Abstract
Voxel-Based Morphometry (VBM) has been used for several years to study differences in brain structure between populations. Recently, a longitudinal version of VBM has been used to show changes in gray matter associated with relatively short periods of training. In the present study we use fMRI and three different standard implementations of longitudinal VBM: SPM2, FSL, and SPM5 to assess functional and structural changes associated with a simple learning task. Behavioral and fMRI data clearly showed a significant learning effect. However, initially positive VBM results were found to be inconsistent across minor perturbations of the analysis technique and ultimately proved to be artifactual. When alignment biases were controlled for and recommended statistical procedures were used, no significant changes in grey matter density were found. This work, initially intended to show structural and functional changes with learning, rather demonstrates some of the potential pitfalls of existing longitudinal VBM methods and prescribes that these tools be applied and interpreted with extreme caution.
Introduction
As early as 1960, researchers demonstrated that learning and experience could produce profound changes in gross measures of brain morphology in rats such as brain weight and cortical thickness (Krech et al., 1960; Rosenzweig et al., 1972; Klintsova and Greenough, 1999). Subsequently, it has been demonstrated that many aspects of brain structure and function can be modified by learning — including synaptic density, neural and glial cell size and ratio, vascularization, dendritic branching, fMRI activation, and neurotransmitter concentration (Black, et al., 1990; van Praag et al., 2000; Poldrack, 2000; Floyer-Lea et al., 2006). In some cases, these gross changes in brain structure can be detected after as little as 10 days of training (Kleim et al., 2007).
Recently, an adaptation of voxel-based morphology (VBM) has been introduced that attempts to non-invasively measure longitudinal changes in gray matter density using MRI (Ashburner and Friston, 2000; Draganski et al., 2004). This within-group approach is statistically more powerful and does not require the large number of subjects traditionally used in VBM studies. Using this approach Draganski et al. (2004) reported a localized 3% increase in gray matter density in MT after 3 months of juggling practice. Draganski et al. (2006) reported both increases and decreases in gray matter density associated with 3 months of studying for a medical exam. Most recently Ilg et al. (2008) reported gray matter and functional increases in right occipital cortex associated with 2 weeks of mirror-reading practice.
Many criticisms of VBM have been published since its introduction. Bookstein (2001), Davatzikos (2004) and Crum et al. (2003) are primarily concerned with the automated nonlinear registration technique used by VBM and the potential problems in establishing functional homology between groups. (See Ashburner and Friston (2001) for a rebuttal to some of these issues.) Although these critiques are focused on between group studies, they illustrate that relatively minor differences in brain anatomy or other initial conditions can have significant effects on final results. While a difference in brain anatomy is not relevant in a longitudinal design, sensitivity to initial conditions such as field inhomogeneities may prove problematic for both cross-sectional and longitudinal studies.
VBM analysis has recently been applied to diffusion tensor MRI (DT-MRI) where authors have also found reason to be skeptical of the reproducibility of the technique. Jones et al. (2005) notes the large impact of different smoothing kernel sizes in the analysis of DT-MRI data. They also note the lack of normality in the residual images hampering the ability to make statistical inferences using parametric methods. Most recently, Jones et al. (2007) demonstrated that ten different groups analyzing a common DT-MRI dataset using voxel-based methods drew widely disparate conclusions. This current state of controversy limits the conclusions one can draw from VBM-based results in isolation. Because of these controversies, it is critical to demonstrate the reproducibility and convergence with other methods to establish the validity of VBM-based results.
In this work we use longitudinal VBM in combination with fMRI to explore functional and structural changes associated with learning a simple visual–motor task. In contrast to other longitudinal VBM studies, subjects participated in both the control and the learning phase of the experiment thus controlling for false positives due to group differences. Since each subject serves as his or her own control, this within-group approach provides an opportunity to explore the robustness and sensitivity of longitudinal VBM that is independent from the problems inherent in establishing homology between groups. We also explore the consistency of longitudinal VBM results by comparing its implementation in three different software packages: SPM2, FSL, and SPM5. We hypothesized that the regions which show learning related changes in functional activation should also demonstrate changes (either increases or decreases) in gray matter density. We also hypothesized that these results should be consistent across implementations of VBM.
Methods
Subjects
Twelve healthy, right-hand dominant subjects were included in this study. (mean age: 32.5 years, range: 23–40, 6 men, 6 women). All gave informed consent according to a protocol approved by the NIH IRB. Five other subjects were excluded due to poor quality scans or missing scan sessions. All of the subjects analyzed participated in both the control and learning phases of the experiment.
Training paradigm
The experimental paradigm is illustrated in Fig. 1. Subjects initially underwent a baseline structural MRI scan (scan 1). After a two-week control period the subjects underwent a second structural MRI (scan 2) as well as four fMRI scans during which they alternatively performed the mirror task and the control task (Fig. 1). The control task required the subject to follow a randomly moving white dot on the screen using a joystick held in the right hand. The mirror task was identical to the control task except that the left–right axis of the joystick was reversed. After scan 2, subjects were trained on the mirror task for a total of 2.5 h over 2 weeks (six 25-minute training sessions). At the end of the training subjects received both structural and functional MRI scans identical to those in scan 2. Eight subjects performed the experiment in a continuous four-week block. Four subjects had an interval of 2 to 12 weeks (mean 44.5 days) between the control and learning phase of the experiment. An extra scanning session was conducted on these subjects at the beginning of the two-week learning phase to serve as a baseline for the learning comparison. Thus, all comparisons on all subjects were over a two-week period.
Fig. 1.
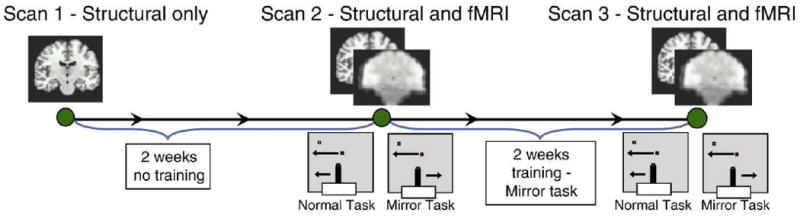
Subjects initially underwent a baseline structural MRI scan (scan 1). After a two-week control period the subjects underwent a second structural MRI scan (scan 2) as well as four fMRI scans during which they alternated performing the mirror task and the control task. The control task required the subject to follow a randomly moving white dot on the screen using a joystick held in the right hand. The mirror task was identical to the control task except that the left–right axis of the joystick is reversed. After scan 2, subjects were trained on the mirror task for a total of 2.5 h over 2 weeks (six 25-minute training sessions). At the end of the training subjects received both structural and functional MRI scans identical to scan 2.
Scanning protocol
All scans were collected on a 3 T General Electric (GE, HDx 14M3) Scanner using a GE eight-channel head coil. Structural scans consisted of two FSPGR scans (256 × 256, 124 slices, 0.85 × 0.85 × 1.2 mm voxels, TI = 400 ms, TE = ∼5 ms). Functional scans consisted of four axial EPI time series (64 × 64, 38 slices, 3.2 × 3.2 × 3.2 mm voxels, TR = 2.5, TE = 30). Each of the four 6-minute scans employed a block design of alternating 30-second periods of performing one of the two tasks with 30 seconds of fixation. Order of task presentation was counterbalanced across subjects.
Functional analyses
The first level of fMRI analysis combined like sessions and produced parameter estimates for each subject's activation compared to baseline in each task. This was carried out using FEAT (FMRI Expert Analysis Tool) Version 5.63, part of FSL (FMRIB's Software Library, www.fmrib.ox.ac.uk/fsl). A second higher level analysis performed a paired T-test to compare subject's activation before and after training. This was carried out using FLAME (FMRIB's Local Analysis of Mixed Effects) stage 1 only (Beckmann et al., 2003, Woolrich et al., 2004). Z-statistic images were thresholded using clusters determined by Z>3.5 and a (corrected) cluster significance threshold of p = 0.05 (Worsley et al., 1992).
Structural analyses
Pre-processing
For each scanning session the two FSPGR images were rigidly aligned using a motion-correction algorithm from either SPM2, FSL, or SPM5 and averaged together. In order to correct for image inhomogeneities, intensity bias correction was performed on the average images using four iterations of the N3 algorithm (MINC Tools, Sled et al., 1998).
SPM2
The SPM2 analysis was performed in Matlab 7.4 using the VBM2 toolbox (v1.07, http://dbm.neuro.uni-jena.de/vbm/) which implements an optimized VBM pipeline (Ashburner and Friston 2000; Good et al., 2001). The first scan of each subject was used as the “baseline” to create a custom gray matter template. All structural scans for a given subject were fed through the longitudinal analysis pipeline of the VBM2 toolbox such that the first scan was used as the source for spatial normalization (piecewise linear). The analysis was also repeated using the scan immediately before training as the baseline for the template and the source for spatial normalization. A third iteration was performed using the subject's mean image aligned to the halfway point between scans 1 and 2 as the baseline. The halfway point was determined by taking the square root of the alignment transformation (rigid, 6 DOF) between scans 1 and 2. This manipulation has been previously utilized in other structural analysis packages such as SIENA (Smith et al., 2002).
Note that for the longitudinal stream the VBM2 toolbox does not “modulate” images (i.e. multiply by the Jacobian determinate, Good et al., 2001). Default options were used for hidden Markov random field (HMRF) weighting, bias correction, cutoff spatial normalization, nonlinear regularization, and number of nonlinear iterations. Normalized gray matter images were smoothed using default settings: an 8 mm full-width half-max (FWHM) Gaussian kernel. An absolute intensity threshold mask of 0.2 was used to remove regions with minimal grey matter intensity. Pseudo paired T-tests were used to compare scan sessions one and two (the control period) and two and three (the training period). Cluster-based inference in VBM is complicated by the lack of stationarity or uniform smoothness (Worsley et al., 1999). Hayasaka et al. (2004) prescribe either non-stationarity correction or permutation-based methods (depending on degrees of freedom) in order to draw statistically valid inferences on VBM data. Both methods are employed here using either the nS toolbox from Hayasaka et al. (2004) or the SnPM3 toolbox from Nichols and Holmes (2002). For all analyses an uncorrected height threshold of p<0.001 was applied and an extent threshold of p = 0.05, corrected across space. A 1 mm FWHM variance smoothing kernel was used for non-parametric analyses. See Supplemental Fig. 1 for a flowchart of steps.
FSL
FSL-VBM (http://www.fmrib.ox.ac.uk/fsl/fslvbm/, version 1.0) has not been specifically designed for longitudinal analysis but can be employed for this purpose with a simple modification to the processing stream. The pre-processed structural scans for all scanning sessions for a given subject were first rigidly aligned to the first scan using FLIRT and a mean image was created. The analysis was run a second time using the halfway point between scans 1 and 2 as the baseline (see above). In both cases, the default settings for FLIRT's 3D rigid body alignment were used (6 DOF) which includes a trilinear interpolation algorithm. The average images were brain-extracted using BET (Smith 2002). Next, tissue-type segmentation was carried out on the subject mean image using FAST (Zhang et al., 2001). The resulting gray-matter partial volume images were then aligned to MNI152 standard space using the affine registration of the IRTK (Rueckert et al., 1999, www.doc.ic.ac.uk/∼dr/software). The resulting images were averaged to create a study-specific template. The brain-extraction and segmentation steps were then repeated on the rigidly aligned structural scans from each scanning session. The segmented native gray matter images were then non-linearly registered to the template using the transformations calculated from the averaged images. The segmented images were then smoothed with an isotropic Gaussian kernel with a sigma of 3.5 mm (analogous to an 8 mm FWHM). Paired T-tests were used to compare scan sessions one and two (the control period) and two and three (the training period). Permutation-based, non-parametric testing was used (Randomise, http://www.fmrib.ox.ac.uk/fsl/randomise/, version 2.0 and 2.1, see Results), with a height threshold of p<0.001 and testing clusters for significance at p<0.05, corrected for multiple comparisons across space. A 0.35 intensity threshold mask was chosen to create a mask similar in size and shape to the SPM2 mask and a 1 mm FWHM variance smoothing kernel was used. See Supplemental Fig. 2 for a flowchart of steps.
SPM5
The SPM5 analysis was performed using the VBM5 toolbox (v1.15, http://dbm.neuro.uni-jena.de/vbm/). The unified segmentation algorithm in SPM5 warps template images into the space of the image to be segmented so no custom template is needed. As in the SPM2 and FSL analyses, spatial normalization was estimated using either the baseline scan or a subject mean image aligned to the halfway point between scans 1 and 2. This nonlinear spatial normalization was then applied to the segmented grey matter images. Defaults were used for all toolbox options. SnPM5 was used for permutation-based non-parametric testing forming clusters at p<0.001 and testing clusters for significance at p<0.05, corrected for multiple comparisons across space. An absolute intensity threshold mask of 0.2 and a 1 mm FWHM variance smoothing kernel were used. All results reported are corrected on the cluster level. See Supplemental Fig. 3 for a flowchart of steps.
Results
Behavior
Performance on the tracking task was measured by the average distance of the joystick cursor from the randomly moving dot during the six-minute scans. For the mirror-tracking task, a paired T-test comparing average distance before and after training showed a significant decrease (p<0.0001). For the normal tracking task there was no significant difference in cursor distance before and after training (p = 0.52, Fig. 2).
Fig. 2.
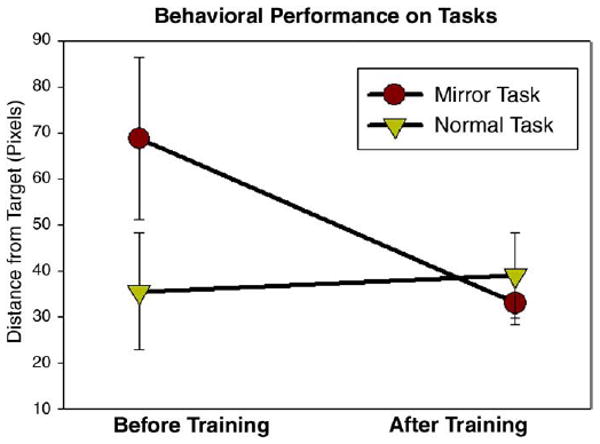
All subjects significantly improved on the mirror-tracking task as measured by average distance of the joystick cursor from the tracking dot (paired T-test, p<0.0001). For the normal tracking task there was no significant difference in cursor distance before and after training (p = 0.52).
Functional results (fMRI)
Comparing functional scans associated with performance of the mirror-tracking task before and after training showed several clusters of increased and decreased activation (Fig. 3 and Table 1). The areas showing decreased activation included the middle frontal gyrus and a large expanse of parietal cortex extending from precuneus to lateral parietal cortex. These regions have been implicated in visual tracking tasks and mental rotation, respectively (Luna et al., 1998; Cohen et al., 1996). Increased activity was seen in the medial frontal cortex and cingulate cortex. These areas fall within the “default” or “resting state” network. Regions in this network commonly show decreased activation during difficult tasks and a relative increase in activation or no change during rest or simple tasks (Supplementary Fig. 4; Raichle et al., 2001). This is consistent with our results as these regions showed greater decreases in activation relative to baseline before training than after.
Fig. 3.
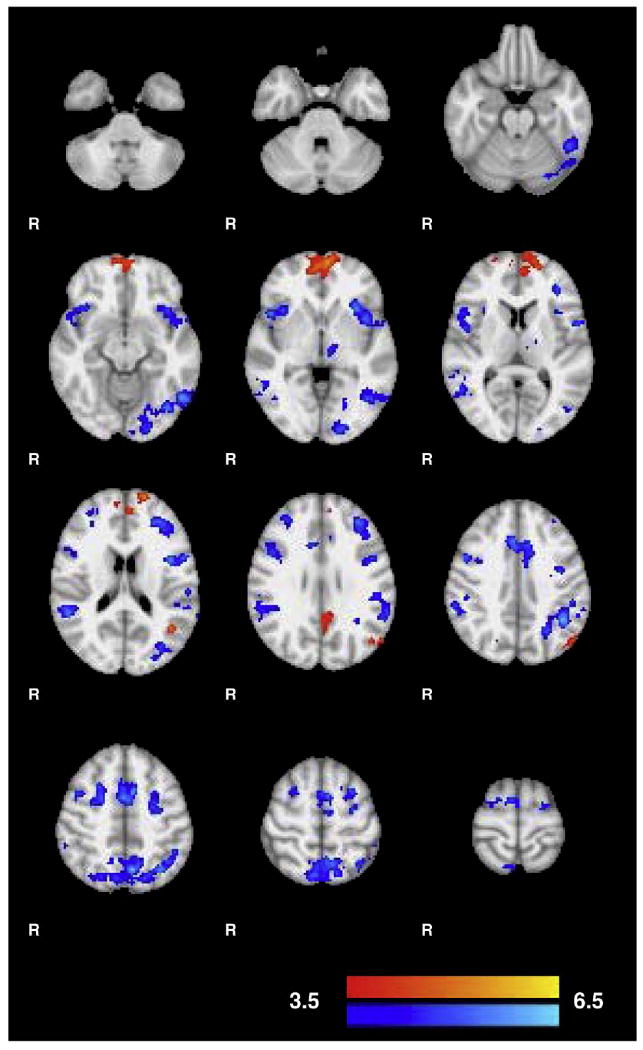
Clusters of increased and decreased activation after training on the mirror-tracking task. Z-stats of increased activity are shown in red and decreased activity in blue. Areas of decreased activity include task specific regions such as the middle frontal gyrus and posterior parietal cortex. Increased functional activity occurred primarily in the medial frontal cortex which has been implicated in response inhibition and the resting state network. Clusters are overlaid on the MNI152. See Table 1 for a list of all clusters.
Table 1.
Clusters showing functional change after training.
| P-value | # Voxels | x | y | z | Area |
|---|---|---|---|---|---|
| Increases | |||||
| <0.0001 | 1291 | −2 | 60 | 2 | Medial frontal |
| <0.0001 | 226 | −48 | −69 | 31 | L. Sup. occiptal |
| 0.002 | 130 | −4 | −52 | 29 | Cingulate gyrus |
| Decreases | |||||
| <0.0001 | 2987 | −6 | −64 | 50 | L. Precunuous/parietal |
| <0.0001 | 1698 | 39 | 10 | 25 | R. Inf. frontal gyrus |
| <0.0001 | 1335 | −35 | −73 | −11 | L. Occipital fusiform gyrus |
| <0.0001 | 1237 | −42 | 4 | 20 | L. Precentral gyrus |
| <0.0001 | 1217 | −1 | 8 | 47 | Paracingulate gyrus |
| <0.0001 | 1004 | 54 | −44 | 20 | R. Supramarginal gyrus |
| <0.0001 | 672 | −35 | 36 | 22 | L. Middle frontal gyrus |
| <0.0001 | 604 | −28 | −4 | 51 | L. Middle frontal gyrus |
| <0.0001 | 244 | 32 | 41 | 25 | R. Frontal pole |
| <0.0001 | 231 | −33 | −80 | 19 | L. Sup. occipital |
| 0.002 | 124 | −10 | −17 | 0 | L. Thalamus |
Clusters showing a change in functional activity after 2 weeks of training on the mirror-tracking task. Voxel coordinates are reported in MNI space and area labels were determined using the Harvard–Oxford Structural Atlas in FSLview (Flitney et al., 2007).
Structural results (VBM)
In the first run of the SPM2 analysis in which we used the baseline scan as the source for spatial normalization, we found no significant increases or decreases in gray matter density during the control period. During the learning period, one cluster of increased gray matter density was found on the ventro-medial edge of primary visual cortex in the left hemisphere (Fig. 4A). Another small cluster of decreased grey matter was found in the right pre-central gyrus. In the second run of the SPM2 analysis in which we used the pre-training scan as the source for spatial normalization, a small cluster of increased grey matter density was found in the right medial frontal gyrus during the control period (Fig. 4B). During the learning period, one cluster of decreased grey matter density was found in the left cerebellum.
Fig. 4.
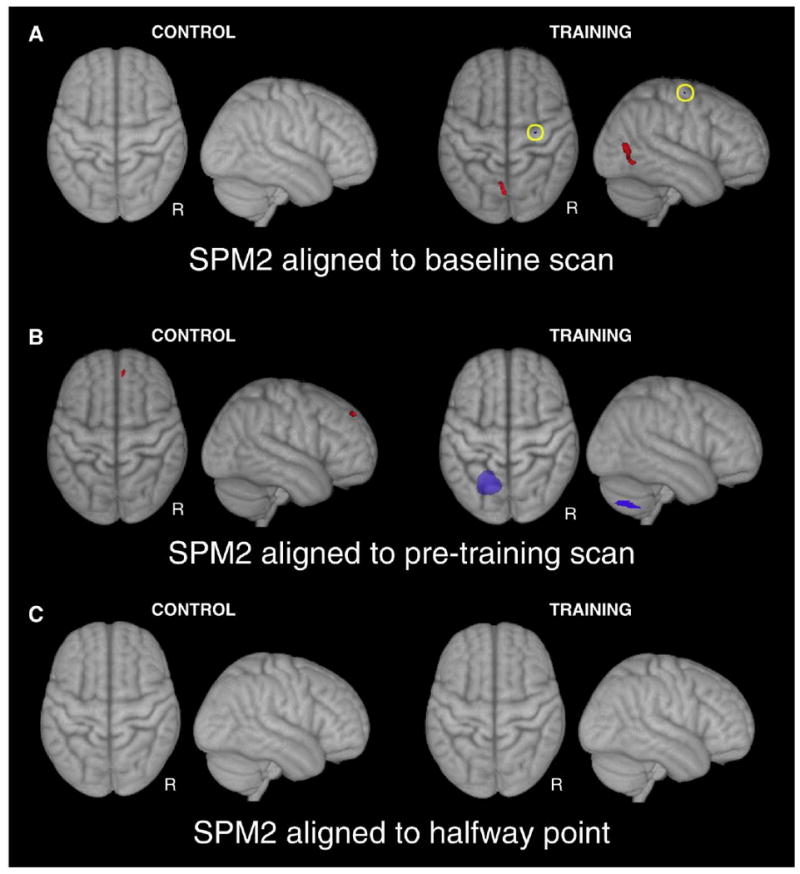
Maximum intensity projections of gray matter increases (red) and decreases (blue) found in the SPM2 VBM analyses. Background image is the MNI152 3D rendered in AFNI (Cox, 1996). Clusters of grey matter changes found with scans aligned to the baseline session (A) were not consistent with clusters found when scans were aligned to the pre-training session (B). (C) When scans were aligned to a halfway point between the two scans no structural changes were found. See Supplemental Table 1 for a list of areas and coordinates.
The variability of our results depending on which scan was used as the source of spatial normalization and the presence of a significant cluster in the control period led us to re-examine the analysis pipeline used here and in other published longitudinal VBM studies (see Discussion). We hypothesized that using any one of the scans as the target of the rigid alignment and the source of the spatial normalization could bias the segmentation results of that scan. We attempted to address this potential confound by aligning to the halfway point between scans 1 and 2 by taking the square root of the alignment transformation matrix. Using this method no significant clusters of grey matter change were found during the control or training period (Fig. 4C).
We also noted that the non-stationarity correction method of statistical inference has been demonstrated to be anticonservative for degrees of freedom less than 30 as is the case here (Hayasaka et al., 2004). Hayasaka et al. recommend the use of permutation-based methods for analyses with relatively small degrees of freedom. Therefore, we re-ran the analysis using SnPM3 for statistical inference. Using this method resulted in no significant clusters of grey matter change regardless of which alignment technique was used.
A very similar pattern of results was found in the FSL-VBM analysis. We began by using the initial scan as the target of the rigid alignment and the source of the spatial normalization. In this analysis we found three clusters of decreased gray matter density and two clusters of increased gray matter density during the control period (Fig. 5A). During the training period five clusters of decreased gray matter were found. Re-running the analysis using the halfway alignment technique resulted in no significant clusters of grey matter change during the control period and a single small cluster of decreased grey matter density on the inferior edge of right temporal cortex (Fig. 5B).
Fig. 5.
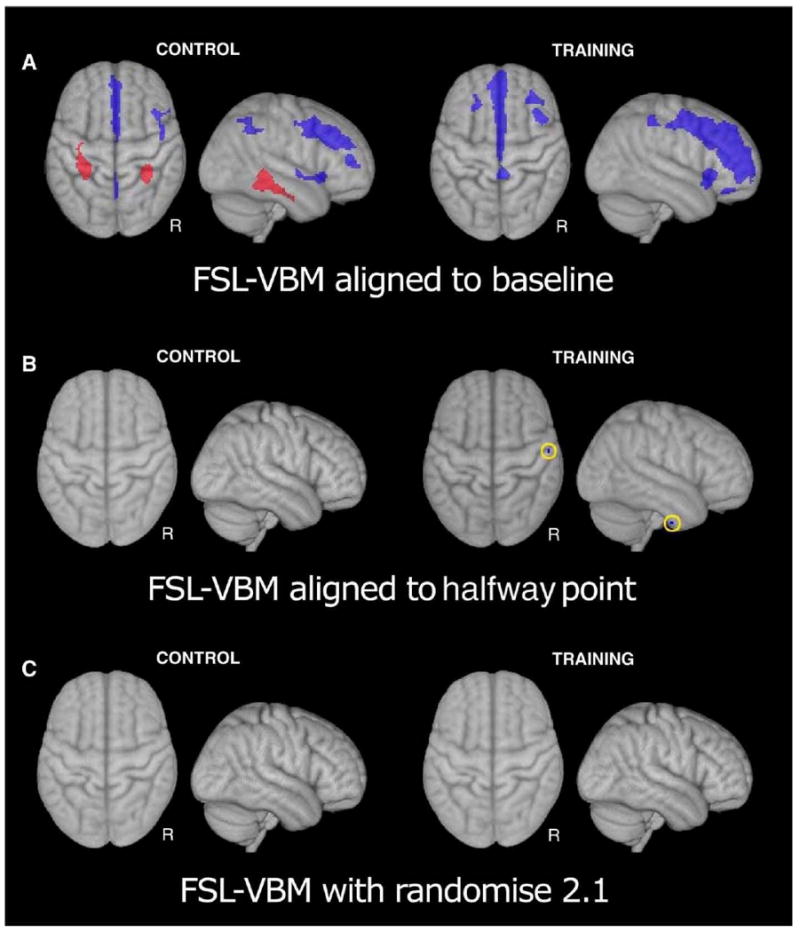
Maximum intensity projection of regions of gray matter change in the FSL-VBM analyses. Clusters of grey matter change found when scans were aligned to the first session (A) were not found when scans were aligned to a halfway point between scans 1 and 2 (B). C) Corrections to the FSL permutation procedure eliminated all significant clusters of grey matter change. See Supplemental Table 1 for a list of areas and coordinates.
Although all FSL-VBM analyses are conducted using permutation-based methods for statistical inference, FSL's “Randomise” program was recently updated to change the method of dealing with confounds in permutation. The method previously used was demonstrated to be very anticonservative (Nichols et al., 2008). Using this updated version of Randomise (2.1) no significant clusters were found in either condition (Fig. 5C).
In the SPM5 analysis no significant clusters of grey matter change were found in either condition regardless of whether scans were aligned to the initial scan or to a halfway point.
Discussion
In this study we have explored longitudinal structural changes as measured by VBM and some of the potential pitfalls in the analyses of these data. Our approach to this question is unique in that we have used the same pool of subjects for both the control and learning phase of the experiment. Contrary to previously published studies, we found no statistically significant grey matter changes associated with 2 weeks of training on a visuo-motor task even though significant changes were found in fMRI activation and behavioral performance. We have suggested modifications to the structural analysis stream used in previously published longitudinal VBM studies. We have also carried out the longitudinal analyses using three different software packages to evaluate the consistency of the structural results.
Initial analyses in both SPM2 and FSL revealed clusters in both learning and control conditions that were ultimately determined to be artifactual. These clusters were determined to be due to two factors. First by aligning all scan sessions to the initial baseline scan a difference in interpolation is introduced, biasing the comparison. Scans that were interpolated are slightly smoothed before segmentation while the baseline scan is not. This leads to artificial differences in apparent grey matter density. When scans were aligned to a halfway point between the first two scans no significant clusters were found. Secondly, the non-stationarity correction method of statistical inference used in SPM2 is anticonservative for analyses in which the degrees of freedom are less than thirty (Hayasaka et al., 2004). Performing the analysis using permutation-based methods in SPM2 resulted in no significant clusters of grey matter change. Also, version 2.0 of FSL's permutation tool, Randomise, used an anticonservative method of handling confounds (Nichols et al., 2008). The corrected method used in Randomise 2.1 results in no significant clusters of grey matter change.
Analysis of the change in BOLD signal with training revealed several regions of decreased activity with learning including inferior parietal, precuneus, middle cingulate cortex, and bilateral middle frontal gyrus as well as several regions of increased activity including the medial frontal cortex. The large cluster spanning parietal and precuneus has been implicated in tasks involving spatial transformations while the middle cingulate cortex has been associated with response inhibition (Cohen et al., 1996; Ridderinkhof et al., 2004). Training on the mirror-tracking task enhances both of these skills. The changes in the activation of the middle frontal cortical areas are possibly related to the motor transformations required for inverting the movement of the joystick. Although our results on fMRI changes related to learning are statistically strong, they are challenging to interpret due to potential changes in the subjects' strategy or changes in the relative difficulty of the task. Most fMRI studies of learning suffer from similar difficulties (Poldrack, 2000).
The central goal of this study was to determine if functional and structural measures of plasticity overlapped. We also sought to determine whether structural measures of plasticity were consistent across VBM implementations. Ultimately no significant structural changes were found, however it is still important to note there are significant differences in currently available longitudinal VBM implementations which are illustrated in the processing flow charts in Supplementary Figs. 1–3. Note that SPM2 performs the segmentation step after the brain has been spatially normalized whereas SPM5 and FSL perform segmentation in the subject's native space. However we believe the most important source of variance is the segmentation itself. Fig. 6 shows the segmented gray matter images and cumulative image histograms from a single subject using each of the three packages. The SPM2 segmented image differs from the others in that a much higher proportion of voxels are classified as 100% gray matter density. The FSL segmentation classifies voxel more continuously between 0 and 100% grey matter. In the SPM5 segmentation, the distribution of grey matter density appears to lie somewhere between the sharp distinction of SPM2 and the more gradual curve of FSL. Note that SPM5 uses the “grand unified segmentation” algorithm that does not rely on study-specific templates (Ashburner and Friston, 2005).
Fig. 6.
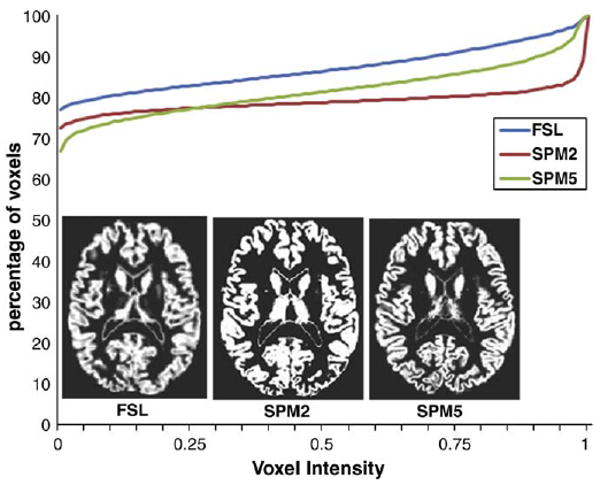
Comparison of a single subject's segmented gray matter images between FSL, SPM2 and SPM5. The line graph shows cumulative percentage of pixels as a function of voxel intensity. SPM2 produces a segmented image with a harder edge and many more voxels assigned to an intensity value of 1 (i.e. 100% gray matter). The FSL (FAST) segmentation produces a more uniform distribution of intensity values with softer edges. SPM5 falls between these two extremes. Virtually identical histograms were produced when the source images were rigidly aligned and interpolated.
Our results also demonstrated that interpolation related to the rigid alignment step had significant effects on the final results. Interestingly, the histograms in Fig. 6 change very little when they are generated using volumes that were rigidly aligned and interpolated. This illustrates the point that the interpolation does not globally bias the grey matter density in one direction or the other. Rather, at different locations within the volume, artifactual focal changes in grey matter may be introduced in an unpredictable way. The changes balance each other out when averaged over the whole volume, but when an interpolated volume is compared against one that is not interpolated, false positives may be detected.
Currently all published longitudinal VBM studies of which we are aware have used the SPM2 pipeline (Draganski et al., 2004, 2006; May et al., 2007; Boyke et al., 2008; Ilg et al., 2008; Driemeyer et al., 2008). In the analyses we performed we have made several small but significant changes from the processing stream of these studies. First, rather than aligning each scan to the initial scan, we align to a halfway point between the two scans being compared. Note that this point is significant regardless of whether subjects are used as their own controls or a separate group of controls is used. Second, we used non-parametric methods for all statistical testing and multiple comparison correction as the non-stationarity correction method has been demonstrated as anticonservative for analyses with relatively small degrees of freedom (Hayasaka et al., 2004). We have demonstrated that these changes have a significant effect on the ultimate results across different software packages. This provides an important demonstration of the point made by Ridgway et al. (2008) that very detailed explanations of VBM method are required for experiments to be replicable.
It should be emphasized that the animal literature leaves little doubt that it is possible for the mammalian brain to undergo large scale changes in its structure (cortical thickness, synaptic and capillary density, etc.) on a time scale of days to weeks (Klintsova and Greenough, 1999). Most of these studies have used histological techniques to measure these changes, though some imaging work has been conducted measuring changes in cortical vascular in rats in response to exercise (Pereira et al., 2007; Swain et al., 2003). Thus despite the methodological concerns we have raised here, previously published studies may have provided more favorable conditions for detecting grey matter changes. Several of these studies were conducted on larger groups of subjects and employed longer periods of training that may have resulted in more dramatic grey matter changes. Nonetheless the contrary findings reported here and the demonstrated susceptibility of longitudinal VBM to false positives warrants a more careful examination of these methods. The development of a standard and robust method of investigating within-subject structural brain change remains an important challenge.
In an effort to help produce such a standard, all of the raw data used in this experiment will be made available on the SUMS database at Washington University (Dickson et al., 2001; http://sumsdb.wustl.edu/sums/directory.do?id=6694686). We encourage other researchers to download these data and reanalyze them with novel methods. It is possible that multivariate analysis techniques may prove more sensitive than traditional univariate analysis. Some of these methods have already been employed on cross-sectional VBM data (Kloppel et al., 2008; Kawasaki et al., 2007). Future studies might employ focused, higher resolution scanning or more targeted pulse sequences to obtain a more detailed picture of underlying changes (Swain et al., 2003; van der Kouwe et al., 2008). We are confident that these techniques in combination with rigorous statistical controls will one day make in vivo measurement of human brain structure possible, opening up an entirely new technique in the study of learning and memory.
Supplementary Material
Acknowledgments
This work was supported by the NIMH Intramural Research Program. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). The authors would like to acknowledge comments and assistance from Rasmus Birn, Gang Chen, Dan Handwerker, David McMahon, Kevin Murphy, Allison Nugent, and Regina Nuzzo.
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2009.05.097.
References
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Why voxel-based morphometry should be used. NeuroImage. 2001;14:1238–1243. doi: 10.1006/nimg.2001.0961. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Jenkinson M, Smith SM. General multilevel linear modeling for group analysis in FMRI. NeuroImage. 2003;20:1052–1063. doi: 10.1016/S1053-8119(03)00435-X. [DOI] [PubMed] [Google Scholar]
- Black JE, Isaacs KR, Anderson BJ, Alcantara AA, Greenough WT. Learning causes synaptogenesis, whereas motor activity causes angiogenesis, in cerebellar cortex of adult rats. Proc Natl Acad Sci U S A. 1990;87:5568–5572. doi: 10.1073/pnas.87.14.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein FL. “Voxel-based morphometry” should not be used with imperfectly registered images. NeuroImage. 2001;14:1454–1462. doi: 10.1006/nimg.2001.0770. [DOI] [PubMed] [Google Scholar]
- Boyke J, Driemeyer J, Gaser C, Buechel C, May A. Training-induced brain structure changes in the elderly. J Neurosci. 2008;28:7031–7035. doi: 10.1523/JNEUROSCI.0742-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Kosslyn SM, Breiter HC, DiGirolamo GJ, Thompson WL, Anderson AK, Bookheimer SY, Rosen BR, Belliveau JW. Changes in cortical activity during mental rotation — a mapping study using functional MRI. Brain. 1996;119:89–100. doi: 10.1093/brain/119.1.89. [DOI] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Crum WR, Griffin LD, Hill DL, Hawkes DJ. Zen and the art of medical image registration: correspondence, homology, and quality. NeuroImage. 2003;20:1425–1437. doi: 10.1016/j.neuroimage.2003.07.014. [DOI] [PubMed] [Google Scholar]
- Davatzikos C. Why voxel-based morphometric analysis should be used with great caution when characterizing group differences. NeuroImage. 2004;23:17–20. doi: 10.1016/j.neuroimage.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Dickson J, Drury H, Van Essen DC. ‘The surface management system’ (SuMS) database: a surface-based database to aid cortical surface reconstruction, visualization and analysis. Philos Trans R Soc Lond, B Biol Sci. 2001;356:1277–1292. doi: 10.1098/rstb.2001.0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draganski B, Gaser C, Busch V, Schuierer G, Bogdahn U, May A. Neuroplasticity: changes in grey matter induced by training — newly honed juggling skills show up as a transient feature on a brain-imaging scan. Nature. 2004;427:311–312. doi: 10.1038/427311a. [DOI] [PubMed] [Google Scholar]
- Draganski B, Gaser C, Kempermann G, Kuhn HG, Winkler J, Buchel C, May A. Temporal and spatial dynamics of brain structure changes during extensive learning. J Neurosci. 2006;26:6314–6317. doi: 10.1523/JNEUROSCI.4628-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driemeyer J, Boyke J, Gaser C, Buchel C, May A. Changes in gray matter induced by learning—revisited. PLoS ONE. 2008;3:e2669. doi: 10.1371/journal.pone.0002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flitney D, Webster M, Patenaude B, Seidman L, Goldstein J, Tordesillas Gutierrez D, Eickhoff S, Amunts K, Zilles K, Lancaster J, Haselgrove C, Kennedy D, Jenkinson M, Smith S. Anatomical brain atlases and their application in the FSLView visualisation tool. Thirteenth Annual Meeting of the Organization for Human Brain Mapping; Toronto. 2007. [Google Scholar]
- Floyer-Lea A, Wylezinska M, Kincses T, Matthews PM. Rapid modulation of GABA concentration in human sensorimotor cortex during motor learning. J Neurophysiol. 2006;95:1639–1644. doi: 10.1152/jn.00346.2005. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. NeuroImage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. NeuroImage. 2004;22:676–687. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Ilg R, Wohlschlager AM, Gaser C, Liebau Y, Dauner R, Woller A, Zimmer C, Zihl J, Muhlau M. Gray matter increase induced by practice correlates with task-specific activation: a combined functional and morphometric magnetic resonance imaging study. J Neurosci. 2008;28:4210–4215. doi: 10.1523/JNEUROSCI.5722-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Symms MR, Cercignani M, Howard RJ. The effect of filter size on VBM analyses of DT-MRI data. NeuroImage. 2005;26:546–554. doi: 10.1016/j.neuroimage.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Jones DK, Chitnis XA, Job D, Khong PL, Leung LT, Marenco S, Smith SM, Symms MR. What Happens When Nine Different Groups Analyze the Same DT-MRI Data Set Using Voxel-Based Methods?. Proceeds of ISMRM 15th Annual Meeting; Berlin. 2007. p. 1. [Google Scholar]
- Kawasaki Y, Suzuki M, Kherif F, Takahashi T, Zhou SY, Nakamura K, Matsui M, Sumiyoshi T, Seto H, Kurachi M. Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. NeuroImage. 2007;34:235–242. doi: 10.1016/j.neuroimage.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Kleim JA, Markham JA, Vij K, Freese JL, Ballard DH, Greenough WT. Motor learning induces astrocytic hypertrophy in the cerebellar cortex. Behav Brain Res. 2007;178:244–249. doi: 10.1016/j.bbr.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klintsova AY, Greenough WT. Synaptic plasticity in cortical systems. Curr Opin Neurobiol. 1999;9:203–208. doi: 10.1016/s0959-4388(99)80028-2. [DOI] [PubMed] [Google Scholar]
- Kloppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Jr, Ashburner J, Frackowiak RS. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krech D, Rosenzweig MR, Bennett EL. Effects of environmental complexity and training on brain chemistry. J Comp Physiol Psychol. 1960;53:509–519. doi: 10.1037/h0045402. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N, Simmons WK, Bellgowan PS, Baker CI. Circular analysis in systems neuroscience: the dangers of double dipping. Nat Neurosci. 2009;12:535–540. doi: 10.1038/nn.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna B, Thulborn KR, Strojwas MH, McCurtain BJ, Berman RA, Genovese CR, Sweeney JA. Dorsal cortical regions subserving visually guided saccades in humans: an fMRI study. Cereb Cortex. 1998;8:40–47. doi: 10.1093/cercor/8.1.40. [DOI] [PubMed] [Google Scholar]
- May A, Hajak G, Ganssbauer S, Steffens T, Langguth B, Kleinjung T, Eichhammer P. Structural brain alterations following 5 days of intervention: dynamic aspects of neuroplasticity. Cereb Cortex. 2007;17:205–210. doi: 10.1093/cercor/bhj138. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TE, Ridgway GR, Webster M, Smith S. GLM Permutation — Nonparametric Inference for Arbitrary General Linear Models. 14th Annual Human Brain Mapping Meeting; Melbourne, Australia. 2008. [Google Scholar]
- Pereira AC, Huddleston DE, Brickman AM, Sosunov AA, Hen R, McKhann GM, Sloan R, Gage FH, Brown TR, Small SA. An in vivo correlate of exercise-induced neurogenesis in the adult dentate gyrus. Proc Natl Acad Sci U S A. 2007;104:5638–5643. doi: 10.1073/pnas.0611721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA. Imaging brain plasticity: conceptual and methodological issues—a theoretical review. NeuroImage. 2000;12:1–13. doi: 10.1006/nimg.2000.0596. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridderinkhof KR, Ullsperger M, Crone EA, Nieuwenhuiss S. The role of the medial frontal cortex in cognitive control. Science. 2004;306:443–447. doi: 10.1126/science.1100301. [DOI] [PubMed] [Google Scholar]
- Ridgway GR, Henley SMD, Rohrer JD, Scahill RI, Warren JD, Fox NC. Ten simple rules for reporting voxel-based morphometry studies. NeuroImage. 2008;40:1429–1435. doi: 10.1016/j.neuroimage.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Rosenzweig MR, Bennett EL, Diamond MC. Chemical and anatomical plasticity of brain: replications and extensions. Macromol Behav. 1972:205–277. [Google Scholar]
- Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. Medical Imaging, IEEE Transactions on. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. Medical Imaging, IEEE Transactions on. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Zhang YY, Jenkinson M, Chen J, Matthews PM, Federico A, De Stefano N. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. NeuroImage. 2002;17:479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- Swain RA, Harris AB, Wiener EC, Dutka MV, Morris HD, Theien BE, Konda S, Engberg K, Lauterbur PC, Greenough WT. Prolonged exercise induces angiogenesis and increases cerebral blood volume in primary motor cortex of the rat. Neuroscience. 2003;117:1037–1046. doi: 10.1016/s0306-4522(02)00664-4. [DOI] [PubMed] [Google Scholar]
- van der Kouwe AJW, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. Neuroimage. 2008;40:559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, Gage FH. Neural consequences of environmental enrichment. Nat Rev, Neurosci. 2000;1:191–198. doi: 10.1038/35044558. [DOI] [PubMed] [Google Scholar]
- Woolrich MW, Behrens TE, Beckmann CF, Jenkinson M, Smith SM. Multilevel linear modelling for FMRI group analysis using Bayesian inference. NeuroImage. 2004;21:1732–1747. doi: 10.1016/j.neuroimage.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Evans AC, Marrett S, Neelin P. A three-dimensional statistical analysis for CBF activation studies in human brain. J Cereb Blood Flow Metab. 1992;12:900–918. doi: 10.1038/jcbfm.1992.127. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Andermann M, Koulis T, MacDonald D, Evans AC. Detecting changes in nonisotropic images. Hum Brain Mapp. 1999;8:98–101. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<98::AID-HBM5>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation–maximization algorithm. IEEE Trans Med Imag. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


