Abstract
While the roles of PPARα and PPARδ (β) in transcriptional regulation of myocardial lipid metabolisms are well established, an essential role of PPARγ in regulating lipid metabolisms in the adult heart remains unclear. In this study, we investigated whether PPARγ is required for normal myocardial lipid metabolism at basal condition in adult mice. We assessed the short-term cardiomyocyte-restricted PPARγ knockout mice with a Tamoxifen inducible Cre-LoxP mediated gene targeting strategy. The expression of PPARγ mRNA and protein in cardiomyocytes of adult mice was substantially reduced after short-term induction. Transcript and protein levels of important proteins in fatty acid uptake and oxidation, such as CD36, heart type-fatty acid binding protein (FABP), and carnitine palmitoyltransferase I (CPT-I) were reduced in the PPARγ deficient hearts. Myocardial fatty acid utilization and cardiac contraction were depressed in PPARγ deficient hearts. The PPARγ deficient hearts exhibited modest cardiac hypertrophy compared with controls. These results indicate that PPARγ is a transcription factor that is required for basal myocardial fatty acid utilization in the adult heart.
Keywords: PPARγ, fatty acid utilization, cardiac function, fatty acid metabolism
Introduction
Peroxisome proliferator-activated receptors [1] are members of the nuclear receptor super-family [2] with three subtypes α, δ, and γ), which are involved in differential regulation of lipid and glucose metabolism, and suppressing inflammatory responses. The liganded PPARs form heterodimers with retinoid X receptor and bind to PPAR responsive element αPPRE), mostly in the promoters of their target genes [3-14]. Cardiac muscle expresses all three PPAR subtypes, but their distinct roles in cardiac structure/function are not completely understood.
Several lines of evidence suggest that all three isoforms modulate cardiac energy metabolism. Most of studies on biological roles of PPARs in the heart focus on PPARα subtype, which mediates lipid-induced activation of FAO genes in vitro and in vivo [15-18]. An essential role of PPARδ in maintaining myocardial fatty acid metabolism has been established [19-24]. Whether PPARγ has similar regulating effects on fatty acid metabolism as PPARα and PPARδ remains unclear. Since all PPARs have a partially overlapping ligand binding profile, it is plausible that PPARγ could mediate similar signals as PPARα and PPARS in cardiomyocytes. Trans-genic overexpression of PPARγ in mouse heart leads to increased fatty acid uptake and metabolism in the heart [25]; however, it remains unknown whether PPARγ is required for myocardial fatty acid utilization.
We previously indicated a PPARγ's role in regulating myocardial oxidative stress by governing transcript expression of the manganese super-oxide dismutase (SOD2). Long-term cardiomyo-cyte-restricted PPARγ deficiency (CR-PPARγ-/-) in mice led to cardiac dysfunction, cardiac hypertrophy and heart failure with increased oxidative injuries [26]. Interestingly, basal transcript expression of known PPAR target genes that are involved in lipid metabolism is not altered in the above long-term PPARγ deficient hearts. The progressive phenotypic changes in CR-PPARγ-/- hearts could have confounding effects that alter gene expression associated with myocardial fatty acid utilization. A short-term induction of PPARγ knockout in the adult heart should help define the essential role of PPARγ signaling in the adult heart.
In the present study, we focused on defining the essential role of PPARγ in the adult heart using an inducible, cardiomyocyte-restricted PPARγ knockout line. The current results indicate the important role of PPARγ in regulating myocardial lipid metabolism in the adult heart.
Materials and methods
Breeding of the conditional PPARγ knockout line
The α-Myosin heavy chain (α-MyHC) promoter driven Mer-Cre-Mer (MCM) overexpression line (C57BL/6J) was mated with PPARγflox/flox mice αC57BL/6J) to generate α-MyHC-MCM/PPARγ flox/flox (MPG) mice. Eight to twelve-week-old male MPG mice were administrated tamoxifen by intraperitoneal injection (80mg/kg/day) for 5 days to remove PPARγ from cardiomyocytes (TMPG). Age- and gender-matched MCM mice were also injected with tamoxifen for 5 days to be used as control group (TMCM). In consistent with our recent report [27], we did not detect overt phenotypic change at basal condition for MCM mice after tamoxifen injection. Animals received standard laboratory chow and water on an ad libitum basis, and lightingwas maintained on a 12-hour cycle in temperature-controlled rooms. All experimental procedures were conducted in accordance with the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
Transcript analysis
Total RNA samples were extracted from left ventricle of mice using an RNA extraction kit (Qiagen). We performed QPCR (quantitative realtime RT-PCR) analyses (ABI and Roche qRT-PCR system) to determine transcript level of target gene. QPCR results from each gene/primer pair was normalized to β-actin.
Protein analysis
Nuclear and cytosolic protein was extracted from left ventricles by using a nuclear protein extraction kit αPierce). Samples were subjected to SDS-PAGE gels and use wet-transfer method to nitrocellulose membranes (Amersham). Western blot image was acquired by using Kodak image station system. We obtained antibody from commercial sources: PPARγ and pan-actin (Sigma Aldrich), CD36 and CPT-II (Abcom Inc.); FATP-1 and H-FABP (Santa Cruz biotechnology). Cardiac total SOD activity was measured with a kit (Cell Technology Inc.) according to manufacturer's instruction with minor modification. SOD2 activity was measured by adding Sodium Cyanide inhibiting SOD1 and SOD1 activity was derived from the difference of total SOD and SOD2 activities.
Echocardiographic measurements
Male TMPG and TMCM mice (10-20 weeks old) was used to detect cardiac structure alterations and cardiac function in vivo by using echocar-diogram (Visualsonic VEVO 770 System) with 35 MHz high resolution probe. Mice were anaesthetized by isoflurane inhalation (1.5-2.5%). Heart rate was maintained at ∼400 BPM and body temperature was maintained at 37°C by placing mice on a heating pad. We obtained IVS (intraventricular septum), LVID (left ventricular internal diameter), LV volume, LVPW (left ventricular posterior wall), EF% and FS% under Long -axis M-mode. Mitral valve Doppler was used to establish MV E/A ratio MV decel time. Pulse Doppler images were collected with the apical four-chamber view to record the mitral Doppler flow spectra. All data and images were saved and analyzed by an Advanced Cardiovascular Package Software.
Isolated working heart preparation
Isolated working heart function was assessed with an isolated working mouse heart apparatus from Harvard Apparatus. A 1.4-F pressure/volume catheter αMillar Instrument) was used to obtain values of pressure and volume, and other parameters. Briefly, mice were anesthetized with isoflurane inhalation, and the hearts were subsequently excised and cannulated via the aorta and left atrium. After equilibrating in the Langendorff mode, hearts were switched to the working heart mode, Mouse isolated hearts were perfused under aerobic conditions with the modified Krebs-Henseleit buffer contained NaCl (118mM), KCl (4.7mM), KH2PO4 (1.2mM), MgSO4 (1.2mM), NaHCO3 (25mM), Glucose (5mM), EDTA (0.5mM), and CaCl2 (2.5mM). Hearts were perfused at constant left atria l preload pressure of 7mmHg and constant aortic after-load pressure of 50 mmHg for 30 minutes. The acquired data were subsequently accessed with the HES software from the manufacturer (Harvard Apparatus).
[1, 3-3H] palmitic acid and [U-14C] glucose oxidation assay
Palmitic acid and glucose oxidation assay were performed by using the isolated working heart system (Harvard Apparatus). Hearts were firstly perfused with the modified Krebs-Henseleit buffer, and subsequently switched to the working buffer containing 3% BSA, 0.8 mmol/L palmitic acid and Insulin 100uU/ml, NaCl (118mM), KCl (4.7mM), KH2PO4 (1.2mM), MgSO4 (1.2mM), NaHCO3 α25mM), Glucose (5mM), EDTA(0.5mM), CaCl2 (2.5mM). [1, 3-3H] palmitic acid (0.1uCi/ml) and [U-14C] glucose (0.lμCi/ml) was used for the working buffer. The coronary effluent that exited from incision of pulmonary artery trunk through right ventricular was collected every 5 minutes. Palmitic acid and glucose oxidation rates were measured through collection of the 3H2O and 14CO2 and assessed 3H and 14C radioactivity by liquid scintillation counting. Dry heart weight (DHW) used as normalizing factor. Myocardial oxygen consumption was measured by using microsensor oxygen meter (PreSens Co.) by placing the needle tip probe at the coronary sinus. Wet heart weight (WHW) was used as normalizing factor.
Statistical analyses
Data for two-group comparison were analyzed using Student's t-test; otherwise the data were analyzed by one factor or mixed, two-factor analysis of Variance (ANOVA) using GraphPad Prism software (GraphPad Software Inc.). Values of quantitative results were expressed as mean±SEM. Differences between groups and treatments were regarded as significant at the P<0.05 probability level.
Results
Tamoxifen induced cardiomyocyte-restricted PPARg deficiency in adult TMPG mice
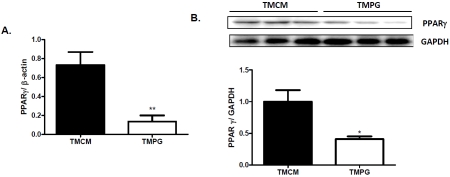
To specifically study the potential role of PPARγ in regulating myocardial lipid and glucose metabolism in the adult heart, we focus on assessing TMPG mice two weeks after the end of tamoxifen induction. The age and gender matched TMCM mice with the same induction were studied in parallel. We confirmed that the transcription and protein levels of PPARγ in TMPG hearts were decreased by about 82% and 60%, respectively, compared with the TMCM hearts (Figure 1 A and B). The expression of PPARα and PPARδ was not altered (data not shown).
Figure 1.
Tamoxifen induced PPARy deletion in adult mouse hearts. (A) Real-time PCR results of PPARy transcript levels in TMCM and TMPG hearts are shown. Heart samples were collected from 12 week old mice 14 days after the end of Tamoxifen treatment. Data are expressed as mean±SEM (n=4, **P<0.01). (B) Western blot analysis of PPARy on nuclear protein samples extracted from the hearts of TMCM and TMPG mice. Relative protein expression of PPARy was measured after normalized to GAPDH. Data are expressed as mean±SEM (n=3, *P<0.05).
PPARγ deficiency in adult hearts leads to decreased expression of key protein involved in fatty acid utilization
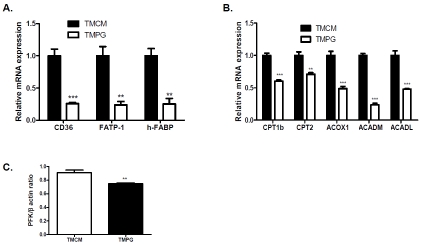
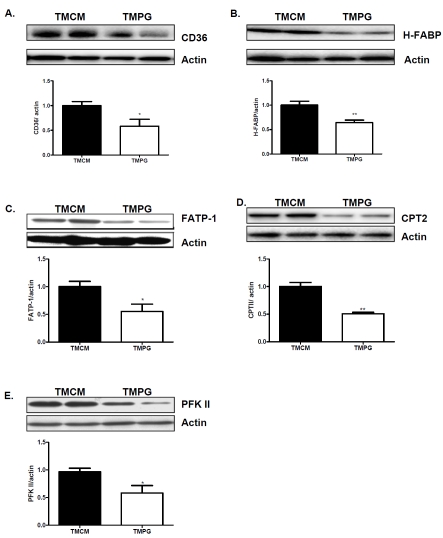
We next investigated whether a short-term induction of the PPARγ deficiency in the adult heart disturbs the expression of important proteins involved in myocardial fatty acid utilization. Real-Time PCR measurement revealed that FA translocase (CD36), heart type fatty acid-binding protein (H-FABP), and Fatty acid transport protein 1 (FATP-1) were downregulated in the TMPG compared with the TMCM heart (Figure 2A). We next measured the transcription levels of fatty acid oxidation genes. Carnitine palmitoyltransferase 1b (CPT1b), carnitine palmitoyltransferase 2 (CPT2), Acyl-Coenzyme A oxidase 1 (ACOX1), acyl-Coenzyme A dehydro-genase, C-4 to C-12 straight chain αACADM), and long-chain 3-hydroxyacyl-coenzyme A dehy-drogenase (ACADL) were markedly decreased in the TMPG adult hearts (Figure 2B). Transcript expression of Fructose-2,6-phosphofructo-2-kinase (PFK 2), a rate limiting enzyme involved in glucose utilization, was downregulated in TMPG vs TMCM hearts (Figure 2C). The protein levels of many of above proteins, such as CD36, h-FABP, FATP-1, CPT2 and PFK 2, were correspondingly decreased in the TMPG relative to the TMCM adult hearts (Figure 3 A to E, respectively).
Figure 2.
Transcript expression of key proteins in fatty acid and glucose utilization. Real Time PCR measurement of important fatty acid uptake (A), fatty acid oxidation (B) and glycolysis (C) genes in samples from left ventricles of 12 week old TMPG and TMCM mice 14 days after the end of tamoxifen treatment. Data are expressed as mean±SEM (n=4, **p<0.01, ***p<0.001).
Figure 3.
Protein expression of key proteins in fatty acid and glucose utilization. Western blot analysis of key protein of fatty acid and glucose utilization on total protein samples extracted from left ventricles of 12 week old TMPG and TMCM mice 14 days after the end of tamoxifen treatment. (A) CD36; (B) Heart type FABP; (C) FATP; (D) CPT2 and E) PFK 2. Data are expressed as mean±SEM (n=4, *P<0.05, **p<0.01).
PPARg deficiency in adult hearts leads to impaired fatty acid utilization
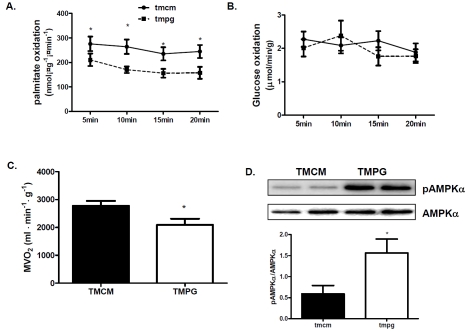
Since the expression of key proteins in fatty acid uptake and oxidation was depressed in the TMPG hearts, we next assess whether cardiac PPARγ deficiency in the adult mice perturbs myocardial fatty acid and glucose oxidation, and compromises cardiac performance. Myocardial fatty acid and glucose oxidation were determined in isolated working hearts. We observed a remarkable repression in the rate of palmitic acid oxidation in the TMPG hearts compared to that of the TMCM hearts (Figure 4A), but the rates of glucose oxidation (Figure 4B) did not changed. With decreased oxidative metabolism, myocardial oxygen (MVO2) consumption was decreased in TMPG hearts compared with TMCM hearts (Figure 4C). With the depressed oxidative metabolism, Western blots demonstrated that AMPK activity was increased with greater phosphorylated AMPKα to AMPKα ratio in TMPG relative to TMCM hearts (Figure 4D). These results demonstrated that myocardial oxidative metabolism was disturbed in the PPAR γ deficient heart.
Figure 4.
oxidative metabolism and AMPK activity. (A) Rate of palmitate oxidation in isolated working hearts from the adult TMPG and TMCM mice14 days after the end of tamoxifen treatment were measured using 3H labeled palmitate. Data are expressed as mean±SEM (n=6, *p<0.05). (B) Rate of glucose oxidation in isolated working hearts from the adult TMPG and TMCM mice 14 days after the end of tamoxifen treatment were measured using 14C labeled glucose. Data are expressed as mean±SEM (n=6, p>0.05). (C) Myocardial oxygen consumption was measured during the isolated heart perfusion. Data are expressed as mean±SEM (n=6, *P<0.05). (D) Western blot analysis of phosphory-lated AMPKα and AMPKα. AMPK activity was estimated by the ratio of pAMPKα to AMPKα. Data are expressed as mean±SEM (n=4, *P<0.05).
PPARγ deficiency in adult heart leads to cardiac hypertrophy and dysfunction
Measurement of isolated heart function revealed that both dLVP/dtmax and dLVP/dtmin were remarkably decreased in TMPG relative to TMCM hearts (Table 1). The value of time to peak (TTpeak) and Contractility index (CI) were substantially declined in the TMPG adult hearts (Table 1). The heart rate and other parameters were not significantly changed in the TMPG compared with the TMCM hearts (Table 1).
Table 1.
Hemodynamic measurement of isolated working heart
| Parameters | TMCM | TMPG |
|---|---|---|
| HR | 377.5±9.7 | 379.5±13.59 |
| LVP(mmHg) | 90±2 | 91±1.2 |
| LVPdia(mmHg) | 7.8±0.6 | 9.4±0.3* |
| LVEDP(mmHg) | 10.7±1 | 12±0.66 |
| dLVPdtmax(mmHg/s) | 5486±443.4 | 4129±247.8* |
| -dLVPdtmin(mmHg/s) | -4044±386 | -3488±227* |
| CI | 108.7±13.43 | 79.95±3.7* |
| TTPeak (ms) | 21.32±1.55 | 28.46±1.67* |
| RT50 (ms) | 32.56±2.16 | 34.74±0.77 |
Abbreviations: Heart rate (HR); LVP (left ventricular pressure); LVPdia (left ventricular diastolic pressure); left ventricular end-diastolic pressure (LVEDP); left ventricular maximal and minimal dP/dt indicate rates of cardiac contractility; contractility index (CI); TTPeak, time to peak and relaxation; 50% relaxation time (RT50).
P<0.05, n=9.
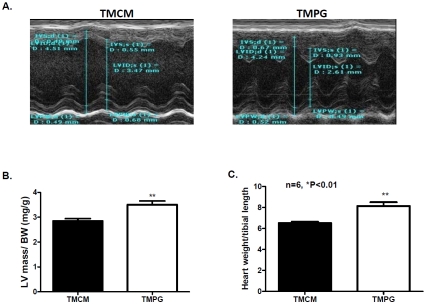
Despite cardiac dysfunction in isolated working heart, there was no major cardiac pathological development in the TMPG hearts after short-term (2 weeks after the end of the tamoxifen induction, data not shown). However, long-term (3 months) PPARγ deficiency in the TMPG heart led to modest cardiac hypertrophy. Echocardiographic measurement (Figure 5A) revealed that interventricular septum (IVS) at end-systolic was increased in TMPG hearts compared with TMCM controls (Table 2). LV mass was also increased in TMPG vs TMCM hearts (Table 2). LV mass/ body weight ratio was increased in the TMPG relative to the TMCM hearts (Figure 5B). However, there was no substantial attenuation in ejection fraction percentage (EF %) and fractional shortening percentage (FS %) in the TMPG heart compared with the TMCM heart (Table 2). We further assessed the heart weight to tibial length ratio and confirmed the echocar-diographic findings (Figure 5C). Despite the modest cardiac hypertrophy, we did not detect major pathological development in histological and ultrastructural levels (data not shown) in the TMPG hearts after 3 months of induced PPARγ deficiency.
Figure 5.
Measurements of cardiac hypertrophy. (A) Representative M mode images of echocardiography from adult TMPG and TMCM mice. (B) Corrected left ventricular (LV) mass values from echocardiographic measurement normalized to body weight. Data are shown as mean ± SEM. (n=6, **p<0.01). (E) Heart weight to tibial length ratio was measured on the adult TMPG and TMCM mice 3 months after the end of tamoxifen treatment. Data are shown as mean ± SEM. (n=6, **p<0.01).
Table 2.
Echocardiography measurement
| Parameters | TMCM | TMPG |
|---|---|---|
| IVS;d (mm) | 0.6±0.03 | 0.67±0.02 |
| IVS;s (mm) | 0.85±0.07 | 0.99±0.06* |
| LVID;d (mm) | 4.51±0.11 | 4.42±0.08 |
| LVID;s (mm) | 3.16±0.14 | 3.07±0.13 |
| LVPW;d (mm) | 0.49±0.06 | 0.52±0.04 |
| LVPW;s (mm) | 0.73±0.06 | 0.67±0.06 |
| Stroke Volume (μL) | 58.93±3.11 | 57.59±2.9 |
| Ejection Fraction (%) | 57±3.6 | 58±3.1 |
| Cardiac Output (ml/min) | 23.75±2 | 25.40±0.78 |
| Corrected LV mass | 65.3±2.3 | 76±4* |
| HR (Beat/Min) | 429±17 | 423±15 |
LVID;s and LVID;d: left ventricular dimension at systole and diastole; LVPW;s and LVPW;d: posterior wall thickness at systole and diastole; IVS;s and IVS; d: interventricular septal wall thickness (diastole and systole); EF%: Ejection fraction; SV: left ventricle stroke volume; HR: heart rate.
P<0.05, n=6.
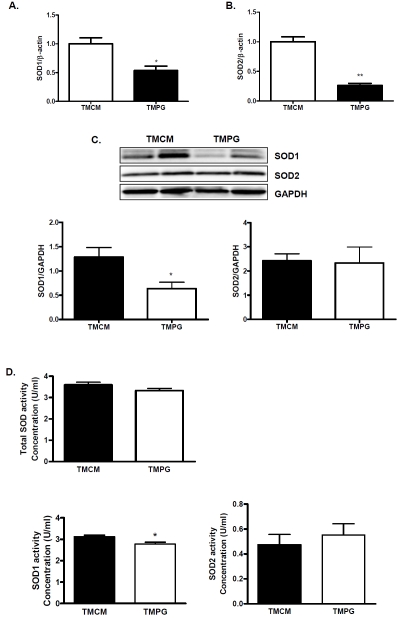
PPARg deficiency in adult hearts leads to decreased transcript expression of SOD1 and SOD2
In our previous study, expression of SOD2 was depressed in cardiomyocyte-restricted PPARγ knockout mice [26]. In TMPG hearts two weeks after induction, the expression of SOD2 was decreased as those previously seen in the long-term PPARγ deficient hearts (Figure 6A). Interestingly, the transcript level of Cu/Zn superox-ide dismutase (SOD1) was also decreased in TMPG heart (Figure 6B). However, Western blot analysis revealed that the protein expression of SOD1, but not SOD2, was correspondingly decreased in the TMPG compared with the TMCM heart (Figure 6C). Whereas the total SOD activity and SOD2 activity were comparable between TMCM and TMPG hearts, SOD1 activity was modestly decreased in TMPG compared with TMCM hearts (Figure 6D). Therefore, it appears that PPARγ is involved in regulating SOD1 and SOD2 expression in the adult heart mainly in the transcript level.
Figure 6.
Transcript and protein expression of SOD1 and SOD2. Real Time PCR analyses of SOD1 (A) and SOD2 (B) transcript in RNA samples extracted from adult heart of the TMPG vs the TMCM mice. Data are expressed as mean±SEM (n=6, * p<0.05, ** p<0.01). (C) Western blot analysis of SOD1 and SOD2 on protein samples extracted from the hearts of TMCM and TMPG mice. Relative protein expression of SOD1 and SOD2 was measured after normalized to GAPDH. Data are expressed as mean±SEM (n=4, *P<0.05). D) Total and subtype SOD activities on protein samples extracted from the hearts of TMCM and TMPG mice. Data are expressed as mean±SEM (n=6, *P<0.05).
Discussion
In this study, we investigate the short-term effect of cardiac PPARγ deficiency in the adult mice. The most important findings are that mice with short-term PPARγ deficiency in their hearts exhibit depressed expression of important lipid metabolic proteins and decreased myocardial fatty acid utilization with subsequent cardiac dysfunction and cardiac hypertrophy.
Cardiac expression of PPARγ is upregulated in patients with ischemic and dilated cardiomyopa-thy [28], and in mice with pressure overload induced cardiac hypertrophy [29]. Accumulating evidence shows that PPARγ may exert anti-inflammation and anti-hypertrophic growth effect in vivo and vitro [30-32]. PPARγ signaling may attenuate cardiac remodeling through pathways not directly involved in controlling lipid and energy metabolism, such as inflammation and/ or anti-oxidative stress [26]. However, whether PPARγ also regulates lipid metabolism in the adult heart is not well established. There are several studies based on mouse models of long-term cardiac-specific PPARγ knockout [26, 33, 34], interestingly, none of these studies have demonstrated depressed transcript expression of fatty acid metabolic genes and overt myocardial lipid disturbances. While all of the above studies demonstrate various degrees of cardiac pathological development in the long-term cardiac-specific PPARγ knockout mice, multiple mechanisms have been proposed. For instant, we found that the PPARγ deficient heart expressed lower SOD2 and developed extensive oxidative damages to mitochondria [26]. Treatment with an SOD mimetic compound protects against cardiac pathology progression. Duan et al showed changes of various signaling pathways in the heart, which attribute to the development of cardiac hypertrophy in the PPARγ deficient heart [33]. Caglayan's study attributes PPARγ's role on regulating basal myocardial inflammation [34]. However, none of these studies identified the role of PPARγ in regulating myocardial lipid metabolism. A recent study on a transgenic mouse model with cardiac-specific transgenic overexpression of the wild type PPAR γ demonstrated that PPARγ may be involved in the transcriptional regulation of genes encoding proteins that are important in cardiac lipid utilization [25]. The increased lipid utilization in the hearts with transgenic PPARγ overexpression leads to lipotoxic cardiomyopathy [25]. However, this finding also brings up a question on why myocardial lipid metabolism does not altered in the long-term PPARγ deficient heart [33, 35] as those in PPARα and PPARδ null hearts [16, 20, 23]. We speculate that compensatory upregulation of metabolic genes might misguide the primary effect of PPARγ deficiency in the heart. The current study using the induc-ible PPARγ knockout strategy enables us to uncover not only the short-term effect of cardiac PPARγ deficiency, but also the effect of cardiac PPARγ deficiency in the adult heart without potential confounding effects during the early life of the animal.
Most interestingly, the short-term PPARγ deficient heart exhibited depressed expression of important lipid metabolic proteins and subsequently led to a depressed myocardial fatty acid oxidation rates without affecting glucose oxidation. Since all PPARs have a partially overlapping ligand binding profile, PPARγ could mediate similar signals to certain degree as PPARα and PPARδ do in cardiomyocytes. All the genes assessed in the current study are well known PPAR target genes. The depressed expression of these lipid metabolic genes and subsequently the rate of myocardial fatty acid oxidation in the PPARγ deficient heart in the adult mice should be primarily due to the impaired transcriptional activity of the PPARγ. With the decrease of fatty acid utilization and the upregulated AMPK activity, the TMPG heart could have switch to increased glucose utilization if the Randle cycle is functioning. However, the diminished expression of PFK-2, an important rate-limiting enzyme in the glycolysis pathway, may compromise glucose utilization despite an upregulating AMPK activity in TMPG hearts. The reduced oxidative metabolism is also manifested by the less consumption of oxygen in TMPG hearts. Even though the expression of other PPAR subtypes was not altered (data not shown), upregulation of activities from other transcription factors is possible. The degree of reduced fatty acid oxidation may not be severe enough to cause major energetic crisis to trigger progressive pathological development in the TMPG heart.
In addition, we confirmed that PPARγ is essential for the full expression of SOD2. It is interesting that the short-term PPARγ deficiency in the adult heart also exhibited a depressed expression of SOD1, another isoforms of SOD. However, protein expression of SOD2 was maintained at the normal levels, possibly by other regulation mechanisms in the adult heart. The modest decrease of SOD1 protein in TMPG hearts did not lead to depressed total SOD activity with no detectable oxidative stress (data not shown). The PPARγ deficient heart shows compromised cardiac performance and modest cardiac hypertrophy. Interestingly, the severity of phenotypic changes was not as progressive as seen in those previously reported (CR-PPARγ-/-) mice with PPARγ deficiency in the ventricles since birth. The modest phenotypic changes in the TMPG hearts may attribute to a potentially less robust PPARγ knockout with the tamoxifen inducible strategy. It is also likely that the adult heart is more adaptive in resistant to cardiac pathological development. The normal protein abundances of SOD1 and SOD2 in the adult TMPG hearts may be the results of these adaptive responses in the adult heart. We have recently demonstrated that PPARδ is essential for not only the constitutive function of fatty acid metabolism and anti-oxidant defense, but is also important in maintaining mitochondrial biogenesis [27]. Because TMPG heart demonstrated normal mitochondrial DNA copy number with normal mitochondrial ultrastructure (data not shown), PPARγ appears to play no essential role in maintaining mitochondrial biogenesis. Therefore, it appears that the disturbing myocardial fatty acid utilization but not increased oxidative stress compromises the capacity of the cardiomyocyte to maintain energy levels in subcellular compartments responsible for contraction and critical homeostatic functions, such as calcium re-uptake. The impaired metabolic flexibility renders the heart susceptible to contractile dysfunction and results in hypertrophy.
Our present study demonstrates for the first time that PPARγ is required for the full expression of fatty acid metabolic genes in the adult heart, hence is an essential determinant of myocardial substrate utilization and cardiac performance.
Acknowledgments
Part of the work was performed in core facilities supported by the Diabetes Research and Training Center (P60 DK079626). This work was supported by grants from National Institute of Health (1R01HL085499 and 1R01HL084456).
References
- 1.Aarenstrup L, Flindt EN, Otkjaer K, Kirkegaard M, Andersen JS, Kristiansen K. HDAC activity is required for p65/RelA-dependent repression of PPARdelta-mediated transactivation in human keratinocytes. J Invest Dermatol. 2008;128:1095–1106. doi: 10.1038/sj.jid.5701146. [DOI] [PubMed] [Google Scholar]
- 2.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 3.Carter ME, Gulick T, Moore DD, Kelly DP. A pleiotropic element in the medium-chain acyl coenzyme A dehydrogenase gene promoter mediates transcriptional regulation by multiple nuclear receptor transcription factors and defines novel receptor-DNA binding motifs. Mol Cell Biol. 1994;14:4360–4372. doi: 10.1128/mcb.14.7.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clabby ML, Robison TA, Quigley HL, Wilson DB, Kelly DP. RXRalpha represses GATA-4-mediated transcription via a retinoid-dependent Interaction with the cardiac-enriched repressor FOG-2. J Biol Chem. 2002 doi: 10.1074/jbc.M208173200. [DOI] [PubMed] [Google Scholar]
- 5.Huss JM, Levy FH, Kelly DP. Hypoxia inhibits the peroxisome proliferator-activated receptor alpha/retinoid X receptor gene regulatory pathway in cardiac myocytes: a mechanism for O2-dependent modulation of mitochondrial fatty acid oxidation. J Biol Chem. 2001;276:27605–27612. doi: 10.1074/jbc.M100277200. [DOI] [PubMed] [Google Scholar]
- 6.Iwaki M, Matsuda M, Maeda N, Funahashi T, Matsuzawa Y, Makishima M, Shimomura I. Induction of adiponectin, a fat-derived antidia-betic and antiatherogenic factor, by nuclear receptors. Diabetes. 2003;52:1655–1663. doi: 10.2337/diabetes.52.7.1655. [DOI] [PubMed] [Google Scholar]
- 7.Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc Natl Acad Sci U S A. 1993;90:2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kliewer SA, Xu HE, Lambert MH, Willson TM. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog Horm Res. 2001;56:239–263. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee R, Jow L, Croston GE, Paterniti JR., Jr Identification, characterization, and tissue distribution of human peroxisome proliferator-activated receptor (PPAR) isoforms PPARgam-ma2 versus PPARgamma1 and activation with retinoid X receptor agonists and antagonists. J Biol Chem. 1997;272:8071–8076. doi: 10.1074/jbc.272.12.8071. [DOI] [PubMed] [Google Scholar]
- 10.Osorio JC, Stanley WC, Linke A, Castellari M, Diep QN, Panchal AR, Hintze TH, Lopaschuk GD, Recchia FA. Impaired myocardial fatty acid oxidation and reduced protein expression of retinoid X receptor-alpha in pacing-induced heart failure. Circulation. 2002;106:606–612. doi: 10.1161/01.cir.0000023531.22727.c1. [DOI] [PubMed] [Google Scholar]
- 11.Schlezinger JJ, Jensen BA, Mann KK, Ryu HY, Sherr DH. Peroxisome proliferator-activated receptor gamma-mediated NF-kappa B activation and apoptosis in pre-B cells. J Immunol. 2002;169:6831–6841. doi: 10.4049/jimmunol.169.12.6831. [DOI] [PubMed] [Google Scholar]
- 12.Schoonjans K, Staels B, Auwerx J. The peroxisome proliferator activated receptors (PPARS) and their effects on lipid metabolism and adipo-cyte differentiation. Biochim Biophys Acta. 1996;1302:93–109. doi: 10.1016/0005-2760(96)00066-5. [DOI] [PubMed] [Google Scholar]
- 13.Steinmetz M, Quentin T, Poppe A, Paul T, Jux C. Changes in expression levels of genes involved in fatty acid metabolism: upregulation of all three members of the PPAR family (alpha, gamma, delta) and the newly described adiponectin receptor 2, but not adiponectin receptor 1 during neonatal cardiac development of the rat. Basic Res Cardiol. 2005;100:263–269. doi: 10.1007/s00395-005-0520-0. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Repa JJ, Gauthier K, Mangelsdorf DJ. Regulation of lipoprotein lipase by the oxys-terol receptors, LXRalpha and LXRbeta. J Biol Chem. 2001;276:43018–43024. doi: 10.1074/jbc.M107823200. [DOI] [PubMed] [Google Scholar]
- 15.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 16.Djouadi F, Weinheimer CJ, Saffitz JE, Pitchford C, Bastin J, Gonzalez FJ, Kelly DP. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator- activated receptor alpha- deficient mice. J Clin Invest. 1998;102:1083–1091. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilde AJ, van der Lee KA, Willemsen PH, Chinetti G, van der Leij FR, van der Vusse GJ, Staels B, van Bilsen M. Peroxisome proliferator-activated receptor (PPAR) alpha and PPARbeta/ delta, but not PPARgamma, modulate the expression of genes involved in cardiac lipid metabolism. Circ Res. 2003;92:518–524. doi: 10.1161/01.RES.0000060700.55247.7C. [DOI] [PubMed] [Google Scholar]
- 20.Cheng L, Ding G, Qin Q, Huang Y, Lewis W, He N, Evans RM, Schneider MD, Brako FA, Xiao Y, Chen YE, Yang Q. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat Med. 2004;10:1245–1250. doi: 10.1038/nm1116. [DOI] [PubMed] [Google Scholar]
- 21.Cheng L, Ding G, Qin Q, Xiao Y, Woods D, Chen YE, Yang Q. Peroxisome proliferator-activated receptor delta activates fatty acid oxidation in cultured neonatal and adult cardiomyocytes. Biochem Biophys Res Commun. 2004;313:277–286. doi: 10.1016/j.bbrc.2003.11.127. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Yin R, Liu J, Wang P, Wu S, Luo J, Zhelyabovska O, Yang Q. Peroxisome proliferator-activated receptor delta regulates mitofusin 2 expression in the heart. J Mol Cell Cardiol. 2009;46:876–882. doi: 10.1016/j.yjmcc.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Cheng L, Qin Q, Liu J, Lo WK, Brako LA, Yang Q. High-fat feeding in cardiomyocyte-restricted PPARdelta knockout mice leads to cardiac overexpression of lipid metabolic genes but fails to rescue cardiac phenotypes. J Mol Cell Cardiol. 2009;47:536–543. doi: 10.1016/j.yjmcc.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burkart EM, Sambandam N, Han X, Gross RW, Courtois M, Gierasch CM, Shoghi K, Welch MJ, Kelly DP. Nuclear receptors PPARbeta/delta and PPARalpha direct distinct metabolic regulatory programs in the mouse heart. J Clin Invest. 2007;117:3930–3939. doi: 10.1172/JCI32578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Son NH, Park TS, Yamashita H, Yokoyama M, Huggins LA, Okajima K, Homma S, Szabolcs MJ, Huang LS, Goldberg IJ. Cardiomyocyte expression of PPARgamma leads to cardiac dysfunction in mice. J Clin Invest. 2007;117:2791–2801. doi: 10.1172/JCI30335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding G, Fu M, Qin Q, Lewis W, Kim HW, Fukai T, Bacanamwo M, Chen YE, Schneider MD, Mangelsdorf DJ, Evans RM, YangQ Cardiac peroxisome proliferator-activated receptor gamma is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc Res. 2007;76:269–279. doi: 10.1016/j.cardiores.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 27.Wang P, Liu J, Li Y, Wu S, Luo J, Yang H, Subbiah R, Chatham J, Zhelyabovska O, Yang Q. Peroxisome proliferator-activated receptor {delta} is an essential transcriptional regulator for mito-chondrial protection and biogenesis in adult heart. Circ Res. 2010;106:911–919. doi: 10.1161/CIRCRESAHA.109.206185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehrabi MR, Thalhammer T, Haslmayer P, Glogar HD, Wieselthaler G, Humpeler S, Marktl W, Ekmekcioglu C. The peroxisome proliferator-activated receptor gamma (PPARgamma) is highly expressed in human heart ventricles. Biomed Pharmacother. 2002;56:407–410. doi: 10.1016/s0753-3322(02)00251-2. [DOI] [PubMed] [Google Scholar]
- 29.Krishnan J, Suter M, Windak R, Krebs T, Felley A, Montessuit C, Tokarska-Schlattner M, Aasum E, Bogdanova A, Perriard E, Perriard JC, Larsen T, Pedrazzini T, Krek W. Activation of a HIF1al-pha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009;9:512–524. doi: 10.1016/j.cmet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Shiomi T, Tsutsui H, Hayashidani S, Suematsu N, Ikeuchi M, Wen J, Ishibashi M, Kubota T, Egashira K, Takeshita A. Pioglitazone, a peroxisome proliferator-activated receptor-gamma agonist, attenuates left ventricular remodeling and failure after experimental myocardial infarction. Circulation. 2002;106:3126–3132. doi: 10.1161/01.cir.0000039346.31538.2c. [DOI] [PubMed] [Google Scholar]
- 31.Asakawa M, Takano H, Nagai T, Uozumi H, Hasegawa H, Kubota N, Saito T, Masuda Y, Kadowaki T, Komuro I. Peroxisome proliferator-activated receptor gamma plays a critical role in inhibition of cardiac hypertrophy in vitro and in vivo. Circulation. 2002;105:1240–1246. doi: 10.1161/hc1002.105225. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto K, Ohki R, Lee RT, Ikeda U, Shimada K. Peroxisome proliferator-activated receptor gamma activators inhibit cardiac hypertrophy in cardiac myocytes. Circulation. 2001;104:1670–1675. doi: 10.1161/hc4001.097186. [DOI] [PubMed] [Google Scholar]
- 33.Duan SZ, Ivashchenko CY, Russell MW, Milstone DS, Mortensen RM. Cardiomyocyte-specific knockout and agonist of peroxisome proliferator-activated receptor-gamma both induce cardiac hypertrophy in mice. Circ Res. 2005;97:372–379. doi: 10.1161/01.RES.0000179226.34112.6d. [DOI] [PubMed] [Google Scholar]
- 34.Caglayan E, Stauber B, Collins AR, Lyon CJ, Yin F, Liu J, Rosenkranz S, Erdmann E, Peterson LE, Ross RS, Tangirala RK, Hsueh WA. Differential roles of cardiomyocyte and macrophage peroxisome proliferator-activated receptor gamma in cardiac fibrosis. Diabetes. 2008;57:2470–2479. doi: 10.2337/db07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding G, Cheng L, Qin Q, Frontin S, Yang Q. PPARdelta modulates lipopolysaccharide-induced TNFalpha inflammation signaling in cultured cardiomyocytes. J Mol Cell Cardiol. 2006;40:821–828. doi: 10.1016/j.yjmcc.2006.03.422. [DOI] [PubMed] [Google Scholar]