Abstract
Genes differentially expressed by tumor cells represent promising drug targets for anti-cancer therapy. Such candidate genes need to be validated in appropriate animal models. This study examined the suitability of rodent models of bladder cancer in B6D2F1 mice and Fischer-344 rats to model clinical bladder cancer specimens in humans. Using a global gene expression approach cross-species analysis showed that 13-34% of total genes in the genome were differentially expressed between tumor and normal tissues in each of five datasets from humans, rats, and mice. About 20% of these differentially expressed genes overlapped among species, corresponding to 2.6 to 4.8% of total genes in the genome. Several genes were consistently dysregulated in bladder tumors in both humans and rodents. Notably, CNN1, MYL9, PDLIM3, ITIH5, MYH11, PCP4 and FM05 were found to commonly down-regulated; while T0P2A, CCNB2, KIF20A and RRM2 were up-regulated. These genes are likely to have conserved functions contributing to bladder carcinogenesis. Gene set enrichment analysis detected a number of molecular pathways commonly activated in both humans and rodent bladder cancer. These pathways affect the cell cycle, HIF-1 and MYC expression, and regulation of apoptosis. We also compared expression changes at mRNA and protein levels in the rat model and identified several genes/proteins exhibiting concordant changes in bladder tumors, including ANXA1, ANXA2, CA2, KRT14, LDHA, LGALS4, SERPINA1, KRT18 and LDHB. In general, rodent models of bladder cancer represent the clinical disease to an extent that will allow successful mining of target genes and permit studies on the molecular mechanisms of bladder carcinogenesis.
Keywords: Human bladder cancer, rodent models, gene expression, proteomics, and cross-species comparison
Introduction
Bladder cancer is one of the most common cancers in the United States, especially in men. In 2009, there will be an estimated 70,980 new cases of bladder cancer with 52,810 cases in males [1]. Approximately 15% of bladder tumors evolve into invasive tumors after infiltration through the basement membrane. Patients with muscle-invasive disease are at high risk for recurrence, progression, and metastasis. Although early stage bladder cancer can be treated surgically, the five-year survival is 45% and 6% for patients with regional and distant recurrences, respectively.
Mice or rats administered N-butyl-N-(4-hydroxybutyl)-nitrosamine (OH-BBN) develop transitional and squamous cell urinary bladder cancers that have significant histopathological similarities to human bladder cancer and are frequently invasive. These rodent models have been used previously to characterize the tumori-genic process for urinary bladder cancer and to assess the efficacy of potential chemopreven-tive agents to inhibit the development of bladder cancers [2-6]. However, to what degree carcinogen-induced rodent models recapitulate molecular features and biological pathways of human bladder cancer has not been characterized.
Many biological systems operate in an evolu-tionarily conserved manner across a large number of species. Cross-species analysis of sequence and gene interaction is often applied to determine the function of new genes. In contrast to these static measurements, microarrays measure the dynamic, condition-specific response of complex biological systems. The recent exponential growth in microarray expression datasets allows researchers to combine expression experiments from multiple species to identify genes that are not only conserved in sequence but also regulated in a similar way across species. In this study we performed cross-species analysis of microarray data for human bladder cancer and carcinogen induced rodent bladder cancer. The major objectives of this study were to identify the degree of cross-species overlap on the single-gene level and to determine the similarity of the biological pathways in the cross-species comparison that may be relevant to the mechanism of bladder car-cinogenesis. Our analysis demonstrates that cross-species comparisons can be used to obtain important information on gene expression and pathway activation that cannot be obtained when analyzing data from a single species.
Materials and Methods
Rodent models of bladder cancer
Female Fischer-344 rats and male B6D2F1 (C57BI/6 × DB/V2 Fl) mice were obtained from Harlan Sprague-Dawley, Inc. (Indianapolis, IN; virus-free colony 202) at 28 days of age and were housed in polycarbonate cages (five per cage). The animals were kept in a lighted room 12 hours each day and maintained at 22 ± 0.5° C. Teklad 4% mash diet (Harlan Teklad, Madison, Wl) and tap water were provided ad libitum. In the mouse study, at 56 days of age, animals received the first of 12 weekly gavage treatments with OH-BBN (TCI America, Portland, OR). Each 7.5-mgdose was dissolved in 0.1 ml ethanol: water (25:75). For the rat study, OH-BBN (150 mg/gavage, 2x/week) was started when the rats were 49 days of age and continued for 8 weeks. The carcinogen vehicle was ethanol:water (20:80) in 0.5 ml. All animals were sacrificed 8 months following the initial OH -BBN treatment.
Bladder tumors were removed and frozen for subsequent molecular assays. All frozen tumor tissues were microdissected to determine the tumor vs normal cell ratio for each specimen. Only microscopic sections from tumor tissues containing more than 80% tumor cells were isolated and stored at -80°C for subsequent RNA isolation. A portion of each tumor was fixed and processed for routine paraffin embedding, cut into 5-μm sections, and mounted for hema-toxylin and eosin (H&E) staining. All bladder tumors used in this study were diagnosed as bladder cancers with a mixed histology showing elements of both transitional and squamous cells. Matching normal epithelia came from the same sex and age-matched controls were also micro-dissected to ensure that specimens consisted of purely normal lung tissue. To isolate bladder epithelia, we separated epithelial cells from the stroma and muscle tissues by cutting the bladder into half and scraping off the epithelium.
Total RNA from normal bladder epithelia and bladder tumors were isolated by Trizol (Invitrogen, Carlsbad, CA) and purified using the RNeasy Mini Kit and RNase-free DNase Set (QIAGEN, Valencia, CA) according to the manufacturer's protocols. In vitro transcription-based RNA amplification was then performed on each sample. cDNAfor each sample was synthesized using a Superscript cDNA Synthesis Kit (Invitrogen) and a T7-(dT)24 primer, 5'-GGCCAGTGAATTGTAATACGACTCACTA-TAGGGAGGCGG-(dT)24-3'. cDNA were cleaned using phase-lock gels (Fisher cat ID E0032005101) and phenol/chloroform extraction. Then, biotin-labeled cRNAwere transcribed in vitro from cDNA using a BioArray High Yield RNA Transcript Labeling Kit (ENZO Biochem, New York, NY) and purified again using the RNeasy Mini Kit. The labeled mouse cRNAwere applied to Affymetrix MGU74Av2 GeneChips and the labeled rat cRNA were applied to Affymetrix Rat230 2.0 GeneChips or Rat Exon 1.0 ST Array (Affymetrix) according to the manufacturer's recommendations. The raw fluorescence intensity data within CEL files from the platform Affymetrix MGU74Av2 and Rat230 2.0 were pre-processed with Robust Multichip Average (RMA) algorithm [7], as implemented with R packages Affy from Bioconductor (http://www.bioconductor.org). This algorithm analyzes the microarray data in three steps: a background adjustment, quantile normalization, and finally summation of the probe intensities for each probe set using a log scale linear additive model for the log transform of (background corrected, normalized) PM intensities. Gene-level signal estimates for the CEL files from the platform Rat Exon 1.0 ST Array were derived by quantile sketch normalization using Iterplier algorithm, as implemented with Expression Console vl.1.1 (http://www.affymetrix.com/products_services/software/specific/expression_console_software.affx).
For ID protein gel electrophoresis, rat samples were solubilized in the following lysis buffers: 25 mM Hepes buffer containing 150 mM NaCI, 10 mM MgCI2, 1% Igepal, 0.25% sodium deoxycho-late, 10% glycerol, 2.5 mM EDTA, and protease/ phosphatase inhibitors. For 2D protein difference electrophoresis, rat samples were solubilized in 100 μL of lysis buffer (30 mM Tris-CI, pH 8.5; 7 M urea, 2 M thiourea, 4% CHAPS) containing protease inhibitor cocktail (Roche, Indianapolis, IN) and phosphatase inhibitor cocktails I and II) (Sigma-Aldrich, St. Louis, MO). After centrifugation at 15,000 rpm for 30 min, the supernatant was recovered as cellular protein for the protein expression study. Protein samples from carcinogen-treated and control rats were labeled with DIGE fluorescent dye (GE Healthcare Amersham Biosciences, Piscataway, NJ), and analysis was performed as described previously [8]. A pooled sample consisting of a mixture of small portions of all protein samples obtained was used as internal control. Briefly, after extraction in lysis buffer, three cyanine dyes, Cy2, Cy3, and Cy5 were used to label the protein samples from normal rats, pooled sample, and carcinogen-treated rats, respectively. The same amount of protein were combined and analyzed on the same gel. For the first-dimension separation, the labeling mixture was applied to Immobiline DryStrips (24 cm long, pH 3 to 10; GE Healthcare). The second dimension was carried out with 10-20% SDS-PAGE gels. The Cy2, Cy3, and Cy5-labeled images were subsequently acquired at the recommended wavelengths using a Typhoon 9400 scanner (GE Healthcare). Comprehensive image analysis was performed using DeCyder-2D Differential Analysis Software 5.0 (GE Healthcare). For spot detection and quantification, the differential in-gel analysis (DIA) module of DeCyder was employed. The biological variance analysis module (BVA) was then used to match the quantified spots of all gels to a chosen master gel. The statistical significance of each expression level was calculated using the Student's t-test in the BVA module. Protein expression levels which showed a statistically significant (p<0.05) increase or decrease were defined as being significant. In addition to log standardized abundances, the matched spot raw volume data of each sample was used in this analysis. A ratio was created by comparing the raw volume of each protein spot to that of its intra-gel internal standard. Selected gel features were excised and digested in situ with trypsin as described previously [8]. The resulting peptide pools were analyzed by tandem mass spectrometry using both MALDI-TOF/TOF instrument (Proteomics 4700, Applied Biosystems, Framingham, MA) and LC-MS/MS (LTQ-FTMS, Thermolelectron, San Jose, CA) performed as described [9]. The peptide fragmentation spectra were processed using Data Explorer, v 4.5 and Analyst software (Applied BioSystems, Framingham, MA and Toronto, Ontario). The processed spectra were used to search protein and conceptually-translated database with MASCOT, v 1.9 (Matrix Sciences, London, UK). Precursor error tolerance was set to 100 ppm and MS/MS fragment error tolerance, 0.8 Da. All the proteins identified should have protein scores greater than 40 and individual ions scores greater than 20 with expected value < 0.05. All the MS/MS spectra were further validated manually. When multiple proteins were identified in a single spot, the proteins with the highest number of peptides were considered as those corresponding to the spot.
Human bladder cancer
The expression data for human clinical specimens from two published study were used. Both of these two studies performed gene expression profiles on the Human Genome U133A human GeneChips containing 22,283 probes representing known genes and expression sequence tags (Affymetrix). One dataset is the study of Dyrskjøt et al. [10] including biopsies of normal bladder mucosa from 9 patients without a bladder cancer history, histologically normal mucosa biopsies from 5 cystectomy specimens, biopsies from 28 superficial transitional cell bladder tumors (13 tumors with surrounding carcinoma in situ and 15 without surroundingcarcinoma in situ) and 13 invasive transitional cell carcinomas. The CEL files were downloaded from Gene Expression Omnibus (GEO accession number: GSE3167). The raw fluorescence intensity data within CEL files were preprocessed with RMA algorithm. Another dataset is the study of San-chez-Carbayo et al. [11], including 49 normal urothelium specimens which were obtained at distant sites from the bladder tumors resected by cystectomy or cystoprostatectomy and 109 bladder cancer (28 superficial and 81 invasive lesions). The expression data of the 157 bladder tissues understudy derived by the Affy-metrix Microarray Suite 5.0 (MAS 5.0)were included in Supplemental table 10 of the original paper (http://ico.ascopubs.org/cgi/content/full/JC0.2005.03.2375/DC1). Basic information of these five microarray datasets used for cross-species gene-expression analysis of bladder cancer is found in Table 1.
Table 1.
Datasets in cross-species gene-expression analysis of bladder cancer
| Datasets | No. of normal | No. of tumor | GEO | PubMed |
|---|---|---|---|---|
| Human_1 | 14 | 41 | GSE3167 | 15173019 |
| Human_2 | 49 | 109 | N/A | 16432078 |
| Rat_1 | 5 | 5 | N/A | 17401461 |
| Rat_2* | 7 | 11 | N/A | N/A |
| Mouse | 4 | 4 | N/A | 15548366 |
These are new microarray data from Rat Exon 1.0 ST arrays used for the present study.
Statistical analyses
The probe sets with the average gene expression level of both tumor and normal groups less than 64 derived by MAS 5.0 and Iterplier or less than 6 derived by RMA were excluded in the following statistical analyses. Two-sample student t test was used to identify differentially expressed genes (DEGs) between tumor and normal groups for each dataset. To adjust the multiple testing in the study of high-dimensional microarray data, both tail area-based false discovery rate values (Q values) and local false discovery rate (LFDR) were estimated [12], which was implemented in R package fdrtool (http://www.r-project.org/). The DEGs were defined as genes with Q value < 0.01, LFDR < 0.05 and fold change > 1.5 between two groups.
Gene set enrichment analysis (GSEA) was per-formed to analyze the pattern of differential gene expression in each dataset respectively. GSEA is a computational method that determines whether a set of genes shows statistically significant differences in expression between two biological states, which has proved successful in discovering molecular pathways involved in human diseases (http://www.broad.mit.edu/gsea). Using the Kolmogorov-Smirnov statistic, GSEA assesses the degree of “enrichment” of a set of genes (e.g. a pathway) in the entire range of the strength of associations with the pheno-type of interest. It was used to identify a priori defined sets of genes that were differentially expressed [13, 14]. We used curated gene sets (c2) which contain genes on certain molecular pathways, and GO gene sets (c5) which consist of genes annotated by the same GO terms in the Molecular Signature Database (MSigDB, http://www.broad.mit.edu/gsea/msigdb/msigdb_index.html). For datasets of rodent models, because of small sample size, GSEA with gene permutation option was performed. Selected gene sets identified from GSEA were then visualized with MetaCore™ (http://www.genego.com/).
Quantitative Real-time PCR
Using the samples from rat model of bladder cancer, the relative expressions of eight random selected genes associated with survival were determined by QRT-PCR. RNA was isolated using Trizol reagent per manufacturer's instructions (Invitrogen). One micrograms of total RNA per sample were converted to cDNA using the Im Prom-II RT kit (Promega Co. Madison, WI) for RT-PCR (Invitrogen). Primers for QRT-PCR analysis (Table 2) were designed using Primer Express software version 2.0 (Applied Biosystems, Foster City, CA). Amplification of each target DNA was performed with SYBR Green PCR Supermix (BIO-RAD, Hercules, CA) in BIO-RAD Single Color Real-Time PCR Detection system according to the protocols provided. One microliter of cDNA was added to a 25 mL total volume reaction mixture containing water, SYBR Green PCR Su-perMix, and primers. Each real-time assay was done in triplicate on a BioRad MylQ thermal cycler. Data were collected and analyzed with iQ5 optical system software, version 2.1 software. The internal control gene GAPDH and target genes were amplified with equal efficiencies. The method for assessing if two amplicons have the same efficiency is to look at how ΔCT (CT,target - CT,GAPDH, CT is cycle number at which the fluorescence signal exceeds background) varies with template dilution. Smaller ΔCT indicates higher gene expression. The fold change of gene expression in the tumor tissues relative to the normal tissues was calculated as 2ΔΔCT (ΔΔCT = ΔCT tumor - ΔCT normal). The differences in expression between two groups were determined by two-tailed Student's t-test. A P-value of less than 0.05 was considered to indicate statistical significance.
Table 2.
Oligonucleotide primers and probes used for quantitative real-time PCR Analysis
| Gene | Sense primer | Anti-sense primer |
|---|---|---|
| GAPDH | GACATGCCGCCTGGAGAA | CTCGGCCGCCTGCTT |
| ANXA2 | GACATTGCCTTCGCCTACCA | ACCAGACAAGGCCGACTTCA |
| TUBB5 | TCCGTTCGCTCAGGTCCTT | CTGCCCCAGACTGACCAAAA |
| NDN | GGACAGAGTCGCGCTGAAC | TCACATAGATGAGACTCAGGATCATGA |
| KIF2c | GGAAGGTATTTGATCTGCTCAACAA | CAACCTGCACCTGCTGCTT |
| KIF22 | CCCAGAAATTAAGCCTCTTACAGAA | CCCAGCAAACGTTCCATACTC |
| CCNA2 | ACAGTATGCGGGCCATCCT | AGCCAAATGCAGGGTCTCAT |
| INHBA | GGCAGGAGGGCCGAAAT | CCTGACTCGGCAAAGGTGAT |
| E2F8 | ACTTTCCCCAAACCACAGGAT | CGACGCCACTGGGATCA |
Results
Differentially expressed bladder cancer genes in each species
Five microarray datasets were used for cross-species gene-expression analysis of bladder cancer including two human datasets, two rat datasets and one mouse dataset (Table 1). One of two rat datasets is our new microarray data from Rat Exon 1.0 ST arrays. We applied a cutoff of Q value < 0.01, LFDR < 0.05 and fold change > 1.5 to detect genes differentially regulated between tumors and normal bladder tissues in all datasets. Table 3 listed total numbers and proportion of genes differentially regulated between tumor and normal bladder tissues in each dataset. We first randomly chose eight DEGs and evaluated the microarray gene expression results from the cross-species analysis. The relative expression of these candidate bladder cancer genes was determined by QRT-PCR analysis using the samples from the rat model. We confirmed the expression results for all of the eight selected genes (CCNA2, TUBB5, KIF22, INHBA, E2F8, ANXA2, KIF2C and NDN) (p< 0.05) (Figure 1). This warrant further investigation and comparison of DEGs among different species.
Table 3.
Total number and proportion of genes differentially regulated between tumor and normal bladder tissues*
| Up-regulated in tumor | Down-regulated in tumor | |||
|---|---|---|---|---|
| Number | Proportion (%) | Number | Proportion (%) | |
| Human_1 | 2729 | 14.7 | 2233 | 12.0 |
| Human_2 | 3104 | 24.5 | 1352 | 10.7 |
| Rat_1 | 816 | 6.5 | 789 | 6.3 |
| Rat_2 | 1069 | 6.8 | 1137 | 7.2 |
| Mouse | 514 | 6.5 | 621 | 7.8 |
The DEGs were defined as genes with Q value < 0.01, LFDR < 0.05 and fold change > 1.5 between two groups.
Figure 1.
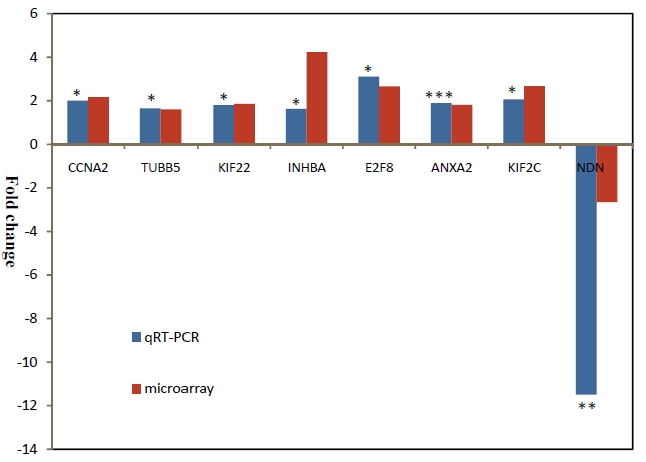
QRT-PCR validation of several candidate bladder cancer genes. Columns represent fold changes for the selected genes with differential expression between normal and tumor samples.. *: P<0.05; **: P<O.O1; ***: P<0.005
Approximately, 26 to 34% of genes were identified as DEGs in the two human datasets while only 13 to 14% were in the rodent models. About 14% of DEGs were identified in both human datasets, including 891 up-regulated genes and 328 down-regulated genes. 7.9% of DEGs were identified in both rat datasets, including 181 up-regulated genes and 232 down-regulated genes. About 20% of these DEGs overlapped among species, corresponding to 2.6 to 4.8% of total genes in the genome (Table 4). This suggests that although a large number of genes were identified to be differentially expressed between bladder tumor and normal tissues in each species, most of these genes are not common to the models. These genes may result from the cancer rather than being causally related.
Table 4.
Cross-species overlap of single differentially expressed genes as absolute number or proportion (in brackets) of overlapping orthologous genes between data sets*
| Human_l | Human_2 | Rat_l | Rat_2 | Mouse | |
|---|---|---|---|---|---|
| Human_l | 891 (10.4%) | 165 (2.9%) | 177 (2.9%) | 154 (3.1%) | |
| Human_2 | 328 (3.8%) | 112 (2.2%) | 103(2.1%) | 131 (3.0%) | |
| Rat_l | 72 (1.3%) | 21 (0.4%) | 181 (3.5%) | 96 (2.4%) | |
| Rat_2 | 115 (1.9%) | 56 (1.2%) | 232 (4.4%) | 86 (2.2%) | |
| Mouse | 112 (2.2%) | 44 (1.0%) | 69 (1.7%) | 82 (2.1%) |
Up triangle, up-regulated genes in tumors; down triangle , down-regulated gene in tumors.
The top 100 DEGs in each dataset were compared. 91 genes were consistently dysregulated in at least two datasets, including 54 down-regulated genes and 37 up-regulated genes (Table 5 and 6). Several genes were consistently dysregulated in bladder tumors in both humans and rodents. These genes are likely to have conserved functions contributing to bladder carcinogenesis. These include, CNN1 (calponin 1, basic, smooth muscle), MYL9 (myosin, light chain 9, regulatory), PDLIM3 (PDZ and LIM domain 3), ITIH5 (inter-alpha (globulin) inhibitor H5), MYH11 (myosin, heavy chain 11, smooth muscle), PCP4 (Purkinje cell protein 4) and FM05 (flavin containing monooxygenase 5) which were found to be commonly down-regulated; while T0P2A (topoisomerase (DNA) II alpha 170kDa), CCNB2 (cyclin B2), KIF20A (kinesin family member 20A) and RRM2 (ribonucleotide reductase M2 polypeptide) were found to be commonly up-regulated.
Table 5.
Down-regulated genes observed in at least two datasets*
| Genes | Description | Datasets |
|---|---|---|
| CASQ2 | calsequestrin 2 (cardiac muscle) | humani, human2 |
| DES | desmin | humani, human2 |
| DMD | dystrophin (muscular dystrophy, Duchenne and Becker types) | humani, human2 |
| DMN | desmuslin | humani, human2 |
| FHL1 | four and a half LIM domains 1 | humani, human2 |
| FOXF1 | forkhead box F1 | humani, human2 |
| KIAA0367 | KIAA0367 | humani, human2 |
| LMOD1 | leiomodin 1 (smooth muscle) | humani, human2 |
| PLN | phospholamban | humani, human2 |
| PTGIS | prostaglandin 12 (prostacyclin) synthase | humani, human2 |
| REEP1 | receptor accessory protein 1 | humani, human2 |
| SORBS1 | sorbin and SH3 domain containing 1 | humani, human2 |
| TNS1 | tensin 1 | humani, human2 |
| TPM2 | tropomyosin 2 (beta) | humani, human2 |
| CNN1 | calponin 1, basic, smooth muscle | humani, human2, mouse |
| MYL9 | myosin, light chain 9, regulatory | humani, human2, mouse |
| PDLIM3 | PDZ and LIM domain 3 | humani, human2, mouse |
| ITIH5 | inter-alpha (globulin) inhibitor H5 | humani, mouse |
| MYH11 | myosin, heavy chain 11, smooth muscle | human2, mouse |
| PCP4 | Purkinje cell protein 4 | human2, mouse |
| FMO5 | flavin containing monooxygenase 5 | rat1, mouse |
| ACE2 | angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 | rat1, rat2 |
| AHRR | aryl-hydrocarbon receptor repressor | rat1, rat2 |
| CAPN13 | calpain 13 | rat1, rat2 |
| CASKIN1 | CASK interacting protein 1 | rat1, rat2 |
| CYP11A1 | cytochrome P450, family 11, subfamily A, polypeptide 1 | rat1, rat2 |
| CYP1A1 | cytochrome P450, family 1, subfamily A, polypeptide 1 | rat1, rat2 |
| CYP3A18 | cytochrome P450, family 3, subfamily A, polypeptide 18 | rat1, rat2 |
| DPYD | dihydropyrimidine dehydrogenase | rat1, rat2 |
| EGFL6 | EGF-like-domain, multiple 6 | rat1, rat2 |
| FNDC5 | fibronectin type III domain containing 5 | rat1, rat2 |
| HSD11B2 | hydroxysteroid (11-beta) dehydrogenase 2 | rat1, rat2 |
| HTR4 | 5-hydroxytryptamine (serotonin) receptor 4 | rat1, rat2 |
| IGF2BP3 | insulin-like growth factor 2 mRNA binding protein 3 | rat1, rat2 |
| PAK3 | p21 (CDKNIA)-activated kinase 3 | rat1, rat2 |
| PCBP3 | poly(rC) binding protein 3 | rat1, rat2 |
| PLAG1 | pleiomorphic adenoma gene 1 | rat1, rat2 |
| PLCE1 | phospholipase C, epsilon 1 | rat1, rat2 |
| PRKCQ | protein kinase C, theta | rat1, rat2 |
| RPS6KA2 | ribosomal protein S6 kinase, 90kDa, polypeptide 2 | rat1, rat2 |
| SHANK2 | SH3 and multiple ankyrin repeat domains 2 | rat1, rat2 |
| SLC15A2 | solute carrier family 15 (H+/peptide transporter), member 2 | rat1, rat2 |
| SLC23A1 | solute carrier family 23 (nucleobase transporters), member 1 | rat1, rat2 |
| TRPV4 | transient receptor potential cation channel, subfamily V, member 4 | rat1, rat2 |
| TSHR | thyroid stimulating hormone receptor | rat1, rat2 |
| KRT20 | keratin 20 | rat1, rat2, mouse |
| HSD17B2 | hydroxysteroid (17-beta) dehydrogenase 2 | rat1, rat2, mouse |
| SH3GL2 | SH3-domain GRB2-like 2 | rat1, rat2, mouse |
| SLC16A7 | solute carrier family 16, member 7 (monocarboxylic acid transporter 2) | rat1, rat2, mouse |
| FLJ45455 (RGD1564618) | FLJ45455 protein (similar to novel protein) | rat2, human2 |
| HIP1R | huntingtin interacting protein 1 related | rat2, mouse |
| NDRG2 | NDRG family member 2 | rat2, mouse |
| UPK2 | uroplakin 2 | rat2, mouse |
| MFSD4 | major facilitator superfamily domain containing 4 | rat2, mouse |
The DEGs were defined as genes with Q value < 0.01, LFDR < 0.05 and fold change > 1.5 between two groups; the top 100 DEGs in each dataset were compared.
Table 6.
Up-regulated genes observed in at least two datasets*
| Genes | Description | Datasets |
|---|---|---|
| CDH1 | cadherin 1, type 1, E-cadherin (epithelial) | humani, human2 |
| EIF2AK1 | eukaryotic translation initiation factor 2-alpha kinase 1 | humani, human2 |
| KPNA2 | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | humani, human2 |
| KRT7 | keratin 7 | humani, human2 |
| LSR | lipolysis stimulated lipoprotein receptor | humani, human2 |
| MLF1IP | MLF1 interacting protein | humani, human2 |
| PRC1 | protein regulator of cytokinesis 1 | humani, human2 |
| SLC38A1 | solute carrier family 38, member 1 | humani, human2 |
| SPINT1 | serine peptidase inhibitor, Kunitztype 1 | humani, human2 |
| TH1L | TH1-like (Drosophila) | humani, human2 |
| TOP2A | topoisomerase (DNA) II alpha 170kDa | humani, human2, mouse |
| CCNB2 | cyclin B2 | rat1, human1 |
| KIF20A | kinesin family member 20A | rat1, human1 |
| RRM2 | ribonucleotide reductase M2 polypeptide | rat1, human1, mouse |
| CEACAM1 | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | rat1, mouse |
| CTGF | connective tissue growth factor | rat1, mouse |
| MMP7 | matrix metallopeptidase 7 (matrilysin, uterine) | rat1, mouse |
| S100A9 | S100 calcium binding protein A9 | rat1, mouse |
| SERPINB2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | rat1, mouse |
| DDIT4 | DNA-damage-inducible transcript 4 | rat1, rat2 |
| KCNN4 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | rat1, rat2 |
| MALL | mal, T-cell differentiation protein-like | rat1, rat2 |
| MMP3 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) | rat1, rat2 |
| PROM1 | prominin 1 | rat1, rat2 |
| RGD1563692 | rat1, rat2 | |
| TNFRSF12A | tumor necrosis factor receptor superfamily, member 12A | rat1, rat2 |
| TRPV2 | transient receptor potential cation channel, subfamily V, member 2 | rat1, rat2 |
| TSPAN1 | tetraspanin 1 | rat1, rat2 |
| VWA1 | von Willebrand factor A domain containing 1 | rat1, rat2 |
| VWF | von Willebrand factor | rat1, rat2 |
| WIF1 | WNT inhibitory factor 1 | rat1, rat2 |
| FGFBP1 | fibroblast growth factor binding protein 1 | rat1, rat2, mouse |
| GPX2 | glutathione peroxidase 2 (gastrointestinal) | rat1, rat2, mouse |
| MMP12 | matrix metallopeptidase 12 (macrophage elastase) | rat1, rat2, mouse |
| ANXA8 | annexin A8 | rat2, mouse |
| BHLHB2 | basic helix-loop-helix domain containing, class B, 2 | rat2, mouse |
| MMP13 | matrix metallopeptidase 13 (collagenase 3) | rat2, mouse |
The DEGs were defined as genes with Q value < 0.01, LFDR < 0.05 and fold change > 1.5 between two groups; the top 100 DEGs in each datasetwere compared.
Proteomic analysis
We then evaluated the concordance between gene expression and protein abundance in rodent models of bladder cancer. Ninety-six proteins that were significantly changed between normal and tumor in rat bladder samples were identified using a process of two-dimensional differential gel electrophoresis (2D-DIGE), in situ gel digestion, tandem mass spectrometry, and database searching (Table 7). Compared with the results from the transcriptomic analyses, 21 genes exhibited concordant changes at the mRNA and protein level in at least one rat mRNA expression dataset (Table 8). Among them, ANXA1 (annexin A1), ANXA2 (annexin A2), CA2 (carbonic anhydrase II), KRT14 (keratin complex 1, acidic, gene 14), LDHA (lactate de-hydrogenase A), LGALS4 (lectin, galactoside-binding, soluble, 4) and SERPINA1 (serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1) were increased in both rat gene expression datasets and concordant changes were observed at the protein level. KRT18 and LDHB were decreased in both rat datasets and showed concordant changes at the protein level (Table 9). The remainder did not show significant differences in gene expression, or showed discordant changes. These results suggest that many proteins which are changed in tumor might involve post-translational modification and the expression changes cannot be observed at the tran-scriptional level.
Table 7.
Protein changed signficantly between rat tumors and normal bladder tissues
| Gene Symbol | Protein Name | Gene Bank gi | Fold |
|---|---|---|---|
| AK2 | adenylate kinase 2 | 13591872 | -2.5 |
| AK3 | adenylate kinase 3 | 6978479 | 3.2 |
| AKR1A1 | aldo-keto reductase family 1, member A1 | 13591894 | 6.2 |
| ALB | Albumin | 55391508 | 4.6 |
| ALDH2 | mitochondrial aldehyde dehydrogenase precursor | 45737868 | -5.8 |
| ALDH3A1 | aldehyde dehyrogenase family 3, member A1 | 47482124 | 4.6 |
| ALDH3B1 | fatty aldehyde dehydrogenase-like | 55742838 | -3.8 |
| ANXA1 | Lipocortin I [Rattus sp.] | 235879 | 3.4 |
| ANXA2 | Calpactin I heavy chain | 312253 | 7.1 |
| APOA4 | Apolipoprotein A-IV | 60552712 | -3.5 |
| BANF1 | barrier to autointegration factor 1 | 16758438 | 4.9 |
| CA2 | Ca2 protein (carbonic anhydrase) | 41388872 | 3.4 |
| CA3 | carbonic anhydrase 3 | 31377484 | -5.5 |
| CACYBP | calcyclin binding protein | 51948388 | -2.5 |
| CCT2 | chaperonin containing TCP1, subunit 2 (beta) | 54400730 | 4 |
| CCT4 | chaperonin delta subunit | 33149357 | 8.5 |
| CKB | creatine kinase | 203474 | 4.9 |
| CNN1 | calponin | 313818 | 3 |
| DCTN2 | dynactin 2 | 51948450 | -4.3 |
| DES | desmin | 38197676 | -2.2 |
| DYNC1LI2 | lumican | 13591983 | 4.2 |
| ENTPD5 | ectonucleoside triphosphate dephosphorylase | 40786479 | -13.1 |
| FAM65B | Liver-regeneration-related protein LRRG069 (Ab2-162) | 33086566 | -9.5 |
| GC | vitamin D binding protein prepeptide | 203927 | 3.8 |
| GDA | guanine deaminase | 7533042 | 3.8 |
| GSTA4 | similar to GST 8 (8-8) | 27720723 | 4.2 |
| GSTA5 | glutathione-S-transferase, alpha type2 | 51036637 | 9.9 |
| GSTM5 | glutathione transferase (EC 2.5.1.18) | 204501 | 4 |
| GSTP1 | GST pi2 | 25453412 | 5.4 |
| HIBCH | 3-hydroxyisobutyryl-Coenzyme A hydrolase (predicted) | 61556993 | 6.2 |
| HSPB1 | heat shock protein 27 | 204665 | 3.4 |
| KA11 | type I keratin KA11 | 57012432 | 4.2 |
| KRT10 | keratin 10 | 57012436 | 1.3 |
| KRT13 | keratin 13 | 51591909 | 4.3 |
| KRT14 | keratin complex 1, acidic, gene 14 (predicted) | 56912233 | 4.4 |
| KRT15 | type I keratin KA15 | 51591903 | -1.8 |
| KRT16 | type I keratin KA16 | 56847618 | -2 |
| KRT18 | keratin complex 1, acidic, gene 18 | 60688216 | -18.9 |
| KRT19 | keratin complex 1, acidic, gene 19 | 42409519 | -12.6 |
| KRT20 | keratin 20 | 27465585 | 5.9 |
| KRT4 | type II keratin Kb4 | 57012360 | 7.6 |
| KRT5 | type II keratin 5 [Mus musculus] | 16303309 | 5.7 |
| KRT7 | similar to keratin complex 2 (predicted) | 34868200 | -5.1 |
| KRT73 | type II keratin Kb36 | 57012358 | -5 |
| KRT8 | cytokeratin-8 [Rattus sp.] | 30352203 | -10.6 |
| LDHA | lactate dehydrogenase A | 8393706 | 3.3 |
| LDHB | lactate dehydrogenase B | 6981146 | -3.5 |
| LGALS4 | lectin, galactoside-binding, soluble, 4 (galectin 4) | 6981152 | 4.3 |
| LZIC | leucine zipper and CTNNBIP1 domain containing | 61557405 | 9.8 |
| MDH2 | malate dehydrogenase, mitochondrial | 42476181 | 4 |
| MYL9 | myosin regulatory light chain, isoform C [Rattus sp.] | 998522 | 5 |
| NAGA | N-acetyl galactosaminidase, alpha (predicted) | 58865810 | 5.9 |
| NIT1 | Nit 1 protein | 56268926 | -2.4 |
| NME2 | RBL-NDP kinase 18kDa subunit (p18) | 206580 | 8 |
| NQO2 | NAD(P)H dehydrogenase, quinone 2 | 51948400 | 2.4 |
| PA2G4 | proliferation associated 2G4, 38kDa | 51948384 | 6 |
| PDIA6 | CaBP1 | 488838 | -7.7 |
| PDLIM1 | LIM protein | 8393153 | 3.9 |
| PECR | peroxisomal trans-2-enoyl-CoA reductase | 18959236 | 4.3 |
| PGRMC1 | progesterone receptor membrane component 1 | 11120720 | 3.7 |
| PKLR | L-type pyruvate kinase | 297533 | 4.2 |
| PKM2 | Unnamed protein product | 56929 | 4.2 |
| PPM1F | protein phosphatase 1F (PP2C domain containing) | 28461153 | 5.9 |
| PPP1R7 | Protein phosphatase 1, regulatory (inhibitor) subunit 7 | 57032943 | -34.9 |
| PRDX2 | peroxiredoxin 2 | 8394432 | 8 |
| PRELP | proline arginine-rich end leucine-rich repeat protein | 16758116 | 8.5 |
| PRKCDBP | protein kinase C, delta binding protein | 19745164 | -3.5 |
| PROSC | similar to Proline synthetase associated(predicted) | 62662820 | -6.1 |
| PSMC4 | proteasome 26S ATPase subunit 4 | 25742677 | -3.9 |
| PTGR1 | Leukotriene B4 12-hydroxydehydrogenase | 59809128 | 8.1 |
| PZP | alpha-1-macroglobulin | 202857 | -3.5 |
| RPSA | laminin receptor 1 | 8393693 | -4.3 |
| RUVBL1 | RuvB-like protein 1 | 22208848 | 4 |
| RUVBL2 | RuvB-like 2 | 70794778 | -15.7 |
| S100A4 | S100 A4 (calvasculin) | 6981326 | 5.3 |
| S100A6 | S100 calcium binding protein A6 (calcyclin) | 16758986 | 4.9 |
| SAE1 | Ubiquitin-like 1 (sentrin) activating enzyme E1A | 50925905 | -3.5 |
| SELENBP1 | selenium binding protein 2 | 18266692 | 4 |
| SERPINA1 | serine protease inhibitor alpha 1 | 51036655 | 2.5 |
| SERPINH1 | serpinh 1 protein | 55824765 | 4.6 |
| SNCG | synuclein, gamma [Mus musculus] | 6755592 | -3.2 |
| STRAP | serine/threonine kinase receptor associated protein | 4063383 | -18.5 |
| TALDO1 | transaldolase | 12002054 | 4.1 |
| TCAG7.1260 | aldoketoreductase family 1 | 27465603 | 8.8 |
| TGM2 | transglutaminase 2, C polypeptide | 42476287 | 5.1 |
| TKT | transketolase | 1729977 | 6 |
| TPM1 | tropomyosin alpha isoform | 14134104 | 4 |
| TUBA1C | tubulin, alpha 6 (predicted) | 58865558 | -2.8 |
| TXN | thioredoxin | 16758644 | 10.4 |
| UBE2V2 | Ubiquitin-conjugating enzyme E2 variant 2 | 24817674 | 4.3 |
| UCHL1 | ubiquitin carboxy-terminal hydrolase L1 | 61098212 | 3.4 |
| VDAC1 | voltage dependent anion channel | 4105605 | 6.5 |
| VIM | vimentin | 57480 | 4.1 |
| YARS | Yars predicted protein | 68534287 | 6 |
| YWHAE | Tyrosine 3-monooxygenase | 30583161 | -3.8 |
Table 8.
Genes exhibited concordant changes at both gene expression and protein levels
| mRNA expression arrays* | Protein expression arrays† | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Symbol | Gene Name | probe_set | Fold | Q-value | LFDR | Dataset | Protein Name | Gene Bank gi | Fold |
| Anxal | annexin A1 | 1367614_at | 12.3 | 3.2E-05 | 3.6E-05 | Rat1 | Lipocortin 1 [Rattus sp.] | 235879 | 3.4 |
| Anxa2 | annexin A2 | 1367584_at | 2.7 | 5.2E-04 | 7.9E-04 | Ratl | Calpactin 1 heavy chain | 312253 | 7.1 |
| Car2 | carbonic anhydrase II | 1367733_at | 3.4 | 1.4E-03 | 2.7E-03 | Ratl | Ca2 protein (carbonic anhydrase) | 41388872 | 3.4 |
| Car2 | carbonic anhydrase II | 1386922_at | 3.1 | 2.5E-03 | 6.4E-03 | Ratl | Ca2 protein (carbonic anhydrase) | 41388872 | 3.4 |
| Gda | guanine deaminase | 1387659_at | 3.1 | 2.0E-03 | 4.3E-03 | Rat1 | guanine deaminase | 7533042 | 3.8 |
| Krt14 | keratin 14 | 1371895_at | 22.5 | 3.0E-05 | 3.6E-05 | Rat1 | keratin complex 1, acidic, gene 14 (predicted) | 56912233 | 4.4 |
| Krt18 | keratin 18 | 1388155_at | -4.0 | 3.4E-03 | 7.9E-03 | Rat1 | keratin complex 1, acidic, gene 18 | 60688216 | -18.9 |
| Krt5 | keratin 5 | 1370863_at | 2.6 | 2.8E-03 | 6.6E-03 | Ratl | type II keratin 5 [Mus musculus] | 16303309 | 5.7 |
| Ldha | lactate dehydrogenase A | 1367586_at | 2.8 | 7.9E-04 | 1.4E-03 | Ratl | lactate dehydrogenase A | 8393706 | 3.3 |
| Ldhb | lactate dehydrogenase B | 1370218_at | -2.1 | 2.6E-03 | 6.6E-03 | Ratl | lactate dehydrogenase B | 6981146 | -3.5 |
| Lgals4 | lectin, galactoside-binding, soluble, 4 | 1368269_at | 12.7 | 3.5E-03 | 7.9E-03 | Ratl | lectin, galactoside-binding, soluble, 4 (galectin 4) | 6981152 | 4.3 |
| Nitl | nitrilase 1 | 1398049_at | -1.5 | 3.5E-03 | 7.9E-03 | Ratl | Nit 1 protein | 56268926 | -2.4 |
| S100a4 | S100 calcium-binding protein A4 | 1367846_at | 2.6 | 1.6E-04 | 2.0E-04 | Ratl | S100 A4 (calvasculin) | 6981326 | 5.3 |
| Serpinhl | serine (or cysteine) peptidase inhibitor, clade H, member 1 | 1371310_s_at | 4.1 | 3.1E-03 | 6.6E-03 | Ratl | serpinh 1 protein | 55824765 | 4.6 |
| Vim | vimentin | 1367574_at | 2.6 | 6.9E-03 | 1.8E-02 | Ratl | vimentin | 57480 | 4.1 |
| Anxa1 | annexin A1 | 7060990 | 5.7 | 4.3E-07 | 1.6E-06 | Rat2 | Lipocortin 1 [Rattus sp.] | 235879 | 3.4 |
| Anxa2 | annexin A2 | 7337587 | 1.8 | 2.6E-05 | 1.4E-04 | Rat2 | Calpactin 1 heavy chain | 312253 | 7.1 |
| Banf1 | barrier to autointegration factor 1 | 7059728 | 1.8 | 4.6E-03 | 1.9E-02 | Rat2 | barrier to autointegration factor 1 | 16758438 | 4.9 |
| Car2 | carbonic anhydrase II | 7206103 | 3.5 | 5.0E-04 | 2.3E-03 | Rat2 | Ca2 protein (carbonic anhydrase) | 41388872 | 3.4 |
| Ka11 | type 1 keratin KA11 | 7082629 | 11.1 | 1.9E-09 | 3.5E-09 | Rat2 | type 1 keratin KA11 | 57012432 | 4.2 |
| Krt14 | keratin 14 | 7082623 | 11.1 | 1.4E-09 | 3.4E-09 | Rat2 | keratin complex 1, acidic, gene 14 (predicted) | 56912233 | 4.4 |
| Krt18 | keratin 18 | 7191382 | -4.7 | 2.3E-06 | 9.5E-06 | Rat2 | keratin complex 1, acidic, gene 18 | 60688216 | -18.9 |
| Ldha | lactate dehydrogenase A | 7031732 | 2.6 | 7.0E-05 | 3.1E-04 | Rat2 | lactate dehydrogenase A | 8393706 | 3.3 |
| Ldhb | lactate dehydrogenase B | 7270937 | -2.4 | 2.1E-07 | 5.3E-07 | Rat2 | lactate dehydrogenase B | 6981146 | -3.5 |
| Lgals4 | lectin, galactoside-binding, soluble, 4 | 7030215 | 16.0 | 1.5E-03 | 7.4E-03 | Rat2 | lectin, galactoside-binding, soluble, 4 (galectin 4) | 6981152 | 4.3 |
| Nme2 | non-metastatic cells 2, protein (NM23B) expressed in | 7081660 | 1.8 | 2.0E-03 | 8.7E-03 | Rat2 | RBL-NDP kinase 18kDa subunit (p18) | 206580 | 8 |
| Prose | proline synthetase co-transcribed homolog (bacterial) | 7152637 | -1.7 | 3.5E-06 | 1.4E-05 | Rat2 | similar to Proline synthetase associated(predicted) | 62662820 | -6.1 |
| Serpina1 | serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | 7309988 | 5.8 | 9.7E-03 | 4.0E-02 | Rat2 | serine protease inhibitor alpha 1 | 51036655 | 2.5 |
| Txn1 | thioredoxin 1 | 7286798 | 1.5 | 1.8E-03 | 8.0E-03 | Rat2 | thioredoxin | 16758644 | 10.4 |
| Vdac1 | voltage-dependent anion channel 1 | 7067512 | 1.9 | 7.2E-03 | 2.9E-02 | Rat2 | voltage dependent anion channel | 4105605 | 6.5 |
The DEGs were defined as genes with Q value < 0.01, LFDR < 0.05 and fold change > 1.5 between two groups.
Protein expression levels which showed a statistically significant (p<0.05) increase or decrease in tumors as compared with their matched normal bladder.
Table 9.
Genes exhibited concordant changes at both mRNA expression and protein levels at both rat bladder cancer datasets
| Gene | mRNA (Rat_1) | mRNA (Rat_2) | Protein | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| probe_set | Normal | Tumor | Fold | probe_set | Normal | Tumor | Fold | Protein Name | Fold | |
| Anxa1 | 1367614_at | 9.0 | 12.6 | 12.3 | 7060990 | 1106.1 | 6285.0 | 5.7 | Lipocortin I [Rattus sp.] | 3.4 |
| Anxa2 | 1367584_at | 10.9 | 12.3 | 2.7 | 7337587 | 3077.4 | 5579.3 | 1.8 | Calpactin I heavy chain | 7.1 |
| Krtl4 | 1371895_at | 8.2 | 12.7 | 22.5 | 7082623 | 633.3 | 7004.7 | 11.1 | keratin complex 1, acidic, gene 14 | 4.4 |
| Ldha | 1367586_at | 9.8 | 11.3 | 2.8 | 7031732 | 641.3 | 1646.2 | 2.6 | lactate dehydrogenase A | 3.3 |
| Lgals4 | 1368269_at | 6.8 | 10.5 | 12.7 | 7030215 | 135.2 | 2162.5 | 16.0 | lectin, galactoside-binding, soluble, 4 | 4.3 |
| Serpinh1 | 1371310_s_at | 7.3 | 9.3 | 4.1 | 7309988 | 33.4 | 194.2 | 5.8 | serine protease inhibitor alpha 1 | 2.5 |
| Krtl8 | 1388155_at | 12.3 | 10.3 | -4.0 | 7191382 | 626.6 | 133.0 | -4.7 | keratin complex 1, acidic, gene 18 | -18.9 |
| Ldhb | 1370218_at | 11.4 | 10.4 | -2.1 | 7270937 | 2438.2 | 1010.8 | -2.4 | lactate dehydrogenase B | -3.5 |
Molecular pathways identified by gene enrichment analysis (GSEA)
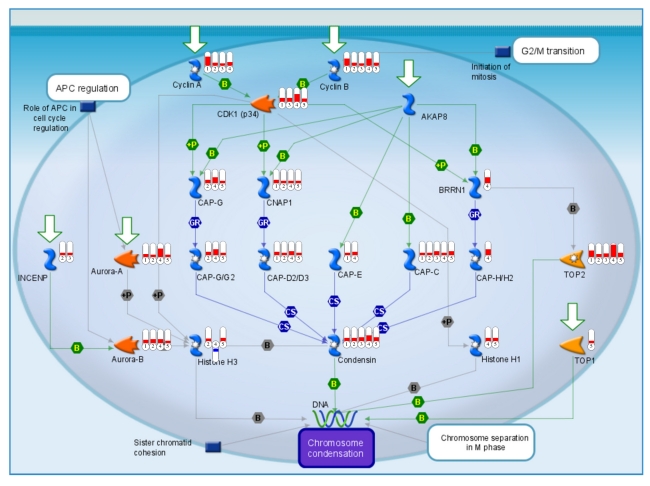
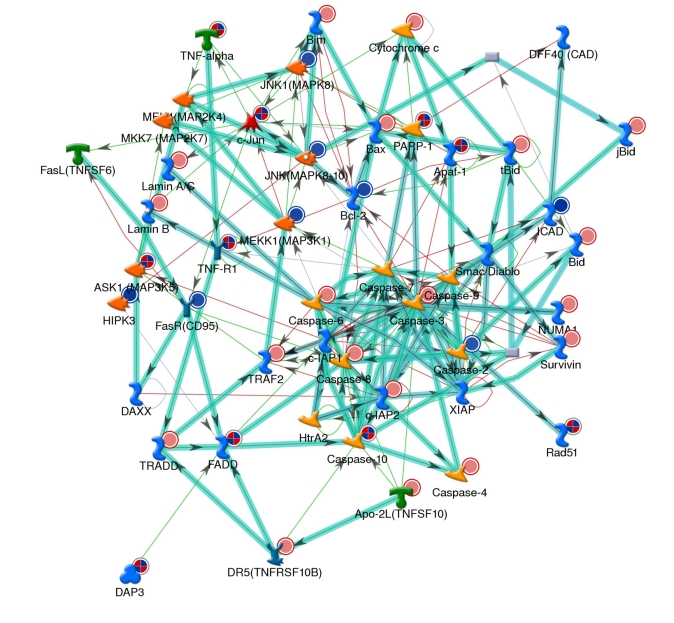
GSEA was performed using the curated gene sets to identify molecular pathways involved in bladder carcinogenesis. A false discovery rate (FDR) of < 0.25 was used for defining GSEA enriched gene sets [13, 14]. Using the curated gene sets c2, GSEA detected 74 enriched gene sets in bladder tumors in all of the five datasets from three different species (Table 10). These pathways include cell cycle, HIF-1 (hypoxia-inducible factor 1) and MYC pathways which were activated in bladder tumors. GSEA also detected additional 14 enriched gene sets among all of the five datasets using the curated gene sets c5 (Table 10). Several pathways are particularly interesting, including apoptosis, and the mitotic (M) phase of the cell cycle (Figure 2 and 3). Dysregulated genes are significantly over-represented in these pathways during bladder neoplastic transformation and progression in both humans and rodents.
Table 10.
Molecular pathways identified by gene enrichment analysis in all five datasets
| Gene sets | Rat_1 | Rat_2 | Mouse | Human_1 | Human_2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Size | FDR | Size | FDR | Size | FDR | Size | FDR | Size | FDR | |
| The curated gene sets c2 | ||||||||||
| ADIP_DIFF_CLUSTER4 | 32 | 0.012 | 24 | 0.000 | 18 | 0.006 | 32 | 8E-06 | 32 | 1E-04 |
| ADIP_DIFF_CLUSTER5 | 33 | 0.039 | 32 | 3E-05 | 16 | 0.007 | 35 | 0.000 | 35 | 8E-06 |
| BASSO_REGULATORY_HUBS | 105 | 0.235 | 103 | 0.020 | 77 | 0.043 | 138 | 0.036 | 138 | 0.000 |
| BENNETT_SLE_UP | 20 | 7E-05 | 19 | 0.010 | 17 | 0.001 | 28 | 0.021 | 28 | 0.009 |
| BRCA_ER_NEG | 583 | 0.002 | 655 | 2E-05 | 494 | 0.000 | 867 | 0.003 | 867 | 0.001 |
| BRCA_PROGNOSIS_NEG | 62 | 0.001 | 62 | 0.008 | 41 | 0.001 | 94 | 4E-05 | 94 | 0.000 |
| BRENTANI_CELL_CYCLE | 65 | 0.182 | 58 | 6E-05 | 49 | 0.005 | 80 | 3E-04 | 80 | 0.002 |
| CANCER_NEOPLASTIC_META_UP | 53 | 0.088 | 40 | 0.084 | 37 | 0.028 | 60 | 0.000 | 60 | 0.000 |
| CANCER_UNDIFFERENTIATED_META_UP | 59 | 0.003 | 41 | 3E-05 | 37 | 0.000 | 67 | 0.000 | 67 | 0.000 |
| CANTHARIDIN_DN | 35 | 0.042 | 32 | 0.018 | 30 | 0.071 | 49 | 4E-04 | 49 | 0.000 |
| CELL_CYCLE | 53 | 0.225 | 51 | 7E-05 | 45 | 0.002 | 74 | 0.000 | 74 | 0.000 |
| CELL_CYCLE_KEGG | 58 | 0.222 | 58 | 4E-04 | 48 | 0.002 | 80 | 0.000 | 80 | 8E-06 |
| CHANG_SERUM_RESPONSE_UP | 108 | 0.241 | 107 | 0.013 | 92 | 0.013 | 143 | 0.002 | 143 | 0.000 |
| CMV_ALL_UP | 73 | 0.239 | 68 | 0.057 | 59 | 0.154 | 90 | 0.167 | 90 | 0.006 |
| CMV_IE86_UP | 34 | 0.072 | 38 | 0.003 | 26 | 0.088 | 49 | 0.000 | 49 | 0.000 |
| CROMER_HYPOPHARYNGEAL_MET_VS_NON_DN | 53 | 0.222 | 52 | 4E-04 | 42 | 0.003 | 80 | 0.020 | 80 | 0.000 |
| CROONQUIST_IL6_STARVE_UP | 27 | 0.011 | 24 | 4E-05 | 20 | 2E-04 | 33 | 0.000 | 33 | 8E-06 |
| DER_IFNA_UP | 40 | 0.010 | 44 | 0.074 | 37 | 0.019 | 65 | 0.042 | 65 | 0.001 |
| DER_IFNB_UP | 64 | 0.047 | 63 | 0.184 | 55 | 0.063 | 91 | 0.139 | 91 | 0.001 |
| DNA_REPLICATION_REACTOME | 38 | 0.012 | 29 | 7E-05 | 24 | 0.132 | 44 | 0.000 | 44 | 0.000 |
| DOX_RESIST_GASTRIC_UP | 25 | 0.043 | 28 | 1E-04 | 15 | 0.001 | 43 | 0.000 | 43 | 0.000 |
| GAY_YY1_DN | 212 | 0.004 | 193 | 1E-04 | 154 | 0.001 | 220 | 7E-06 | 220 | 0.051 |
| GREENBAUM_E2A_UP | 29 | 0.001 | 27 | 3E-05 | 22 | 2E-04 | 32 | 0.000 | 32 | 1E-05 |
| HCC_SURVIVAL_GOOD_VS_POOR_DN | 85 | 0.065 | 89 | 0.021 | 78 | 0.026 | 121 | 4E-04 | 121 | 0.000 |
| HESS_HOXAANMEIS1_DN | 61 | 0.222 | 46 | 0.006 | 41 | 0.051 | 55 | 0.093 | 55 | 0.004 |
| HESS_HOXAANMEIS1_UP | 61 | 0.234 | 46 | 0.005 | 41 | 0.061 | 55 | 0.087 | 55 | 0.003 |
| HG_PROGERIA_DN | 19 | 0.095 | 20 | 4E-04 | 16 | 0.005 | 25 | 3E-04 | 25 | 0.012 |
| HIF1_TARGETS | 31 | 0.030 | 29 | 0.001 | 31 | 0.003 | 35 | 0.021 | 35 | 0.005 |
| HOFFMANN_BIVSBII_BI_TABLE2 | 226 | 0.037 | 150 | 4E-05 | 117 | 0.001 | 184 | 7E-06 | 184 | 2E-05 |
| HOFMANN_MDS_CD34_LOW_AND_HIGH_RISK | 21 | 0.204 | 30 | 0.018 | 26 | 0.004 | 46 | 0.042 | 46 | 0.017 |
| HSA03050_PROTEASOME | 22 | 0.236 | 20 | 0.007 | 19 | 0.135 | 22 | 0.002 | 22 | 0.000 |
| HSA04110_CELL_CYCLE | 82 | 0.210 | 79 | 3E-04 | 70 | 0.001 | 102 | 0.000 | 102 | 0.000 |
| HSA04115_P53_SIGNALING_PATHWAY | 47 | 0.217 | 43 | 0.002 | 39 | 0.002 | 62 | 0.020 | 62 | 3E-04 |
| IDX_TSA_UP_CLUSTER3 | 78 | 3E-04 | 66 | 0.000 | 42 | 0.000 | 83 | 0.000 | 83 | 0.000 |
| IFN_ALPHA_UP | 26 | 0.100 | 28 | 0.214 | 24 | 0.027 | 40 | 0.239 | 40 | 0.020 |
| IFNA_HCMV_6HRS_UP | 33 | 0.001 | 32 | 0.191 | 25 | 0.002 | 53 | 0.011 | 53 | 0.030 |
| IFNALPHA_HCC_UP | 23 | 0.012 | 18 | 0.157 | 23 | 0.005 | 29 | 0.047 | 29 | 0.005 |
| IFNALPHA_NL_UP | 18 | 0.018 | 17 | 0.218 | 18 | 0.012 | 27 | 0.013 | 27 | 0.002 |
| INOS_ALL_UP | 45 | 0.177 | 40 | 0.036 | 35 | 0.159 | 52 | 0.005 | 52 | 0.000 |
| KENNY_WNT_UP | 34 | 0.046 | 34 | 0.200 | 29 | 0.025 | 46 | 0.129 | 46 | 0.005 |
| LE_MYELIN_UP | 105 | 0.000 | 63 | 0.000 | 50 | 0.002 | 82 | 3E-05 | 82 | 0.000 |
| LEE_MYC_E2F1_UP | 42 | 0.001 | 44 | 0.122 | 38 | 3E-04 | 55 | 0.015 | 55 | 0.051 |
| LEE_TCELLS2_UP | 587 | 0.082 | 669 | 0.005 | 489 | 0.011 | 939 | 0.037 | 939 | 0.004 |
| LEE_TCELLS3_UP | 53 | 0.010 | 63 | 3E-05 | 34 | 3E-04 | 93 | 0.000 | 93 | 1E-05 |
| LEI_MYB_REGULATED_GENES | 239 | 0.032 | 234 | 0.023 | 200 | 0.001 | 317 | 0.057 | 317 | 0.000 |
| LI_FETAL_VS_WT_KIDNEY_DN | 114 | 0.051 | 106 | 0.003 | 85 | 0.001 | 159 | 0.000 | 159 | 0.000 |
| MYC_ONCOGENIC_SIGNATURE | 105 | 0.217 | 123 | 0.096 | 96 | 0.062 | 173 | 0.021 | 173 | 0.005 |
| MYC_TARGETS | 38 | 0.222 | 34 | 0.001 | 37 | 0.010 | 39 | 0.104 | 39 | 0.003 |
| OLDAGE_DN | 38 | 0.040 | 33 | 2E-05 | 29 | 0.001 | 47 | 0.000 | 47 | 0.000 |
| P21_ANY_DN | 26 | 0.004 | 27 | 0.002 | 22 | 0.008 | 32 | 0.000 | 32 | 2E-04 |
| POD1_KO_UP | 256 | 0.166 | 282 | 0.070 | 210 | 2E-04 | 339 | 0.011 | 339 | 0.037 |
| PRMT5_KD_UP | 158 | 0.082 | 130 | 0.013 | 99 | 4E-04 | 166 | 0.007 | 166 | 0.000 |
| PROTEASOME | 17 | 0.095 | 15 | 0.021 | 16 | 0.115 | 17 | 1E-04 | 17 | 2E-05 |
| PROTEASOME_DEGRADATION | 29 | 0.192 | 20 | 0.019 | 27 | 0.213 | 31 | 0.015 | 31 | 9E-06 |
| RADAEVA_IFNA_UP | 33 | 0.004 | 32 | 0.147 | 37 | 0.001 | 50 | 0.003 | 50 | 0.001 |
| RADIATION_SENSITIVITY | 19 | 0.075 | 19 | 0.095 | 19 | 0.046 | 24 | 0.063 | 24 | 0.005 |
| SANA_IFNG_ENDOTHELIAL_UP | 34 | 0.020 | 43 | 0.004 | 31 | 0.000 | 60 | 0.161 | 60 | 0.196 |
| SCHUMACHER_MYC_UP | 43 | 0.081 | 35 | 0.020 | 31 | 0.023 | 50 | 0.002 | 50 | 0.000 |
| SERUM_FIBROBLAST_CELLCYCLE | 83 | 1E-04 | 90 | 0.000 | 53 | 0.000 | 110 | 0.000 | 110 | 0.000 |
| SERUM_FIBROBLAST_CORE_UP | 123 | 0.132 | 138 | 0.010 | 105 | 0.049 | 174 | 2E-04 | 174 | 0.000 |
| SHEPARD_BMYB_MORPHOLINO_DN | 128 | 0.023 | 129 | 0.010 | 99 | 0.009 | 152 | 4E-05 | 152 | 0.020 |
| SHEPARD_CRASH_AND_BURN_MUT_VS_WT_DN | 100 | 0.120 | 109 | 0.115 | 81 | 0.056 | 137 | 8E-05 | 137 | 0.011 |
| SHEPARD_GENES_COMMON_BW_CB_MO | 48 | 0.004 | 49 | 0.072 | 37 | 0.064 | 60 | 0.000 | 60 | 0.008 |
| SHIPP_FL_VS_DLBCL_DN | 29 | 0.103 | 27 | 0.190 | 28 | 0.056 | 34 | 3E-05 | 34 | 0.000 |
| STEMCELL_EMBRYONIC_UP | 1162 | 0.189 | 909 | 0.083 | 717 | 0.075 | 1165 | 0.050 | 1165 | 0.000 |
| TARTE_PLASMA_BLASTIC | 244 | 0.038 | 221 | 5E-05 | 198 | 0.001 | 305 | 0.000 | 305 | 0.000 |
| UVB_NHEK3_ALL | 316 | 0.084 | 280 | 0.001 | 279 | 0.004 | 390 | 0.237 | 390 | 0.000 |
| UVB_NHEK4_6HRS_UP | 21 | 0.157 | 19 | 0.075 | 19 | 0.043 | 27 | 0.067 | 27 | 0.001 |
| VANTVEER_BREAST_OUTCOME_GOOD_VS_POOR_DN | 51 | 0.001 | 45 | 0.006 | 37 | 0.002 | 63 | 4E-05 | 63 | 0.001 |
| WIELAND_HEPATITIS_B_INDUCED | 63 | 0.000 | 66 | 0.018 | 60 | 3E-05 | 106 | 0.001 | 106 | 0.050 |
| YU_CMYC_UP | 43 | 0.000 | 28 | 0.003 | 20 | 0.002 | 27 | 0.000 | 27 | 0.000 |
| ZELLER_MYC_UP | 23 | 0.047 | 19 | 0.014 | 23 | 0.057 | 23 | 0.030 | 23 | 0.001 |
| ZHAN_MMPC_SIMAL | 36 | 0.239 | 37 | 0.011 | 34 | 0.008 | 47 | 0.071 | 47 | 0.011 |
| ZUCCHI_EPITHELIAL_UP | 30 | 0.095 | 30 | 0.072 | 30 | 3E-04 | 41 | 0.034 | 41 | 5E-04 |
| The curated gene sets c5 | ||||||||||
| APOPTOSIS_GO | 307 | 0.136 | 322 | 0.003 | 269 | 0.062 | 380 | 0.131 | 380 | 0.020 |
| CELL_CYCLE_PHASE | 115 | 0.217 | 121 | 0.002 | 83 | 0.018 | 145 | 0.000 | 145 | 0.009 |
| CELL_CYCLE_PROCESS | 133 | 0.209 | 136 | 0.002 | 94 | 0.025 | 165 | 0.000 | 165 | 0.001 |
| INTERPHASE | 49 | 0.232 | 43 | 0.056 | 35 | 0.174 | 58 | 0.011 | 58 | 0.006 |
| M_PHASE | 74 | 0.181 | 83 | 0.002 | 53 | 0.006 | 95 | 0.000 | 95 | 0.081 |
| M_PHASE_OF_MITOTIC_CELL_CYCLE | 54 | 0.178 | 61 | 9E-05 | 42 | 0.001 | 70 | 0.000 | 70 | 0.007 |
| MITOSIS | 52 | 0.129 | 60 | 1E-04 | 42 | 0.001 | 67 | 0.000 | 67 | 0.009 |
| MITOTIC_CELL_CYCLE | 102 | 0.141 | 110 | 0.002 | 78 | 0.019 | 128 | 0.000 | 128 | 3E-04 |
| PROGRAMMED_CELL_DEATH | 307 | 0.141 | 323 | 0.003 | 270 | 0.059 | 381 | 0.131 | 381 | 0.017 |
| PROTEOLYSIS | 121 | 0.178 | 138 | 0.116 | 117 | 0.003 | 162 | 0.246 | 162 | 0.078 |
| REGULATION_OF_APOPTOSIS | 245 | 0.161 | 255 | 0.004 | 213 | 0.050 | 300 | 0.082 | 300 | 0.029 |
| REGULATION_OF_DNA_METABOLIC_PROCESS | 33 | 0.134 | 34 | 0.001 | 20 | 0.217 | 38 | 0.130 | 38 | 0.115 |
| REGULATION_OF_MITOSIS | 30 | 0.137 | 28 | 4E-04 | 20 | 0.016 | 35 | 0.000 | 35 | 0.049 |
| REGULATION_OF_PROGRAMMED_CELL_DEATH | 245 | 0.172 | 256 | 0.004 | 214 | 0.046 | 301 | 0.078 | 301 | 0.023 |
Figure 2.
Cell cycle genes consistently altered across species during bladder carcinogenesis. Red and blue indicates overexpressed and underexpressed genes in tumor samples, respectively. 1, dataset Rat_1; 2, Rat_2; 3, Mouse; 4, Human_1; and 5, Human_2.
Figure 3.
Network analyses of apoptosis, programmed cell death and cell death. Multiple genes in this network are dysregulated in bladder tumors. Red and blue indicates overexpressed and underexpressed genes in tumor samples, respectively.
Discussion
Rodent models represent a powerful tool in the research of cancer mechanisms, prevention and therapy. Determination of the extent as to which findings in animal models can be translated to human disease is an important step. In this study we examined whether global gene expression profiling can assist in determining the suitability of rodent models of bladder cancer for the detection of cancer-related genes and prediction of cancer prevention effects.
The present study found that rodent models of bladder cancer (in OH-BBN treated B6D2F1 mice and Fischer-344 rats) accurately represent the clinical situation to an extent that will allow successful mining of target genes. About 20% of DEGs in bladder tumors overlapped among species, corresponding to 2.6 to 4.8% of total genes in the genome. Several genes that were concordantly regulated across species are of particular interest. Among these genes is ribonucleotide reductase M2 polypeptide (RRM2) which was increased in tumors across three species. RRM2 is an enzyme that catalyzes the formation of deoxyribonucleotides from ribonu-cleotides [15]. Deoxyribonucleotides in turn are used in the synthesis of DNA. The reaction catalyzed by RRM2 is strictly conserved in all living organisms [16]. Furthermore RRM2 plays a critical role in regulating the total rate of DNA synthesis so that DNA to cell mass is maintained at a constant ratio during cell division and DNA repair [17]. RRM2 plays an important role in tumor angiogenesis and growth through regulation of the expression of TSP-1 and VEGF [18]. TOP2A was consistently increased across data-sets and encodes a DNA topoisomerase, an enzyme that controls and alters the topologic states of DNA during transcription. TOP2A plays an important role in checkpoint activation and the maintenance of genomic stability [19]. Increased TOP2A correlated with advanced his-tological grading, microvascular invasion, and an early age onset of the hepatocellular malignancy [20]. T0P2A has been reported to be over-expressed in pancreatic adenocarci-noma [21], renal medullary carcinomas [22], ovarian cancer [23], acute lymphocytic leukemia [24], colorectal cancer [25], gastric carcinoma [26] and laryngeal squamous cell carcinoma [27]. CCNB2 (Cyclin B2), found commonly upregulated, is a member of the cyclin family, specifically the B-type cyclins. Cyclin B2 also binds to transforming growth factor beta Rll and thus cyclin B2/cdc2 may play a key role in transforming growth factor beta-mediated cell cycle control [28]. These commonly regulated genes across mouse, rat and human may be functionally important regulators of bladder tumorigenesis.
To integrate the expression changes observed in multiple datasets we utilized pathway analysis algorithms to identify functional classifications that were altered in bladder tumors compared to normal bladder epithelium of mouse, rat and human. Biological processes and molecular functions that were enriched in tumors included apoptosis, cell cycle, and DNA replication. In particular, TO3P2A was consistently activated in the biological classification of regulation of programmed cell death. Several genes including CCNB2, KIF20A and T0P2A have previously been reported to be up-regulated in bladder carcinogenesis [29]. CCNB2 was also found to be up-regulated in the cell cycle. Additional pathways enriched in bladder tumors included those for interphase and proteolysis. Our data suggest that human bladder cancer and carcinogen-induced rodent models may show more common similarity at global cellular pathway levels than at the single-gene levels previously described.
The proportion of dysregulated orthologous genes overlapped in two species is low which may be partly explained by the histological difference in bladder cancer among mouse, rat and human. Histopathology of B6D2F1 mouse bladder cancers have previously shown that these urinary bladder carcinomas had either transitional cell differentiation alone or in combination with either squamous or glandular differentiation or both squamous and glandular differentiation [2]. These patterns were also observed in highly invasive variants of human transitional cell bladder carcinoma [30]. Immu-nohistochemical staining of intermediate filament types showed that OH-BBN-induced rat bladder tumors had marked quantitative and qualitative differences from the most common, purely transitional, human bladder carcinomas [31]. Smaller lesions were similar to human urothelial dysplasia both histologically and im-munohistochemically. Progression of the lesions demonstrated large exophytic papillomas with extensive endophytic epithelial growth into abundant stroma and these lesions showed increasing predominance of squamous over transitional elements. Immunohistochemical findings confirmed these results and also demonstrated that morphologically indistinct cells, even in early lesions, express heavier cytokeratins characteristic of keratinizing squamous epithelium [30, 31]. The proportion of invasive transitional cell carcinomas in human bladder cancer biopsies from Sanchez-Carbayo et al. (81/108) is much higher than that from Dyrskjøt et al. (13/41) [10] and this may explain why there was more gene expression overlap between the rodent models and human bladder cancer from Sanchez-Carbayo et al. than those from Dyrskjøt et al. (4.1-4.8% vs. 2.6-4.0%). These data also suggest that histology information should be taken into account when analyzing animal models of bladder cancer.
Acknowledgments
The authors thank Alan Davis, Petra Erdmann-Gilmore and Julia Gross for expert technical assistance. This work was supported, in part, by NCI Contract Number HHSN-261200433008C (N01-CN43308) and the National Centers of Research Resources of the National Institutes of Health (P41-RR00954).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CA Cancer J Clin. 2009 doi: 10.3322/caac.20006. caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Becci PJ, Thompson HJ, Strum JM, Brown CC, Sporn MB, Moon RC. N-Butyl-N-(4-Hydroxybutyl)nitrosamine-induced Urinary Bladder Cancer in C57BL/6 × DBA/2 Fl Mice as a Useful Model for Study of Chemoprevention of Cancer with Retinoids. Cancer Res. 1981;41:927–932. [PubMed] [Google Scholar]
- 3.Grubbs CJ, Lubet RA, Koki AT, Leahy KM, Masferrer JL, Steele VE, Kelloff GJ, Hill DL, Seibert K. Celecoxib Inhibits N-Butyl-N-(4-hydroxybutyl)-nitrosamine-induced Urinary Bladder Cancers in Male B6D2F1 Mice and Female Fischer-344 Rats. Cancer Res. 2000;60:5599–5602. [PubMed] [Google Scholar]
- 4.Grubbs CJ, Moon RC, Squire RA, Farrow GM, Stinson SF, Goodman DG, Brown CC, Sporn MB. 13-cis-Retinoic acid: inhibition of bladder carcinogenesis induced in rats by N-butyl-N-(4-hydroxybutyl)nitrosamine. Science. 1977;198:743–744. doi: 10.1126/science.910158. [DOI] [PubMed] [Google Scholar]
- 5.McCormick DL, Ronan SS, Becci PJ, Moon RC. Influence of total dose and dose schedule on induction of urinary bladder cancer in the mouse by N-butyl-N-(4-hydroxy-butyl)nitrosamine. Carcinogenesis. 1981;2:251–254. doi: 10.1093/carcin/2.3.251. [DOI] [PubMed] [Google Scholar]
- 6.Moon RC, Kelloff GJ, Detrisac CJ, Steele VE, Thomas CF, Sigman CC. Chemoprevention of OH -BBN-induced bladder cancer in mice by piroxicam. Carcinogenesis. 1993;14:1487–1489. doi: 10.1093/carcin/14.7.1487. [DOI] [PubMed] [Google Scholar]
- 7.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix Ge-neChip probe level data. Nucleic Acids Res. 2003;31:el5. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredemeyer AJ, Lewis RM, Malone JP, Davis AE, Gross J, Townsend RR, Ley TJ. A proteomic approach for the discovery of protease substrates. Proc Natl Acad Sci U S A. 2004;101:11785–11790. doi: 10.1073/pnas.0402353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King JB, Gross J, Lovly CM, Rohrs H, Piwnica-Worms H, Townsend RR. Accurate mass-driven analysis for the characterization of protein phosphorylation. Study of the human Chk2 protein kinase. Anal Chem. 2006;78:2171–2181. doi: 10.1021/ac051520l. [DOI] [PubMed] [Google Scholar]
- 10.Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, Orntoft TF. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 12.Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elledge SJ, Zhou Z, Allen JB. Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem Sci. 1992;17:119–123. doi: 10.1016/0968-0004(92)90249-9. [DOI] [PubMed] [Google Scholar]
- 16.Torrents E, Aloy P, Gibert I, Rodriguez-Trelies F. Ribonucleotide reductases: divergent evolution of an ancient enzyme. J Mol Evol. 2002;55:138–152. doi: 10.1007/s00239-002-2311-7. [DOI] [PubMed] [Google Scholar]
- 17.Herrick J, Sclavi B. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol. 2007;63:22–34. doi: 10.1111/j.1365-2958.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Hu S, Wu J, Chen L, Lu J, Wang X, Liu X, Zhou B, Yen Y. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: implication of RRM2 in angiogenesis. Mol Cancer. 2009;8:11. doi: 10.1186/1476-4598-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo K, Yuan J, Chen J, Lou Z. Topoisomerase llalpha controls the decatenation checkpoint. Nat Cell Biol. 2009;11:204–210. doi: 10.1038/ncb1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong N, Yeo W, Wong WL, Wong NL, Chan KY, Mo FK, Koh J, Chan SL, Chan AT, Lai PB, Ching AK, Tong JH, Ng HK, Johnson PJ, To KF. T0P2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124:644–652. doi: 10.1002/ijc.23968. [DOI] [PubMed] [Google Scholar]
- 21.Baiocchi GL, Villanacci V, Rossi E, Gheza F, Portolani N, Giulini SM. HER-2/neu and topoi-somerase-II-alpha expression and genie amplification in pancreatic adenocarcinoma. Dig Dis Sci. 2009;54:2049–2051. doi: 10.1007/s10620-009-0897-y. [DOI] [PubMed] [Google Scholar]
- 22.Albadine R, Wang W, Brownlee NA, Toubaji A, Billis A, Argani P, Epstein JI, Garvin AJ, Cousi R, Schaeffer EM, Pavlovich C, Netto GJ. Topoisomerase II alpha status in renal medullary carcinoma: immuno-expression and gene copy alterations of a potential target of therapy. J Urol. 2009;182:735–740. doi: 10.1016/j.juro.2009.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faggad A, Darb-Esfahani S, Wirtz R, Sinn B, Sehouli J, Konsgen D, Lage H, Weichert W, Noske A, Budczies J, Muller BM, Buckendahl AC, Roske A, Eldin Elwali N, Dietel M, Denkert C. Topoisomerase llalpha mRNA and protein expression in ovarian carcinoma: correlation with clinicopa-thological factors and prognosis. Mod Pathol. 2009;22:579–588. doi: 10.1038/modpathol.2009.14. [DOI] [PubMed] [Google Scholar]
- 24.Wang YH, Takanashi M, Tsuji K, Tanaka N, Shiseki M, Mori N, Motoji T. Level of DNA topoisomerase llalpha mRNA predicts the treatment response of relapsed acute leukemic patients. Leuk Res. 2009;33:902–907. doi: 10.1016/j.leukres.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Coss A, Tosetto M, Fox EJ, Sapetto-Rebow B, Gorman S, Kennedy BN, Lloyd AT, Hyland JM, O'Donoghue DP, Sheahan K, Leahy DT, Mulcahy HE, O'Sullivan JN. Increased topoisomerase llalpha expression in colorectal cancer is associated with advanced disease and chemotherapeutic resistance via inhibition of apoptosis. Cancer Lett. 2009;276:228–238. doi: 10.1016/j.canlet.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Liang Z, Zeng X, Gao J, Wu S, Wang P, Shi X, Zhang J, Liu T. Analysis of EGFR, HER2, and T0P2A gene status and chromosomal polysomy in gastric adenocarcinoma from Chinese patients. BMC Cancer. 2008;8:363. doi: 10.1186/1471-2407-8-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shvero J, Koren R, Shvili I, Yaniv E, Sadov R, Hadar T. Expression of human DNA Topoisomerase ll-alpha in squamous cell carcinoma of the larynx and its correlation with clinicopathologic variables. Am J Clin Pathol. 2008;130:934–939. doi: 10.1309/AJCPROG61USKCBEI. [DOI] [PubMed] [Google Scholar]
- 28.Bellanger S, de Gramont A, Sobczak-Thepot J. Cyclin B2 suppresses mitotic failure and DNA re-replication in human somatic cells knocked down for both cyclins B1 and B2. Oncogene. 2007;26:7175–7184. doi: 10.1038/sj.onc.1210539. [DOI] [PubMed] [Google Scholar]
- 29.Yu D, Cozma D, Park A, Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann N Y Acad Sci. 2005;1059:145–159. doi: 10.1196/annals.1339.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koss LG. Tumors of the urinary bladder. In: Firminger HI, editor. Atlas of Tumor Pathology. Washington, DC: Armed Forces Institute of Pathology; 1975. [Google Scholar]
- 31.Herman CJ, Vegt PD, Debruyne FM, Vooijs GP, Ramaekers FC. Squamous and transitional elements in rat bladder carcinomas induced by N-butyl-N-4-hydroxybutyl-nitrosamine (BBN). A study of cytokeratin expression. Am J Pathol. 1985;120:419–426. [PMC free article] [PubMed] [Google Scholar]