | Belgium |
| F1 | 93BR020_1 | AF005494 | BR | Brazil | F1 | 93BR020_1 | AF005494 | BR | Brazil |
| F1 | FIN9363 | AF075703 | FI | Finland | F1 | FIN9363 | AF075703 | FI | Finland |
| F1 | MP411 | AJ249238 | FR | France | F1 | MP411 | AJ249238 | FR | France |
| F2 | MP255 | AJ249236 | CM | Cameroon | F2 | MP255 | AJ249236 | CM | Cameroon |
| F2 | MP257 | AJ249237 | CM | Cameroon | F2 | MP257 | AJ249237 | CM | Cameroon |
| G | DRCBL | AF084936 | BE | Belgium | G | DRCBL | AF084936 | BE | Belgium |
| G | HH8793 | AF061641 | KE | Kenya | G | HH8793 | AF061641 | KE | Kenya |
| G | 92NG083 | U88826 | NG | Nigeria | G | 92NG083 | U88826 | NG | Nigeria |
| G | SE6165 | AF061642 | SE | Sweden | G | SE6165 | AF061642 | SE | Sweden |
| H | VI991 | AF190127 | BE | Belgium | H | VI991 | AF190127 | BE | Belgium |
| H | VI997 | AF190128 | BE | Belgium | H | VI997 | AF190128 | BE | Belgium |
| H | 90_056 | AF005496 | CF | Central African Republic | H | 90_056 | AF005496 | CF | Central African Republic |
| J | SE7887 | AF082394 | SE | Sweden | J | SE7887 | AF082394 | SE | Sweden |
| J | SE7022 | AF082395 | SE | Sweden | J | SE7022 | AF082395 | SE | Sweden |
| K | EQTB11C | AJ249235 | CD | Congo | K | EQTB11C | AJ249235 | CD | Congo |
| K | MP535 | AJ249239 | CM | Cameroon | K | MP535 | AJ249239 | CM | Cameroon |
| 01_AE | 90CF11697 | AF197340 | CF | Central African Republic | 01_AE | 90CF11697 | AF197340 | CF | Central African Republic |
| 01_AE | 90CF402 | U51188 | CF | Central African Republic | 01_AE | 90CF402 | U51188 | CF | Central African Republic |
| 01_AE | 90CF4071 | AF197341 | CF | Central African Republic | 01_AE | 90CF4071 | AF197341 | CF | Central African Republic |
| 01 AE | CM240 | U54771 | TH | Thailand | 01_AE | CM240 | U54771 | TH | Thailand |
| 02_AG | MP807 | AJ286133 | CM | Cameroon | 02_AG | DJ263 | AF063223 | FR (DJ) | Djibouti |
| 02_AG | DJ264 | AF063224 | FR (DJ) | Djibouti | 02_AG | DJ264 | AF063224 | FR (DJ) | Djibouti |
| 02_AG | IBNG | L39106 | NG | Nigeria | 02_AG | IBNG | L39106 | NG | Nigeria |
| 02_AG | SE7812 | AF107770 | SE | Sweden | 02_AG | SE7812 | AF107770 | SE | Sweden |
| 06_cpx | BFP90 | AF064699 | AU | Australia | 06_cpx | BFP90 | AF064699 | AU | Australia |
| 06_cpx | 95ML127 | AJ288982 | ML | Mali | 06_cpx | 95ML127 | AJ288982 | ML | Mali |
| 06_cpx | 95ML84 | AJ245481 | ML | Mali | 06_cpx | 95ML84 | AJ245481 | ML | Mali |
| 06_cpx | 97SE1078 | AJ288981 | SN | Senegal | 06_cpx | 97SE1078 | AJ288981 | SN | Senegal |
| 09_cpx | 96SN1795 | AY093603 | SN | Senegal | 09_cpx | 96SN1795 | AY093603 | SN | Senegal |
| 09_cpx | 95SN7808 | AY093604 | SN | Senegal | 09_cpx | 95SN7808 | AY093604 | SN | Senegal |
| 09_cpx | 96GH2911 | AY093605 | GH | Ghana | 09_cpx | 96GH2911 | AY093605 | GH | Ghana |
| 09_cpx | 99DE4057 | AY093607 | US | USA | 09_cpx | 99DE4057 | AY093607 | US | USA |
| 09_cpx | 00IC_10092 | AJ866553 | CI | Côte d'lvoire | 09_cpx | 00IC_10092 | AJ866553 | CI | Côte d'lvoire |
The current study revealed a broad HIV-1 genetic diversity in Mali, identifying genetic materials of CRF02_AG, CRF06_cpx, CRF09_cpx, and subsubtype A3. We also report the presence of the recently described CRF09_cpx sequences in Mali for the first time. The CRF09_cpx, an intersubtype recombinant composed of CRF02_AG, Z321 and some uncharacterized fragments, was originally identified in the Senegal cohort in 2004.29 While initially thought to have limited spread, subsequent reports of the presence of CRF09_cpx in Côte d'Ivoire, Cameroon, and Nigeria,16,17,19 coupled with our present report of presence of CRF09_cpx in Mali, indicate that the CRF09_cpx may be more broadly distributed and at higher prevalence in West and Central Africa. Furthermore, we have identified two intersubtype recombinants involving CRF02_AG and subsubtype A3 in the pol and env segments. The subsubtype A3 sequences, which we describe in this study for the first time in Mali, were reported earlier to be circulating in other parts of West and Central Africa, where they had also recombined with CRF02_AG.21 The exact prevalence of the A3/CRF02_AG intersubtype recombinants is currently unclear. However, given that the A3-containing recombinants were possibly introduced to the population as early as 1989,22 A3/CRF02_AG could be more broadly distributed over West and Central African countries.
The 1995 study conducted in Mali reported a predominance of subtype A and subtype G virus with a few strains belonging to subtypes C and D.23 The subsequent sequence analysis revealed that some of the subtype G sequences from the 1995 study clustered with CRF06_cpx. In the present study, we have shown that all isolates do indeed cluster with subtypes A and G in the pol and env regions. However, two significant differences emerged. First, non-recombinant subtype A, or “pure” subtype A, viruses were no longer found, and all the subtype A viruses were primarily CRF02_AG. Similarly, all the subtype G viruses were intersubtype recombinant: CRF06_cpx. Second, in addition to the aforementioned CRF02_AG, we found other subtype A sequences, including subsubtype A3 and CRF09_cpx. These findings indicate a shift in the HIV-1 virus population circulating in Mali and demonstrated the recombination-prone nature of HIV-1 in areas such as Mali where CRFs have a high prevalence.
Although the rate of HIV-1 infections in Mali is, at present, fairly low in the general population, compared to other West and Central African countries, a high HIV prevalence in bridge populations, such as truck drivers, street vendors, or commercial sex workers (3.9%, 4.6%, and 31.9%, respectively), has been reported in urban areas of Mali.30 This may foreshadow a wider epidemic and potential HIV diversification in Mali. In the present study, our cohort was restricted to HIV-infected individuals living in the capital city of Mali, Bamako. Extensive surveys covering all regions of the country with a sufficient number of samples will enhance our understanding of viral diversity, which is critical to the development of relevant diagnostic testing, treatment strategies, and vaccine candidates.
Acknowledgments
The authors thank Julia A. Metcalf, Jeanne Warfield, and Jennifer L. Imes for arrangement of blood specimens. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This project was supported in part by the National Institute of Allergy and Infectious Disease, National Institutes of Health, under contract N01-CO-12400.
Disclosure Statement
No competing financial interests exist.
References
- 1.McCutchan FE. Understanding the genetic diversity of HIV-1. AIDS. 2000;14(Suppl. 3):S31–S44. [PubMed] [Google Scholar]
- 2.Robertson DL. Anderson JP. Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 3.Korber B, et al., editors. HIV ImmunologySequence Databases. Los Alamos National Laboratory; Los Alamos NM: [Google Scholar]
- 4.Andersson S. Norrgren H. Dias F. Biberfeld G. Albert J. Molecular characterization of human immunodeficiency virus (HIV)-1 and - 2 individuals from Guinea-Bissau with single or dual infections: Predominance of a distinct HIV-1 subtype A/G recombinant in West Africa. Virology. 1999;267:312–330. doi: 10.1006/viro.1999.9867. [DOI] [PubMed] [Google Scholar]
- 5.Carr J. Laukkanen T. Salminen M, et al. Characterization of subtype A HIV-1 from Africa by full genome sequencing. AIDS. 1999;13:1819–1826. doi: 10.1097/00002030-199910010-00003. [DOI] [PubMed] [Google Scholar]
- 6.Cornelissen M. van Den Burg R. Zorgdrager F. Goudsmit J. Spread of distinct human immunodeficiency virus type 1 AG recombinant lineages in Africa. J Gen Virol. 2000;81:515–523. doi: 10.1099/0022-1317-81-2-515. [DOI] [PubMed] [Google Scholar]
- 7.Montavon C. Toure-kane C. Liegeois F, et al. Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to the prototype AG recombinant virus IBNG. J Acquir Immune Defic Syndr. 2000;23:363–374. doi: 10.1097/00126334-200004150-00001. [DOI] [PubMed] [Google Scholar]
- 8.Osmanov S. Pattou C. Walker N. Schwardländer B. Esparza J. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr. 2002;29:184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- 9.Peeters M. Esu-Williams E. Vergne L, et al. Predominance of subtype A and G HIV type 1 in Nigeria, with geographical differences in their distribution. AIDS Res Hum Retroviruses. 2000;16:315–325. doi: 10.1089/088922200309197. [DOI] [PubMed] [Google Scholar]
- 10.Burda ST. Konings FA. Williams CA, et al. HIV-1 CRF09_cpx circulates in the north west province of Cameroon where CRF02_AG infections predominate and recombinant strains are common. AIDS Res Hum Retroviruses. 2004;20(12):1358–1363. doi: 10.1089/aid.2004.20.1358. [DOI] [PubMed] [Google Scholar]
- 11.Mamadou S. Vidal N. Montavan C, et al. Emergence of complex and diverse CRF02-AG/CRF06-cpx recombinant HIV type 1 strains in Niger, West Africa. AIDS Res Hum Retroviruses. 2003;19:77–82. doi: 10.1089/08892220360474005. [DOI] [PubMed] [Google Scholar]
- 12.Powell RL. Zhao J. Konings FA, et al. Identification of a novel circulating recombinant form (CRF) 36_cpx in Cameroon that combines two CRFs (01_AE and 02_AG) with ancestral lineage of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23:1008–1019. doi: 10.1089/aid.2006.0289. [DOI] [PubMed] [Google Scholar]
- 13.Powell RL. Zhao J. Konings FA, et al. Circulating recombinant form (CRF) 37_cpx: An old strain in Cameroon composed of diverse, genetically distant lineage of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23:923–933. doi: 10.1089/aid.2007.0040. [DOI] [PubMed] [Google Scholar]
- 14.2007 World Population Data Sheet by the Population Reference Bureau. www.prb.org/Publications/Datasheets/2007 www.prb.org/Publications/Datasheets/2007
- 15.UNAIDS: 2006 report on the global AIDS epidemic. 2006.
- 16.Ndongmo CB. Pieniazek D. Holberg-Petersen M, et al. HIV genetic diversity in Cameroon: Possible public health importance. AIDS Res Hum Retroviruses. 2006;22:812–816. doi: 10.1089/aid.2006.22.812. [DOI] [PubMed] [Google Scholar]
- 17.Sankalé JL. Langevin S. Odaibo G, et al. The complexity of circulating HIV type 1 strains in Oyo State, Nigeria. AIDS Res Hum Retroviruses. 2007;23:1020–1025. doi: 10.1089/aid.2006.0304. [DOI] [PubMed] [Google Scholar]
- 18.Montavon C. Toure-Kane C. Nkengasong JN, et al. CRF06_cpx: A new circulating recombinant form of HIV-1 in West Africa involving subtypes A, G, K, and J. J Acquir Immune Defic Syndr. 2002;29:522–530. doi: 10.1097/00126334-200204150-00014. [DOI] [PubMed] [Google Scholar]
- 19.Toni T. Adjé-Touré C. Vidal N, et al. Presence of CRF09_cpx and complex CRF02_AG/CRF09_cpx recombinant HIV type 1 strains in Côte d'Ivoire, West Africa. AIDS Res Hum Retroviruses. 2005;21:667–672. doi: 10.1089/aid.2005.21.667. [DOI] [PubMed] [Google Scholar]
- 20.Tebit DM. Ganame J. Sathiandee K. Nagabila Y. Coulibaly B. Krausslich HG. Diversity of HIV in rural Burkina Faso. J Acquir Immune Defic Syndr. 2006;43:144–152. doi: 10.1097/01.qai.0000228148.40539.d3. [DOI] [PubMed] [Google Scholar]
- 21.Meloni ST. Kim B. Sankalé JL, et al. Distinct human immunodeficiency virus type 1 subtype A virus circulating in West Africa: Sub-subtype A3. J Virol. 2004;78:12438–12445. doi: 10.1128/JVI.78.22.12438-12445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meloni ST. Sankalé; JL. Hamel DJ, et al. Molecular epidemiology of human immunodeficiency virus type 1 subsubtype A3 in Senegal from 1988 to 2001. J Virol. 2004;78:12455–12461. doi: 10.1128/JVI.78.22.12455-12461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peeters M. Koumare B. Mulanga C, et al. Genetic subtypes of HIV type 1 and HIV type 2 strains in commercial sex workers from Bamako, Mali. AIDS Res Hum Retroviruses. 1998;14:51–58. doi: 10.1089/aid.1998.14.51. [DOI] [PubMed] [Google Scholar]
- 24.Imamichi H. Crandall KA. Natarajan V, et al. Human immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J Infect Dis. 2001;183:36–50. doi: 10.1086/317641. [DOI] [PubMed] [Google Scholar]
- 25.Imamichi H. Igarashi T. Imamichi T, et al. Amino acid deletions are introduced into the V2 region of gp120 during independent pathogenic simian immunodeficiency virus/HIV chimeric virus (SHIV) infections of rhesus monkeys generating variants that are macrophage tropic. Proc Natl Acad Sci USA. 2002;99:13813–13818. doi: 10.1073/pnas.212511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou N. Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 28.Lole KS. Bollinger RC. Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCutchan FE. Sankalé JL. M'Boup S, et al. HIV type 1 circulating recombinant form CRF09_cpx from West Africa combines subtypes A, F, G, and may share ancestors with CRF02_AG and Z321. AIDS Res Hum Retroviruses. 2004;20:819–826. doi: 10.1089/0889222041725163. [DOI] [PubMed] [Google Scholar]
- 30.UNAIDS. Report on HIV/AIDS program in Mali. February 2005.
- 31.Adojaan M. Kivisild T. Mannik A, et al. Predominance of a rare type of HIV-1 in Estonia. J Acquir Immune Defic Syndr. 2005;39:598–605. [PubMed] [Google Scholar]
- 32.Felsenstein J. PHYLIP-Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]