Abstract
The Gene Expression Database (GXD) is a community resource of gene expression information for the laboratory mouse. By combining the different types of expression data, GXD aims to provide increasingly complete information about the expression profiles of genes in different mouse strains and mutants, thus enabling valuable insights into the molecular networks that underlie normal development and disease. GXD is integrated with the Mouse Genome Database (MGD). Extensive interconnections with sequence databases and with databases from other species, and the development and use of shared controlled vocabularies extend GXD’s utility for the analysis of gene expression information. GXD is accessible through the Mouse Genome Informatics web site at http://www.informatic s.jax.org/ or directly at http://www.informatics.jax.org/me nus/expression_menu.shtml.
INTRODUCTION
The laboratory mouse has become a pivotal animal model in biomedical research because it is closely related to the human and readily amenable to genetic and molecular analysis. Tissues from many different mouse strains and mutants, and from all developmental and adult stages, are easily accessible for expression analysis. The different methods used to detect gene products vary in sensitivity and spatial resolution and contribute distinct and complementary expression information. The Gene Expression Database (GXD) has been designed as an open-ended system that can integrate many different types of expression data, such as RNA in situ hybridization, immunohistochemistry, northern blot, western blot, RT–PCR, cDNA source and array data (1–5). Thus, as data accumulate, GXD can provide increasingly complete information about what transcripts and proteins are produced by what genes; where, when and in what amounts these gene products are expressed; and how their expression varies in different mouse strains and mutants. Expression patterns reported from assays with differing spatial resolution are described in standardized and integrated form using an extensive, hierarchically structured dictionary of anatomical terms for mouse development built in collaboration with our Edinburgh colleagues (6). Digitized images of original expression data are linked to the respective expression records. GXD is integrated with the Mouse Genome Database (MGD) (7) to enable a combined analysis of genotype, expression and phenotype information, and has comprehensive links to other resources, such as sequence databases (8–12), OMIM, MEDLINE and databases from other species. Such information places gene expression data in the larger biological and analytical context.
GXD is implemented in the Sybase relational database management system. It has been available online since July 1998 and has been updated on a daily basis. Access to data is provided primarily via Web-based query forms. Users interested in direct SQL access may arrange for an SQL account by contacting MGI User support (see below). GXD and its WWW query interface have been described in more detail previously (3,4). Here, we illustrate recent enhancements of GXD from the user’s perspective, by taking the different query forms provided by GXD as an entry point.
The Gene Expression Data index
Using the GXD index, one can rapidly identify publications that contain endogenous developmental gene expression information for specific genes, for particular days of mouse development, from specific assay types or for any combination of these parameters. Additional query fields are provided for bibliographic information (authors, journals, year) and for words (text strings) that occur in the abstract of the respective articles. We continue to keep the GXD index up-to-date. All pertinent journal articles from 1993 to the present and articles from major developmental journals from 1990 to the present are indexed. As of September 28, 2000, the index includes 17 401 entries covering 5635 references and expression information for 3837 genes.
The Gene Expression Data query form
The Gene Expression Data query form provides access to data from RNA in situ hybridization, immunohistochemistry, northern blot, western blot, RT–PCR and RNase protection experiments. Using combinations of search parameters, one can ask increasingly complex expression queries, such as: ‘In what anatomical structures and/or at what developmental stages has a specified gene or a specified set of genes been detected/not detected?’ or ‘What genes have been detected/not detected in a given tissue and/or at a particular time of development using a specified set of expression assays?’ Due to the hierarchical structure of the anatomical dictionary, spatial queries can include anatomical substructures or superstructures. Further, it is possible to correlate gene expression with chromosomal location, a query particularly relevant for hunting candidate genes. The query capabilities for chromosomal location have been refined. It is now possible to search for genes located on a particular chromosome, between specified loci or within a specified distance from a genetic locus. The most significant enhancement of the Gene Expression Data query form is that one can now also search for genes whose products perform a particular ‘molecular function’ (e.g. ‘transcription factor’ or ‘DNA helicase’); are involved in a specified ‘biological process’ (e.g. ‘apoptosis’ or ‘purine metabolism’); or belong to a defined ‘cellular component’ (e.g. ‘nucleus’ or ‘origin recognition complex’). As a member of the Gene Ontology Project (13), we participate in building shared controlled vocabularies for these three categories and in assigning pertinent terms to genes in our database. The addition of these search parameters enables important new queries such as ‘What transcription factors are expressed in the diencephalon from day 11.5–15 of mouse development?’ or ‘What genes involved in apoptosis have been detected in the limb?’ and enhances the utility of the expression data stored in GXD.
The Mouse Anatomical Dictionary Browser
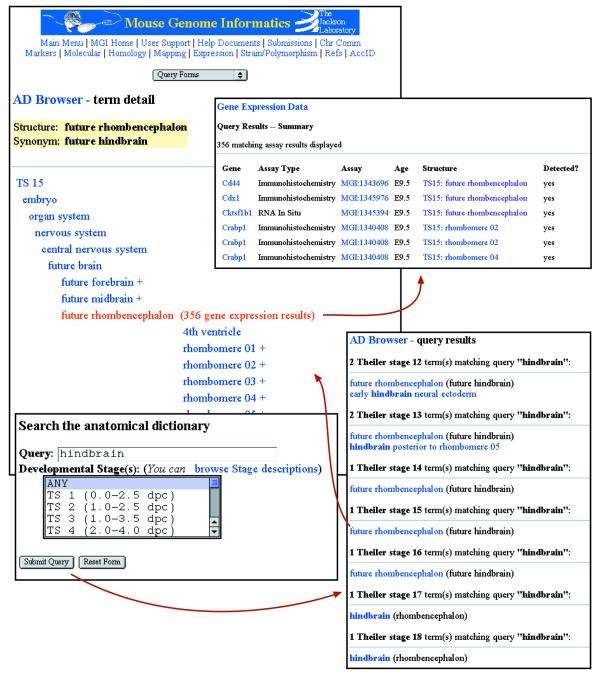
The Mouse Anatomical Dictionary Browser has been added as a new tool to navigate through the extensive dictionary hierarchies for the different developmental stages, to locate specific anatomical structures in the hierarchies and to look up expression results associated with those structures (Fig. 1). Thus, while the Gene Expression Data query form described above enables powerful combinatorial queries (including anatomical structures), the anatomy browser lets users view gene expression data directly from the developmental anatomy perspective. It must be noted that Theiler stages 23–26 have been added only recently and are still under development. So far, only limited expression data have been entered for these stages of mouse development.
Figure 1.
The Mouse Anatomical Dictionary Browser. The hierarchy of anatomical structures may be entered by either browsing from the top level or by searching for specific terms. Searching for a term in the Anatomical Dictionary requires a text string and optional specification of Theiler stages to search (lower left panel). Results of the query (lower right panel) are sorted by stage and each structure is linked to a detail page. The detail page for an anatomical structure displays the structure’s name, synonyms and a view of the structure within the stage hierarchy (upper left panel). All superstructures, structures found at the same level as the selected structure and substructures are displayed. Each structure is linked to its own detail page. A plus sign next to a structure indicates that substructures exist. The number of expression results currently annotated to the selected structure is displayed next to the structure name. Clicking on this link retrieves expression assay result summaries (upper right panel) that lead to detailed expression data.
The cDNA clone query form
This query form is designed for investigating expression information derived from cDNA source data. It allows one to ask questions such as ‘From which tissues or cell lines have cDNAs for a given gene been isolated?’ or ‘What genes located in a particular chromosomal region are, according to cDNA source information, expressed in a specified tissue?’ The cDNA clone query form now offers the same query capabilities for Gene Ontology classifications and chromosomal location as the Gene Expression Data query form. To make these new query parameters meaningful for the analysis of cDNA source data, we improved our means to assign cDNA clones and ESTs putatively to genes in our database. On a daily basis, GXD and MGD are establishing and curating links between genes in MGI and sequence entries in DDBJ/EMBL/Genbank (8–10). In an ongoing collaborative effort with SWISS-PROT (12), we further curate links between genes and protein entries from which additional cross-references to nucleotide sequences can be derived. Based on our curated gene/nucleotide sequence links we, in collaboration with the NCBI UniGene project (14), generate putative links between cDNA clones/ESTs and genes dynamically. By transitivity, these links provide (putative) information about chromosomal location, molecular function, biological process and cellular components for the respective cDNAs.
Electronic data submission: the Gene Expression Notebook
GXD curators continue to annotate expression data from the literature. However, extracting data from the literature and bringing them into a standardized format is a time-intensive process. Further, standard publications normally include only a small portion of the data generated by the authors. Therefore, GXD has been conceptualized from the beginning as an electronic framework for the community to store and analyze expression data. To facilitate electronic submission, we have developed the Gene Expression Notebook (GEN). It can be used as a laboratory notebook for storing expression results, images, molecular probes, specimens, experimental conditions, etc. and data can easily be exported in standardized format for electronic submission to GXD. Implemented in Microsoft Excel™, it is easy to use and to customize on both the Macintosh and the PC platform. The GEN has been described in more detail previously (4). During the last year, we have refined it based on feedback from a number of test laboratories and developed it into a tool that can now be used by the broader community. Users interested in obtaining the GEN may contact gen@informatics.jax.org. We welcome feedback and data submissions. Data submissions will receive accession numbers that can be cited in publications, and they will be subject to several levels of review as described previously (3). The GEN is primarily designed for conventional laboratories that study expression on a gene-by-gene basis. We are also working with groups that generate mouse expression data in a high-throughput fashion. Those laboratories normally maintain their own laboratory databases from which we can download data in bulk.
FUTURE DIRECTIONS
GXD will continue to acquire expression data from the literature and work with laboratories and publishers to obtain more data via electronic submission. The database will be expanded to include array expression data for the laboratory mouse. Obviously, GXD can add significant value to these data by integrating them with other types of expression data, and, via its interconnection with MGD, with genetic and phenotype data for mouse strains and mutants. We will continue to collaborate with others in establishing shared controlled vocabularies and links with external resources in order to provide an adequate and ever-improving framework for the analysis of gene expression information. Eventually, GXD will be coupled with the 3-D atlas/graphical gene expression database for mouse development, being developed by our Edinburgh colleagues, that enables a 3-D graphical storage and analysis of anatomy and in situ expression data (1,15,16).
USER SUPPORT
GXD provides user support through online documentation and dedicated User Support Staff. User Support can be contacted by telephone (+1 207 288 6445), fax (+1 207 288 6132) or email (mgi-help@informatics.jax.org).
CITING GXD
To reference the database itself, please cite this article. For referring to specific GXD data, we suggest the following format: These data were retrieved from the Gene Expression Database (GXD), Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine, USA, World Wide Web (http://www.informatics.jax.org). [Type in date (month, year) when you retrieved the data cited.]
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Jon Beal for his work on the Gene Expression Notebook, Lori Corbani and Prita Mani for help in software development and database maintenance, David Shaw, Paul Szauter and David Garippa for help in user support, John Boddy for artwork and Janice Ormsby for secretarial assistance. It is a pleasure to thank our colleagues Drs Jonathan Bard and Matthew Kaufman at the University of Edinburgh and Drs Duncan Davidson and Richard Baldock at the MRC Human Genetics Unit in Edinburgh for making the Dictionary of Mouse Developmental Anatomy available to us. We also would like to thank the journal Development for making electronic image files available to us on a regular basis. The Gene Expression Database is supported by NIH grant HD33745.
References
- 1.Ringwald M., Baldock,R., Bard,J., Kaufman,M., Eppig,J.T., Richardson,J.E., Nadeau,J.H. and Davidson,D. (1994) A Database for Mouse Development. Science, 265, 2033–2034. [DOI] [PubMed] [Google Scholar]
- 2.Ringwald M., Davis,G.L., Smith,A.G., Trepanier,L.E., Begley,D.A., Richardson,J.E. and Eppig,J.T. (1997) The Mouse Gene Expression Database GXD. Semin. Cell Dev. Biol., 8, 489–497. [DOI] [PubMed] [Google Scholar]
- 3.Ringwald M., Mangan,M.E., Eppig,J.T., Kadin,J.A., Richardson,J.E. and the Gene Expression Database Group. (1999) GXD: a Gene Expression Database for the laboratory mouse. Nucleic Acids Res., 27, 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ringwald M., Eppig,J.T., Kadin,J.A., Richardson,J.E. and the Gene Expression Database Group. (2000) GXD: a Gene Expression Database for the laboratory mouse: current status and recent enhancements. Nucleic Acids Res., 28, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ringwald M., Eppig,J.T. and Richardson,J.E. (2000) GXD: integrated access to gene expression data for the laboratory mouse. Trends Genet., 16, 188–190. [DOI] [PubMed] [Google Scholar]
- 6.Bard J.B.L., Kaufman,M.H., Dubreuil,C., Brune,R.M., Burger,A., Baldock,R.A. and Davidson,D.R. (1998) An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech. Dev., 74, 111–120. [DOI] [PubMed] [Google Scholar]
- 7.Blake J.A., Eppig,J.T., Richardson,J.E., Bult,C.J., Kadin,J.A. and the Mouse Genome Database Group. (2001) The Mouse Genome Database (MGD): Integration Nexus for the Laboratory Mouse. Nucleic Acids Res., 29, 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker W., van den Broek,A., Camon,A., Hingamp,P., Sterk,P., Stoesser,G. and Tuli,M.A. (2000) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 28, 19–23. Updated article in this issue: Nucleic Acids Res. (2001), 29, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tateno Y. Miyazaki,S., Ota,M., Sugawara,H. and Gojobori,T. (2000) DNA Data Bank of Japan (DDBJ) in collaboration with mass sequencing teams. Nucleic Acids Res., 28, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harger C., Chen,G., Farmer.A., Huang,W., Inman,J., Kiphart,D., Schilkey,F., Skupski,M.P. and Weller,J. (2000) The Genome Sequence DataBase. Nucleic Acids Res., 28, 31–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T., Harris,M.A., Hill,D.P., Issel-Tarver,L., Kasarskis,A., Lewis,S., Matese,J.C., Richardson,J.E., Ringwald,M., Rubin,G.M. and Sherlock,G. (2000) Gene Ontology: Tool for the unification of biology. Nature Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuler, G.D. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 15.Davidson D., Bard,J., Brunet,R., Burger,A., Dubreuil,C., Hill,W., Kaufman,M., Quinn,J., Stark,M. and Baldock,R. (1997) The mouse atlas and graphical gene-expression database. Semin. Cell Dev. Biol., 8, 509–517. [DOI] [PubMed] [Google Scholar]
- 16.Brune R.M., Bard,J.B.L., Dubreuil,C., Guest,E., Hill,W., Kaufman,M., Stark,M., Davidson,D., and Baldock,R.A. (1999) A Three-Dimensional Model of the Mouse at Embryonic Day 9. Dev. Biol., 216, 457–468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.