Abstract
CYP2B6 is a highly inducible and polymorphic enzyme involved in the metabolism of an increasing number of clinically important drugs. Significant interindividual variability in CYP2B6 expression has been attributed to either genetic polymorphisms or chemical-mediated induction through the activation of constitutive androstane receptor and/or pregnane X receptor (PXR). It was reported that the −82T→C substitution within the CYP2B6*22 allele creates a functional CCAAT/enhancer-binding protein (C/EBP) binding site and enhances the basal expression of the CYP2B6 gene. Here, we explored whether this polymorphic mutation could affect drug-mediated induction of CYP2B6. Cell-based promoter reporter assays demonstrated that CYP2B6 luciferase activity was synergistically enhanced in the presence of both −82T→C mutation and rifampicin (RIF)-activated PXR. On the other hand, this synergism was attenuated by disrupting the C/EBP binding site or knocking down C/EBPα expression. Mechanistic studies revealed that C/EBPα plays an important role in such synergism by directly interacting with PXR; enhancing RIF-mediated recruitment of PXR to the −82T→C harboring CYP2B6 promoter; and looping the PXR-bound distal phenobarbital-responsive enhancer module toward the proximal C/EBP binding site. Furthermore, the genotype-phenotype association was evaluated in cultured human primary hepatocytes from 44 donors. Interestingly, RIF-mediated induction of CYP2B6 in four −82T/C carriers was higher compared with that in the reference −82T/T homozygotes. Together, our results demonstrate, for the first time, a synergistic interplay between a CYP2B6 polymorphism and PXR-mediated induction, which may contribute to the large individual variations and inducibility of CYP2B6 in humans.
Introduction
Cytochrome P450 2B6 (CYP2B6) participates in the metabolism of a growing number of clinically important drugs, such as cyclophosphamide, ifosfamide, efavirenz, selegiline, bupropion, tamoxifen, and methadone, as well as a wide variety of environmental chemicals (Hidestrand et al., 2001; Lang et al., 2001; Hodgson and Rose, 2007; Wang and Tompkins, 2008). Notably, significant interindividual variations in CYP2B6 expression have been observed previously. In these studies, CYP2B6 exhibits 20- to 280-fold interindividual variability in its gene expression and 25- to 80-fold variations in its enzymatic activity (Code et al., 1997; Ekins et al., 1998; Stresser and Kupfer, 1999; Faucette et al., 2000; Hanna et al., 2000; Hesse et al., 2000, 2004; Lamba et al., 2003). These findings suggest that the remarkable variability in CYP2B6 expression and activity may be a major determinant of the efficacy and toxicity of these widely used drugs that are characterized by narrow therapeutic indices.
During the past several years, numerous investigations have focused on elucidating the predominant mechanisms underlying interindividual variability in CYP2B6 expression. Among the factors studied such as age, gender, ethnics, genetics, and xenobiotics, it seems that receptor-mediated gene regulation and genetic polymorphisms are the major contributors. Currently, it has been well established that CYP2B6 is highly inducible through the activation of nuclear receptors (NRs) such as constitutive androstane receptor (CAR, NR1I3) and pregnane X receptor (PXR, NR1I2) (Sueyoshi et al., 1999; Goodwin et al., 2001). By cross-talking, CAR and PXR coordinately control the induction of CYP2B6 expression via recognizing and binding to two putative xenobiotic response clusters located at −1.7 kb [phenobarbital-responsive enhancer module (PBREM)] and −8.5 kb [xenobiotic-responsive enhancer module (XREM)] of the CYP2B6 promoter (Sueyoshi et al., 1999; Wang et al., 2003). PXR and CAR are considered promiscuous xenobiotic sensors that recognize a broad array of xenobiotics and translate chemical activation into enhanced expression of target genes.
In addition to chemical induction, CYP2B6 is also characterized as a highly polymorphic gene. Presently, 29 alleles of CYP2B6 have been identified (available at http://www.cypalleles.ki.se/) from more than 50 mutations in different combinations. Among these alleles, extensive studies have centered on the nonsynonymous single nucleotide polymorphisms (SNPs), which often result in decreased expression or malfunction of CYP2B6 protein (Lang et al., 2004; Rotger et al., 2007). Comparatively, little is known regarding the function of polymorphisms in the promoter of CYP2B6 gene. Initial genotyping of 83 and 108 human samples led to the identification of 10 and 14 mutations in the promoter of CYP2B6, respectively (Lamba et al., 2003; Hesse et al., 2004). Sequence analysis of these mutations purported that a number of them may influence CYP2B6 expression by altering consensus transcription factor binding sites. Subsequently, Zukunft et al., (2005) reported that the −82T→C, a mutation within the CYP2B6*22 allele, enhances the basal expression of CYP2B6 by introducing a functional CCAAT/enhancer-binding protein (C/EBP) binding site and shifting the transcription start site downstream.
C/EBPs belong to the basic region leucine zipper protein family and play pivotal roles in cellular differentiation, liver regeneration, apoptosis, and liver-specific gene expression (Schrem et al., 2004). An earlier study showed that introduction of C/EBPα to HepG2 cells augments the basal expression of CYP2B6, CYP2C9, and CYP2D6 mRNA (Jover et al., 1998). Disruption of the C/EBPα binding sites in the promoter of CYP3A4 repressed the basal expression of CYP3A4 reporter construct (Rodríguez-Antona et al., 2003). Nevertheless, whether and how the introduction/disruption of C/EBP binding site would affect NR-mediated induction of cytochrome P450 enzymes was largely unexplored.
Given that CYP2B6 expression exhibits dramatic intra- and interindividual variability, we hypothesized that the polymorphic (−82T→C) substitution may affect drug-induced expression of CYP2B6 by facilitating communication between C/EBP and PXR. Here, we provide experimental evidence to show the synergistic effect between this polymorphic mutation and PXR-mediated induction of CYP2B6 gene expression. By using coimmunoprecipitation (CoIP), chromatin immunoprecipitation (ChIP), chromosome conformation capture (3C) assays, and transient luciferase promoter reporter experiments, our studies revealed, for the first time, that C/EBPα directly interplays with xenobiotic receptor PXR and CAR to coregulate the synergistic induction of the CYP2B6 gene. In addition, a pilot genotype-phenotype study showed a possible correlation between this polymorphism and the potent induction of CYP2B6 through PXR activation. Together, these findings reveal a novel role of C/EBP-PXR in the maximal induction of CYP2B6 and may have pharmacological significance in the efficacy and toxicity of drugs as CYP2B6 substrates.
Materials and Methods
Materials.
Phenobarbital (PB), dimethyl sulfoxide (DMSO), RIF, and collagenase type IV were purchased from Sigma-Aldrich (St. Louis, MO). Oligonucleotide primers were synthesized by Integrated DNA technologies (Coralville, IA). The Dual-Luciferase Reporter Assay System was purchased from Promega (Madison, WI). FuGENE 6 and FuGENE HD transfection reagents were from Roche Diagnostics (Basel, Switzerland). Lipofectamine 2000 transfection reagent was from Invitrogen (Carlsbad, CA). Matrigel, insulin, and ITS+ were obtained from BD Biosciences (San Jose, CA). Other cell culture reagents were purchased from Invitrogen or Sigma-Aldrich.
Cell Lines.
HepG2, HepG2-stable expression of hPXR (PXR-HepG2), HepG2-stable expression of hCAR (Yh18), Huh7, and COS1 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The Yh18 cell line was obtained from Dr. Masahiko Negishi (National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC) (Swales et al., 2005). PXR-HepG2 was a cell line generated by stable transfection of hPXR (pCR3-hPXR) expression vector and selected for neomycin resistance. A single cell clone was selected and functionally evaluated (Supplementary Fig. 1).
Human Primary Hepatocytes.
Human liver tissues (15 donors) were obtained after surgical resection by qualified pathology staff after diagnostic criteria were met and prior approval from the Institutional Review Board at the University of Maryland School of Medicine was obtained. Hepatocytes were isolated from human liver specimens by a modification of the two-step collagenase digestion method as described previously (LeCluyse et al., 2005). Another 29 human primary hepatocyte preparations were obtained from Life Technologies Corporation (Durham, NC). Hepatocytes were seeded at 1.5 × 106 cells/well in six-well Biocoat (BD Biosciences) plates in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 4 μg/ml insulin, and 1 μM dexamethasone and then cultured in serum-free Williams' E medium as described previously (Wang et al., 2003). All human primary hepatocytes were treated with RIF (10 μM) or vehicle control (0.1% DMSO) for 24 or 72 h before harvesting for mRNA or CYP2B6 activity analysis, respectively.
Plasmids.
The pSG5-hPXR expression vector was obtained from Dr. Steven Kliewer (University of Texas Southwestern Medical Center, Dallas, TX). The CYP2B6-1.8 kb luciferase reporter vector (Swales et al., 2005) and the pMEX-C/EBPα and pcDNA3-C/EBPβ expression vectors were kindly provided by Dr. Masahiko Negishi (National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC). A 2-kb fragment spanning −1 to −1993 bp of the native CYP2B6 promoter region was polymerase chain reaction (PCR)-amplified using specific primers listed in Table 1. This product was subcloned into the NheI-HindIII site of pGL3-basic vector, resulting in the construct termed CYP2B6-2kb. Polymorphic variant −82T→C and C/EBP disruption −82M were constructed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) (Fig. 1A). The CYP2B6-2kb construct and all of the mutants were sequencing-confirmed. The pRL-TK Renilla reniformis luciferase plasmids used to normalize firefly luciferase activities were from Promega.
TABLE 1.
Primer sequences for PCR assays
| Assay | Gene | Sequence (5′→3″) | Reference |
|---|---|---|---|
| RT-PCR | GAPDH | CCCATCACCATCTTCCAGGAG | Spandidos et al., 2009 |
| GTTGTCATGGATGACCTTGGC | |||
| C/EBPα | TCGGTGGACAAGAACAGCAA | ||
| TTTCAGGAGGCACCGGAATCT | |||
| C/EBPβ | CTTCAGCCCGTACCTGGAG | ||
| GGAGAGGAAGTCGTGGTGC | |||
| CYP2B6 | AGACGCCTTCAATCCTGACC | ||
| CCTTCACCAAGACAAATCCGC | |||
| ChIP | RVprimer3 | CTAGCAAAATAGGCTGTCCC | Song et al., 2004 |
| CYP2B6(PBREM) | GATGCTGATTCAGGGAATGGA | ||
| 3C | CYP2B6(loop) | AGGATAAAAGGCCCAGTTGGA | Inoue et al., 2009 |
| CTAAGATTGGGTGCTCATTGCA | |||
| CYP2B6(control) | GGCCAGGATGGTCTCGAA | ||
| CCAGCACGTTGGGAAGCT | |||
| CYP2B6-2kb | CYP2B6-2kNheI | CTAGCTAGCGGACAATGTAGCCCCAACCC | |
| CYP2B6-2kHindIII | CCCAAGCTTGGTCCTGGTCTGACTGCCCTG |
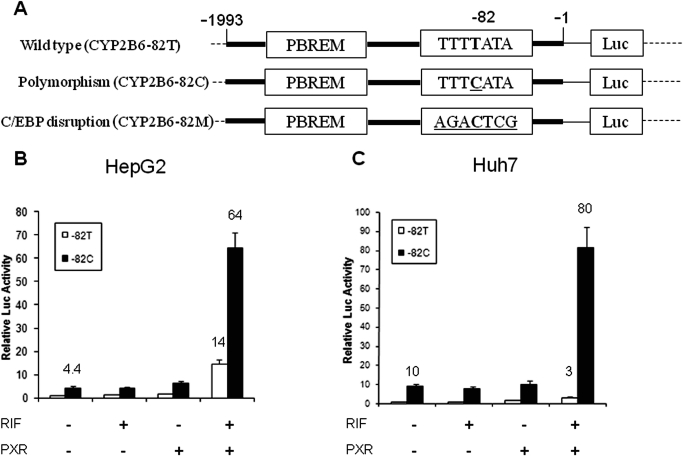
Fig. 1.
Synergistic activation of CYP2B6 reporter in human hepatoma cells. A, three firefly luciferase reporter constructs carrying wild-type (−82T), mutant (−82C), or C/EBP binding site disruptive mutation (−82M) of −2-kb CYP2B6 promoter. The location of the PBREM is also indicated. B and C, representative reporter assays in HepG2 cells and Huh7 cells. Cells were cotransfected with the reporter constructs carrying −82T or −82C, in the presence of PXR expression vector or empty pGS5 vector. Twenty-four hours after transfection, cells were treated with RIF (10 μM) or vehicle (0.1% DMSO) for 24 h. Luciferase activities were measured using the dual-luciferase kit (Promega). Triplicate samples were performed for each treatment.
Transient Transfection and Luciferase Assay.
HepG2 or Huh7 cells in 24-well plates were transfected with hPXR expression vector and CYP2B6-2kb reporter construct using Fugene 6 transfection reagent following the manufacturer's instructions. Twenty-four hours after transfection, cells were treated with solvent (0.1% DMSO) or 10 μM RIF for another 24 h. Thereafter, cell lysates were assayed for firefly activities normalized against the activities of cotransfected R. reniformis luciferase using the Dual-Luciferase kit (Promega). Data are represented as mean ± S.D. of three individual transfections.
Short Interfering RNA.
The predesigned siRNA specific for C/EBPα (mixture of Hs_CEBPA_2 and Hs_CEBPA_4) and a nontargeting siRNA were obtained from QIAGEN (Valencia, CA). To detect the knockdown of endogenous C/EBPα, HepG2 cells plated in 12-well plates were transfected with siRNA-CEBPα (40 pmol) or nontargeting siRNA (40 pmol) using Lipofectamine 2000 (Invitrogen) transfection reagent. Forty-eight hours after transfection, cells were harvested, and total RNA was isolated and reverse-transcribed into cDNA. C/EBP gene expression was measured using SYBR real-time PCR as described under Quantitative RT-PCR. In cell-based reporter assays, after 24 h of siRNA transfection, PXR expression vector and CYP2B6-2kb reporter vector were also transfected in HepG2 cells using FuGENE HD transfection reagent. The double-transfected cells were treated with RIF (10 μM) or vehicle control (0.1% DMSO) for 24 h and then subjected to dual-luciferase assays as described above.
Coimmunoprecipitation Assays.
COS1 cells were transfected with C/EBPα and PXR expression vectors and were treated with RIF (10 μM) for 24 h. Thereafter, cells were washed with phosphate-buffered saline buffer and scraped into the lysis buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 0.5% Nonidet P-40, 0.5 mM phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor cocktail). After incubation on ice for 15 min, the lysate was centrifuged at 13,000g for 15 min at 4°C. The supernatant fractions were collected and incubated with antibodies (anti-PXR rabbit polyclonal antibody and anti-C/EBPα mouse monoclonal antibody; Santa Cruz Biotechnology, Santa Cruz, CA) and protein A-Sepharose beads overnight at 4°C. Corresponding isotype IgG was used as a negative control. The beads were washed three times, and the precipitated protein complexes were analyzed by Western blotting with PXR and C/EBPα antibodies, respectively.
Chromatin Immunoprecipitation Assays.
Experiments were performed using a ChIP assay kit according to the manufacturer's protocol (Millipore Corporation, Billerica, MA). In brief, 2 × 106 PXR-HepG2 cells were seeded into a 10-cm dish, grown to 80% confluence, and transfected with constructs containing −82T, −82C, or −82M. Transfected cells were treated with RIF (10 μM) or DMSO [0.1% (v/v)] for 3 h. Thereafter, cells were cross-linked with 1% formaldehyde for 10 min at 37°C, washed with ice-cold phosphate-buffered saline containing a protease inhibitor cocktail, lysed, and sonicated. Immunoprecipitation was performed overnight at 4°C using 2.5 μg of rabbit anti-human PXR polyclonal antibody or isotype control IgG followed by precipitation using protein A coupled to agarose beads. After de–cross-linking and protease digestion, DNA fragments were recovered by QIAquick PCR purification kit (QIAGEN). Quantitative PCR was performed using specific sets of primers (Table 1) (Song et al., 2004). PCR products were also resolved on a 1.5% agarose gel and visualized by ethidium bromide staining.
3C Assays.
3C assays were performed as described previously with minor modification (Babu et al., 2008; Inoue and Negishi, 2009). In brief, 100 μg of nuclear extract prepared from PXR-HepG2 cells were shaken with 50 ng of the CYP2B6-1.8kb construct containing −82T, −82C, or −82M, respectively, at room temperature for 45 min and were cross-linked with formaldehyde then terminated by the addition of glycine. Thereafter, these protein-DNA complexes were precipitated by ethanol at −20°C. Precipitates were dissolved in 80 μl of Tris-EDTA buffer, digested by XhoI at 37°C for 4 h, and then incubated for 20 min at 70°C to inactivate the enzyme activity of XhoI. Digests were ligated by T4 DNA ligase (Fermentas, Hanover, MD) for 5 min at room temperature. After ligation, protein-DNA complexes were de–cross-linked by adding SDS and NaCl up to 1% and 0.3 M, respectively, and treated overnight at 55°C and then treated with proteinase K for an additional 1 h at 65°C. DNA was purified by PCR purification kit (QIAGEN). Purified DNA was subjected to amplification using two primer sets (Table 1).
Quantitative RT-PCR.
Total RNA was isolated from treated hepatocytes using the RNeasy Mini kit (QIAGEN) and reverse-transcribed using High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) following the manufacturers' instructions. CYP2B6 mRNA expression was normalized against the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Real-time PCR assays were performed in 96-well optical plates on an ABI Prism 7000 Sequence Detection System with SYBR Green PCR Master Mix (Applied Biosystems). Primers for CYP2B6 and GAPDH and C/EBP mRNA detection are shown in Table 1 (Spandidos et al., 2010). Fold induction values were calculated using 2ΔΔCT, where ΔCt represents the differences in cycle threshold numbers between the target gene and GAPDH, and ΔΔCt represents the relative change in these differences between control and treatment groups.
CYP2B6 Activity Assay.
CYP2B6 basal and induced enzymatic activities were determined in situ from cultures of human primary hepatocytes using the probe substrate bupropion. Incubations were performed with 0.5 ml of Hanks' balanced salt solution containing 250 μM bupropion for 15 min for each preparation of hepatocytes after 72-h treatment with 0.1% DMSO or 10 μM RIF. After incubations were complete, supernatants were transferred to deep well blocks and frozen at −80°C for subsequent liquid chromatography/tandem mass spectrometry analysis of hydroxybuprion formation using standard analytical methods as described previously (Lau and Chang, 2009).
Genotyping.
Genomic DNA samples from human primary hepatocytes of 44 liver donors (Supplemental Table 1) were isolated and purified by QIAamp DNA Mini kit (QIAGEN). Genotyping of CYP2B6 −82T→C was undertaken using ABI Prism 7000 Sequence Detection System, and probes and primers were also designed by Applied Biosystems (TaqMan Drug Metabolism SNP Genotyping assay, C_27830964_10). In brief, 50 ng of whole genomic DNA was used as a template for amplification of the CYP2B6 target sequence. The PCR reactions were conducted in a reaction volume of 10 μl (DNA template, 2× genotyping master mix, and 20× mixture of primers and probes) under the following conditions: initial denaturation step of 10 min at 95°C followed by 50 cycles of denaturation 92°C for 15 s, annealing, and extending for 90 s at 60°C. The allelic discrimination was performed using the SDS software (Applied Biosystems).
Statistics.
All reporter assay data represent at least three independent experiments and are expressed as the mean ± S.D. Statistical comparisons were made using Student's t test. Because CYP2B6 activity data were not normally distributed, nonparametric method (rank sum test) was used to compare phenotypic data. Statistical significance was set at p < 0.05.
Results
Synergistic Enhancement of CYP2B6 Reporter Expression by CYP2B6 −82T→C and PXR Activation.
Initial investigation of the potential interplay between CYP2B6 −82T→C (CYP2B6-82C) polymorphisms and PXR activation was carried out using cell-based reporter assays in HepG2 and Huh7 cells, two human hepatoma cell lines. As expected, in HepG2 cells, CYP2B6-82C mutation alone enhanced the basal reporter expression by 4.4-fold over the reference CYP2B6-82T, and RIF-mediated activation of PXR resulted in 14-fold increase in CYP2B6-82T expression (Fig. 1B). Surprisingly, the combination of CYP2B6-82C mutation and activation of PXR led to a 64-fold augmentation of reporter expression over the reference CYP2B6-82T, which is substantially higher than the additive effects of the polymorphism and PXR activation individually (4.4- plus 14-fold). In Huh7 cells, a similar trend but more robust synergistic effect has been observed, in which CYP2B6-82C and PXR activation resulted in 10- and 3-fold increases, respectively, whereas in combination, they enhance the reporter activity by 80-fold over the reference CYP2B6-82T control (Fig. 1C). Notably, without ligand activation, overexpression of PXR had no effects on either of these two CYP2B6 reporter constructs (Fig. 1, B and C), suggesting that ligand-based conformational changes in PXR are required for these observed synergistic enhancement of CYP2B6 reporter expression.
C/EBPα Plays an Important Role in the Observed Synergism.
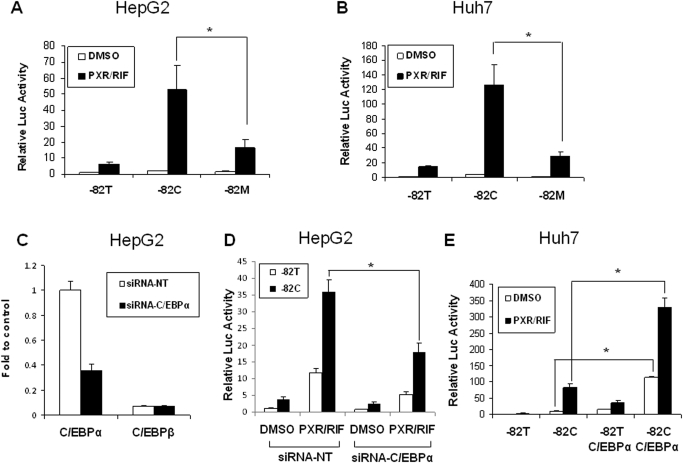
Because the polymorphic −82C was reported to form a new C/EBP binding site, we further evaluated the role of C/EBP in this synergistic effect in HepG2 and Huh7 cells. As depicted in Fig. 1A, a construct containing mutations disrupting the C/EBP binding site without affecting the polymorphic −82C was generated and termed CYP2B6-82M. Reporter assays revealed that PXR-mediated activation of CYP2B6-82M expression was significantly reduced compared with CYP2B6-82C in transfected HepG2 and Huh7 cells (Fig. 2, A and B). To further test the effects of C/EBPα expression on PXR-mediated activation of CYP2B6-82C expression, an siRNA specific for C/EBPα was used to knock down the C/EBPα expression in HepG2 cells. Forty-eight hours after transfection, the gene expression of C/EBPα was down-regulated by 65% compared with the control group transfected with nontargeting siRNA (Fig. 2C). This knockdown of C/EBPα significantly decreased the synergistic effects between CYP2B6-82C and PXR activation (Fig. 2D). On the other hand, overexpression of transfected C/EBPα expression vector led to an enhancement of PXR-mediated activation of CYP2B6-82C (Fig. 2E). Together, these results support the pivotal role of C/EBP in the observed synergism of CYP2B6-82C and PXR activation. In addition, because the major isoform of C/EBP expressed in HepG2 cells is C/EBPα (Fig. 2C), this isoform is the focus of the current studies.
Fig. 2.
Both the −82T→C mutation and C/EBPα protein are required in the synergistic response. Different reporter and/or expression constructs, as well as siRNAs, were transfected into HepG2 or Huh7 cells as described under Materials and Methods. A and B, HepG2 and Huh7 cells were cotransfected with the reporter constructs carrying −82T, − 82C, or −82M, and PXR expression vector or empty vector. Transfected cells were then treated with vehicle or RIF for 24 h. Luciferase activities were measured using the dual-luciferase kit (Promega). C, HepG2 cells were transfected with siRNA-control or siRNA-CEBPα. Expression levels of C/EBPα and C/EBPβ were detected using real-time RT-PCR. D, in a parallel experiment, 24 h after C/EBPα knockdown, HepG2 cells were subsequently cotransfected with cytochrome P450 −82T, or −82C, and PXR expression vector or empty vector. Luciferase activities were measured as described above after RIF treatment. E, similar reporter assays were conducted in Huh7 cells after the initial transfection of C/EBPα expression vector. All experiments were performed in triplicate samples. *, p < 0.05.
Direct Interaction between PXR and C/EBPα.
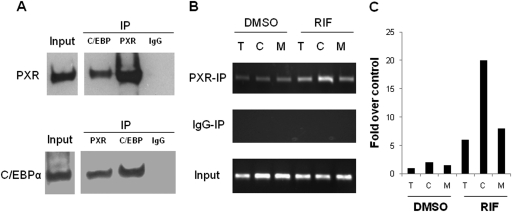
To determine whether the two proteins interact in a cellular context, we performed CoIP assays using COS-1 cells transiently transfected with PXR and C/EBPα expression vectors. As demonstrated in Fig. 3A, immunoprecipitation (IP) of PXR protein using PXR antibody resulted in coimmunoprecipitation of C/EBPα protein, a result that was not observed when normal rabbit IgG was used for IP instead. Alternatively, coimmunoprecipitation of PXR was also observed when C/EBPα antibody was used for IP. These findings suggest that the two proteins are components of an intracellular complex.
Fig. 3.
C/EBPα interacts with PXR, and the −82T→C mutation enhances the recruitment of PXR to the PBREM of CYP2B6. A, direct protein-protein interaction between PXR and C/EBPα was detected by CoIP assay in COS1 cells transiently transfected with PXR and C/EBPα expression vectors. B, PXR recruitment to the PBREM region of CYP2B6 was detected using a modified ChIP assay as described under Materials and Methods. T, −82T wild type; C, −82C mutant; M, the C/EBP binding site disruption mutant. C, quantity of the de–cross-linked DNA was measured by real-time PCR.
−82T→C Mutation Increased Recruitment of PXR to the Promoter of CYP2B6.
To demonstrate whether the presence of the polymorphic −82C mutation would enhance the binding of PXR to the PBREM in the native CYP2B6 promoter, a modified ChIP assay was conducted in PXR-HepG2 cells. CYP2B6 promoter construct CYP2B6-82T, CYP2B6-82C, or CYP2B6-82M was transfected into the PXR-HepG2 cells, respectively, and treated with RIF (10 μM) for 3 h as described under Materials and Methods. To eliminate the interference of the genomic wild-type CYP2B6 inherited in HepG2 cells, a pair of primers specific to the plasmid-expressed PBREM region was designed (Table 1). In particular, the forward primer complements with the backbone sequence of pGL3-basic plasmid but not the CYP2B6 promoter. Using this unique system, we observed that recruitment of PXR to the PBREM in CYP2B6-82C construct was clearly stronger than that to the CYP2B6-82T and CYP2B6-82M after RIF stimulation (Fig. 3, B and C).
Looping the Distal PBREM Toward the Proximal C/EBP Binding Site.
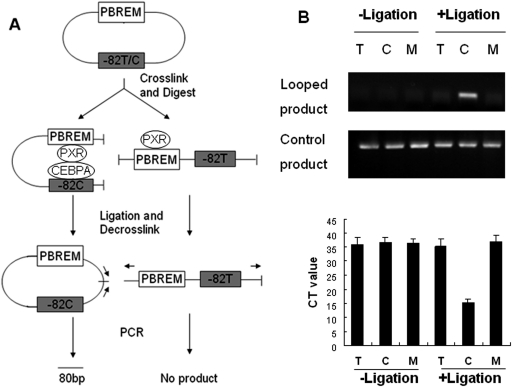
It seems that both the distal PBREM and the −82C formed proximal C/EBPα binding site are required for the synergistic effects of PXR-mediated transcription of CYP2B6. An in vitro 3C assay was used to explore whether a looping mechanism exists by which the PBREM-bound PXR would have physical access to the C/EBPα sitting on −82C generated proximal binding site (Hagège et al., 2007; Inoue and Negishi, 2009). As depicted in Fig. 4A, the −1.8-kb CYP2B6 promoter constructs containing either −82T, −82C, or −82M were cross-linked with nuclear proteins and digested to open the circled plasmids. After ligation, PCR reaction should produce an 80-bp fragment if looping occurs. When the −1.8 kb CYP2B6 promoter was incubated with nuclear proteins prepared from PXR-HepG2 cells, a dramatically increased amplification of the 80-bp fragments was observed in the construct containing −82C, whereas no PCR product can be detected in either the reference −82T or the −82M construct (Fig. 4B).
Fig. 4.
Physical interaction between PXR-bound PBREM and C/EBPα harboring −82C. A, schematic presentation of the chromosome conformation capture (3C) experimental procedure. B, the −1.8-kb CYP2B6 constructs containing −82T (T), −82C (C), or −82M (M) were incubated with nuclear extracts prepared from PXR-HepG2 cell line treated with RIF (10 μM) or vehicle control and subjected to 3C experiments as outlined under Materials and Methods. The degrees of ligation were determined by PCR using primers listed in Table 1. The PCR amplicons were loaded to a 1.5% agarose gel and stained with ethidium bromide.
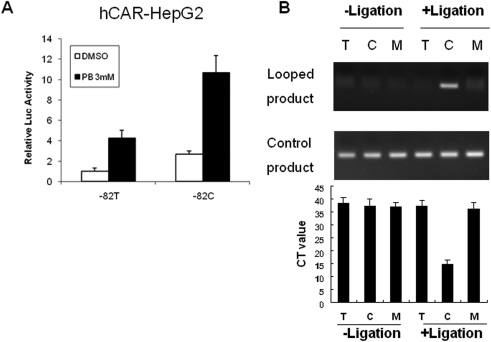
Because CAR induces CYP2B6 through binding to the PBREM, we also examined whether −82T→C mutation could enhance CAR-mediated induction of CYP2B6 in the same manner as with PXR. In cell-based reporter assays, CYP2B6-82C and PB-stimulated CAR activation resulted in 2.5- and 4-fold increases over the reference CYP2B6-82T control, respectively, whereas combined, they enhanced the reporter activity by 10-fold (Fig. 5A). Similar to the results from PXR, a parallel 3C experiment using nuclear extract from the Yh18 cells revealed that CAR-bound PBREM was looped toward the C/EBP-occupied −82C region, which has led the amplification of an 80-bp PCR product in CYP2B6-82C only (Fig. 5B).
Fig. 5.
Interplay between CAR and C/EBPα. A, human CAR-HepG2 stable cell line (Yh18) was transfected with CYP2B6 reporter constructs carrying −82T or −82C, in the presence of pRL-TK vector as internal control. Transfected cells were then treated with vehicle or PB (3 mM) for 24 h. Luciferase activities were measured using the dual-luciferase kit (Promega). Triplicate samples were performed for each treatment. B, similar 3C assays as described above were conductive using nuclear extracts prepared from Yh18 cells.
Genotype-Phenotype Association of CYP2B6-82T/C polymorphism and RIF-Mediated Induction of CYP2B6 in Human Primary Hepatocytes.
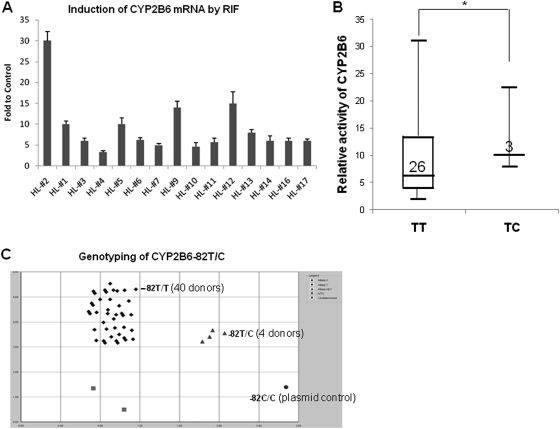
To further characterize the role of the −82T→C mutation in drug-mediated CYP2B6 induction in humans, a collection of fresh human primary hepatocytes from 44 liver donors was treated with RIF for 24 h, and the RIF-mediated induction of CYP2B6 mRNA (15 donors) or bupropion hydroxylation activity (29 donors) was analyzed (Supplemental Table 1). Overall, four donor preparations were identified with the CYP2B6-82T/C genotype, whereas the remaining 40 donors were reference carriers of CYP2B6-T/T (Fig. 6C). Real-time PCR analysis of 15 human hepatocyte preparations from University of Maryland Medical Center showed that RIF induction of CYP2B6 mRNA was most robustly in HL#2 (30-fold), which happens to be the only CYP2B6-82T/C carrier in these 15 donors (Fig. 6A). In addition, CYP2B6 activity (bupropion hydroxylation) was analyzed in 29 human primary hepatocytes from Life Technologies Corporation. As demonstrated in Fig. 6B, the average induction of RIF in three CYP2B6-82T/C heterozygotes (11-fold) was significantly higher than that observed in the 26 donors with reference CYP2B6-82T/T (6.5-fold).
Fig. 6.
Association of CYP2B6-82T/C genotyping with the induction of CYP2B6 by rifampicin treatment. A, 15 human primary hepatocyte preparations were treated with RIF (10 μM) or vehicle control (0.1% DMSO) for 24 h. Real-time RT-PCR was conducted to measure fold induction of CYP2B6 mRNA (HL#2 is −82T/C and all the rest are −82T/T carriers). B, 29 human hepatocyte preparations were treated with RIF (10 μM) or vehicle control (0.1% DMSO) for 72 h. Bupropion hydroxylation was used to detect fold induction of CYP2B6 activity as described under Materials and Methods. C, the genotyping distribution of CYP2B6-82T→C SNP in the 44 liver donors. *, p < 0.05.
Discussion
Remarkable inter- and intraindividual variations in CYP2B6 expression and catalysis have been documented through an array of in vivo and in vitro studies (Code et al., 1997; Ekins et al., 1998; Hanna et al., 2000; Hesse et al., 2000). These variations are relevant to clinical outcomes in context of interindividual drug safety and efficacy with CYP2B6-metabolized drugs such as bupropion, efavirenz, and cyclophosphamide (Gatanaga et al., 2007; Rotger et al., 2007). To date, mounting evidence indicates that both nuclear receptor-mediated induction and genetic variation are important factors in variable interindividual CYP2B6 expression (Sueyoshi et al., 1999; Lang et al., 2001; Lamba et al., 2003; Wang and Tompkins, 2008). Nevertheless, these two pivotal factors have predominantly been considered separately, with a clear lack of studies investigating the interplay between genetic polymorphisms and NR-mediated drug induction for CYP2B6. In this report, we have demonstrated that the SNP −82T→C that creates a functional C/EBP binding site in the proximal promoter of CYP2B6, exerts synergistic effects with PXR on RIF-mediated induction of CYP2B6 expression. Moreover, a novel mechanism has been revealed by which ligand-activated PXR could mediate the synergistic response through interaction with C/EBPα protein harboring the −82T→C introduced C/EBP binding site.
Several previous studies have indicated that polymorphic changes in the promoter of drug-metabolizing genes may affect the basal expression and/or activity of these isozymes. For instance, UGT1A1*28, which introduces an extra TA to the TATA box of this gene, has resulted in a lower UGT1A1 expression and activity (Lampe et al., 1999; Biason et al., 2008); SNPs of −750T→C and −82T→C in the promoter of CYP2B6 have led to decreased and increased basal expression and activity of CYP2B6, respectively (Lamba et al., 2003; Hesse et al., 2004; Zukunft et al., 2005). Interestingly, further analysis of the CYP2B6 promoter by Zukunft et al. (2005) revealed that the −82T→C substitution introduces a functional C/EBP binding site and shifts the transcription start site downstream. Knowing that C/EBP proteins play pleiotropic roles in hepatic gene expression, we initially explored whether the C/EBP site forming SNP (−82T→C) would affect the PXR-mediated CYP2B6 expression. To our surprise, a potent synergism between −82T→C mutation and ligand-activated PXR has been observed in our cell-based reporter experiments using both HepG2 and Huh7 cells. This is further evidenced by the synergistic response attenuated by both site-directed disruption of the C/EBP binding site and by genetic knockdown of the C/EBPα expression by siRNA. On the other hand, overexpression of C/EBPα in Huh7 cells was found to augment this synergistic response. Together, these results and the elevated induction responses observed in primary hepatocyte cultures from −82T/C individuals suggest the existence of a positive interdependence between PXR and C/EBPα, leading to synergism.
It has been well established that induction of CYP2B6 gene expression is predominantly regulated at the transcriptional level. To date, most findings suggest that induction of CYP2B6 gene by xenobiotics is mediated by activation of the nuclear receptors CAR and/or PXR through interactions with the PBREM/XREM located in the upstream of CYP2B6 transcriptional start site (Sueyoshi et al., 1999; Goodwin et al., 2001; Wang et al., 2003). Nevertheless, several significant phenomena regarding CYP2B6 induction could not be fully explained by this simplified model. For instance, in contrast to the potent induction of CYP2B6 gene, relatively moderate induction has been observed for the majority of other PXR/CAR target genes such as CYP2Cs and UGT1A1 (Sugatani et al., 2004; Ferguson et al., 2005; Chen and Goldstein, 2009). Moreover, dramatic variations of PXR/CAR-mediated induction of CYP2B6 occurred among individuals even with the same PBREM/XREM sequences. This suggests that other regulatory factors, in addition to PXR/CAR proteins and the PBREM/XREM sites, may be also involved in the maximal induction of the CYP2B6 gene. In addition to the synergy between the −82T→C mutation and PXR activation in cell-based reporter assays, our coimmunoprecipitation data clearly showed that C/EBPα and PXR cross-coupled together in an intracellular environment. Of importance, recruitment of PXR to the PBREM of CYP2B6 was remarkably enhanced only when the −82T→C was presented in the modified ChIP assays, suggesting that both C/EBP protein and the novel binding site are crucial for the beneficial interaction of PXR with the CYP2B6 promoter. C/EBPs are liver-enriched transcription factors with pleiotropic influences on hepatic gene expression (Schrem et al., 2004). Previous studies indicate that cross-talk with other transcriptional factors exists for C/EBPs in modulating their target gene expression. For example, the formation of a complex between the glucocorticoid receptor and the C/EBPα involves the regulation of glucocorticoids on lymphocytic and mesenchymal cell proliferation (Rüdiger et al., 2002); whereas coupling of C/EBPs and nuclear factor κB affects the expression of genes with putative κ-B enhancer motifs via protein-protein interactions (Stein and Baldwin, 1993; Stein et al., 1993). Recently, Song et al. (2006) illustrated the essential role of C/EBPα in vitamin D receptor-mediated induction of SULT2A1 by directly interacting with each other through adjacent vitamin D-responsive element and C/EBP sites. In contrast, the known PXR/CAR-interacting PBREM and the novel C/EBP binding site in the promoter of CYP2B6 are distally spaced by approximately 1.7 kb.
Insight into the genomic organization of mammals reveals that regulatory elements, enhancers in particular, in the promoter of genes are often dispersed remotely encompassing a long scale of kilobase pairs (Hagège et al., 2007). Proper folding of the sequence at this scale is extremely important for gene regulation. In light of the recent development of an in vitro 3C assay, which allows the detection of physical interactions of protein-bound DNA segments and DNA looping (Hagège et al., 2007; Inoue and Negishi, 2009), we applied this assay to test the hypothesis that PXR-bound PBREM is physically folded toward the proximal C/EBP binding site through a looping mechanism. Our 3C experiments revealed that both PXR- and CAR-bound PBREMs were efficiently looped to the C/EBP harboring −82C site but not to the −82T region (Figs. 4B and 5B). Given that DNA looping is a widely accepted mechanism by which distantly separated cis-acting elements were physically coupled, it is reasonable to speculate that constitutive factors such as C/EBPs and hepatic nuclear factors may interact with xenobiotic factors such as PXR, CAR, aryl hydrocarbon receptor, and vitamin D receptor through the common looping mechanisms to facilitate coregulation of their target genes.
To further investigate the association of −82T→C with RIF-mediated induction of CYP2B6, a pilot genotype-phenotype association study has been conducted in human primary hepatocytes collected from 44 donors. Notably, four CYP2B6-82T/C heterozygous and no CYP2B6-82C/C homozygous carriers were identified from these donors. It is in agreement with an earlier report in that −82T/C heterozygotes were the dominant form of this SNP with no −82C/C being identified, yet the basal expression of CYP2B6 was significantly increased in the −82T/C carriers (Zukunft et al., 2005). In our functional experiments, RIF induction of CYP2B6 mRNA or activity was clearly elevated in hepatocyte cultures from the four −82T/C heterozygous compared with the reference donors (Fig. 6). Although we cannot reach a conclusive correlation between −82T→C SNP and RIF-mediated induction of CYP2B6 in humans at this point because of the limited sample size and the difficult nature of obtaining human primary hepatocytes, these pilot results indicate that the −82T/C carrier might be more sensitive to chemical induction of CYP2B6 gene.
In conclusion, our data suggest that polymorphic −82T >C mutation exerts synergistic effects with PXR in xenobiotic-mediated induction of CYP2B6 gene. This synergy is mediated through a novel mechanism by which the constitutive C/EBPα interplays with xenobiotic-sensitive PXR and led to the enhanced recruitment of PXR to the PBREM and folding of the distal PBREM toward the proximal region of CYP2B6 in which transcription happens. Moreover, the pilot genotype-phenotype association study seems to support the observation that PXR-mediated induction of CYP2B6 is enhanced in donors with such a gain-of-function allele. Overall, these findings bridge the polymorphism of CYP2B6 with the NR-mediated induction of this gene and shed light on a potential explanation of the remarkable inter- and intraindividual variations in CYP2B6 expression. Given the increasing importance of CYP2B6 in drug metabolism and detoxification, these findings may also have clinical implications to drug-drug interactions arising from the induction of CYP2B6-mediated substrates.
Supplementary Material
Acknowledgments
We thank Dr. Masahiko Negishi (National Institute of Environmental Health Sciences/National Institutes of Health, Research Triangle Park, NC) and Dr. Steven Kliewer (University of Texas Southwestern Medical Center, Dallas, TX) for the provision of multiple vectors as indicated in Materials and Methods. We also gratefully acknowledge the liver samples obtained from Life Technologies (Durham, NC) and The University of Maryland Medical Center (Baltimore, MD).
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institute of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant DK061652].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.065185.
- NR
- nuclear receptor
- CAR
- constitutive androstane receptor
- C/EBP
- CCAAT/enhancer-binding protein
- ChIP
- chromatin immunoprecipitation assay
- CoIP
- coimmunoprecipitation assay
- 3C
- chromosome conformation capture assay
- DMSO
- dimethyl sulfoxide
- PB
- phenobarbital
- PBREM
- phenobarbital-responsive enhancer module
- PXR
- pregnane X receptor
- RIF
- rifampicin
- XREM
- xenobiotic-responsive enhancer module
- SNP
- single nucleotide polymorphism
- kb
- kilobase
- bp
- base pair
- siRNA
- short interfering RNA
- PCR
- polymerase chain reaction
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- IP
- immunoprecipitation
- RT-PCR
- reverse transcriptase polymerase chain reaction
- CYP2B6-82C
- CYP2B6 −82T→C.
References
- Babu DA, Chakrabarti SK, Garmey JC, Mirmira RG. (2008) Pdx1 and BETA2/NeuroD1 participate in a transcriptional complex that mediates short-range DNA looping at the insulin gene. J Biol Chem 283:8164–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biason P, Masier S, Toffoli G. (2008) UGT1A1*28 and other UGT1A polymorphisms as determinants of irinotecan toxicity. J Chemother 20:158–165 [DOI] [PubMed] [Google Scholar]
- Chen Y, Goldstein JA. (2009) The transcriptional regulation of the human CYP2C genes. Curr Drug Metab 10:567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Code EL, Crespi CL, Penman BW, Gonzalez FJ, Chang TK, Waxman DJ. (1997) Human cytochrome P4502B6: interindividual hepatic expression, substrate specificity, and role in procarcinogen activation. Drug Metab Dispos 25:985–993 [PubMed] [Google Scholar]
- Ekins S, Vandenbranden M, Ring BJ, Gillespie JS, Yang TJ, Gelboin HV, Wrighton SA. (1998) Further characterization of the expression in liver and catalytic activity of CYP2B6. J Pharmacol Exp Ther 286:1253–1259 [PubMed] [Google Scholar]
- Faucette SR, Hawke RL, Lecluyse EL, Shord SS, Yan B, Laethem RM, Lindley CM. (2000) Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab Dispos 28:1222–1230 [PubMed] [Google Scholar]
- Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. (2005) Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4alpha. Mol Pharmacol 68:747–757 [DOI] [PubMed] [Google Scholar]
- Gatanaga H, Hayashida T, Tsuchiya K, Yoshino M, Kuwahara T, Tsukada H, Fujimoto K, Sato I, Ueda M, Horiba M, et al. (2007) Successful efavirenz dose reduction in HIV type 1-infected individuals with cytochrome P450 2B6 *6 and *26. Clin Infect Dis 45:1230–1237 [DOI] [PubMed] [Google Scholar]
- Goodwin B, Moore LB, Stoltz CM, McKee DD, Kliewer SA. (2001) Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol 60:427–431 [PubMed] [Google Scholar]
- Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nature Protocols 2:1722–1733 [DOI] [PubMed] [Google Scholar]
- Hanna IH, Reed JR, Guengerich FP, Hollenberg PF. (2000) Expression of human cytochrome P450 2B6 in Escherichia coli: characterization of catalytic activity and expression levels in human liver. Arch Biochem Biophys 376:206–216 [DOI] [PubMed] [Google Scholar]
- Hesse LM, He P, Krishnaswamy S, Hao Q, Hogan K, von Moltke LL, Greenblatt DJ, Court MH. (2004) Pharmacogenetic determinants of interindividual variability in bupropion hydroxylation by cytochrome P450 2B6 in human liver microsomes. Pharmacogenetics 14:225–238 [DOI] [PubMed] [Google Scholar]
- Hesse LM, Venkatakrishnan K, Court MH, von Moltke LL, Duan SX, Shader RI, Greenblatt DJ. (2000) CYP2B6 mediates the in vitro hydroxylation of bupropion: potential drug interactions with other antidepressants. Drug Metab Dispos 28:1176–1183 [PubMed] [Google Scholar]
- Hidestrand M, Oscarson M, Salonen JS, Nyman L, Pelkonen O, Turpeinen M, Ingelman-Sundberg M. (2001) CYP2B6 and CYP2C19 as the major enzymes responsible for the metabolism of selegiline, a drug used in the treatment of Parkinson's disease, as revealed from experiments with recombinant enzymes. Drug Metab Dispos 29:1480–1484 [PubMed] [Google Scholar]
- Hodgson E, Rose RL. (2007) The importance of cytochrome P450 2B6 in the human metabolism of environmental chemicals. Pharmacol Ther 113:420–428 [DOI] [PubMed] [Google Scholar]
- Inoue K, Negishi M. (2009) Early growth response 1 loops the CYP2B6 promoter for synergistic activation by the distal and proximal nuclear receptors CAR and HNF4alpha. FEBS Lett 583:2126–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover R, Bort R, Gómez-Lechón MJ, Castell JV. (1998) Re-expression of C/EBP alpha induces CYP2B6, CYP2C9 and CYP2D6 genes in HepG2 cells. FEBS Lett 431:227–230 [DOI] [PubMed] [Google Scholar]
- Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, Fackenthal JD, Rogan PK, Ring B, Wrighton SA, et al. (2003) Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther 307:906–922 [DOI] [PubMed] [Google Scholar]
- Lampe JW, Bigler J, Horner NK, Potter JD. (1999) UDP-glucuronosyltransferase (UGT1A1*28 and UGT1A6*2) polymorphisms in Caucasians and Asians: relationships to serum bilirubin concentrations. Pharmacogenetics 9:341–349 [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Fischer J, Nüssler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, Zanger UM. (2001) Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11:399–415 [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Richter T, Zibat A, Kerb R, Eichelbaum M, Schwab M, Zanger UM. (2004) Multiple novel nonsynonymous CYP2B6 gene polymorphisms in Caucasians: demonstration of phenotypic null alleles. J Pharmacol Exp Ther 311:34–43 [DOI] [PubMed] [Google Scholar]
- Lau AJ, Chang TK. (2009) Inhibition of human CYP2B6-catalyzed bupropion hydroxylation by Ginkgo biloba extract: effect of terpene trilactones and flavonols. Drug Metab Dispos 37:1931–1937 [DOI] [PubMed] [Google Scholar]
- LeCluyse EL, Alexandre E, Hamilton GA, Viollon-Abadie C, Coon DJ, Jolley S, Richert L. (2005) Isolation and culture of primary human hepatocytes. Methods Mol Biol 290:207–229 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Antona C, Bort R, Jover R, Tindberg N, Ingelman-Sundberg M, Gómez-Lechón MJ, Castell JV. (2003) Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma. Mol Pharmacol 63:1180–1189 [DOI] [PubMed] [Google Scholar]
- Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Décosterd L, Blievernicht J, Saussele T, Günthard HF, Schwab M, et al. (2007) Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther 81:557–566 [DOI] [PubMed] [Google Scholar]
- Rüdiger JJ, Roth M, Bihl MP, Cornelius BC, Johnson M, Ziesche R, Block LH. (2002) Interaction of C/EBPalpha and the glucocorticoid receptor in vivo and in nontransformed human cells. FASEB J 16:177–184 [DOI] [PubMed] [Google Scholar]
- Schrem H, Klempnauer J, Borlak J. (2004) Liver-enriched transcription factors in liver function and development. Part II: the C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev 56:291–330 [DOI] [PubMed] [Google Scholar]
- Song CS, Echchgadda I, Seo YK, Oh T, Kim S, Kim SA, Cho S, Shi L, Chatterjee B. (2006) An essential role of the CAAT/enhancer binding protein-alpha in the vitamin D-induced expression of the human steroid/bile acid-sulfotransferase (SULT2A1). Mol Endocrinol 20:795–808 [DOI] [PubMed] [Google Scholar]
- Song X, Xie M, Zhang H, Li Y, Sachdeva K, Yan B. (2004) The pregnane X receptor binds to response elements in a genomic context-dependent manner, and PXR activator rifampicin selectively alters the binding among target genes. Drug Metab Dispos 32:35–42 [DOI] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. (2010) PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 38:D792–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein B, Baldwin AS., Jr (1993) Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-kappa B. Mol Cell Biol 13:7191–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein B, Cogswell PC, Baldwin AS., Jr (1993) Functional and physical associations between NF-kappa B and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol 13:3964–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresser DM, Kupfer D. (1999) Monospecific antipeptide antibody to cytochrome P-450 2B6. Drug Metab Dispos 27:517–525 [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. (1999) The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 274:6043–6046 [DOI] [PubMed] [Google Scholar]
- Sugatani J, Yamakawa K, Tonda E, Nishitani S, Yoshinari K, Degawa M, Abe I, Noguchi H, Miwa M. (2004) The induction of human UDP-glucuronosyltransferase 1A1 mediated through a distal enhancer module by flavonoids and xenobiotics. Biochem Pharmacol 67:989–1000 [DOI] [PubMed] [Google Scholar]
- Swales K, Kakizaki S, Yamamoto Y, Inoue K, Kobayashi K, Negishi M. (2005) Novel CAR-mediated mechanism for synergistic activation of two distinct elements within the human cytochrome P450 2B6 gene in HepG2 cells. J Biol Chem 280:3458–3466 [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. (2003) A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem 278:14146–14152 [DOI] [PubMed] [Google Scholar]
- Wang H, Tompkins LM. (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Current drug metabolism 9:598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukunft J, Lang T, Richter T, Hirsch-Ernst KI, Nussler AK, Klein K, Schwab M, Eichelbaum M, Zanger UM. (2005) A natural CYP2B6 TATA box polymorphism (−82T–> C) leading to enhanced transcription and relocation of the transcriptional start site. Mol Pharmacol 67:1772–1782 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.