Abstract
Ewing's family tumors are characterized by a specific t(11;22) chromosomal translocation that results in the formation of EWS-Fli1 oncogenic fusion protein. To investigate the effects of EWS-Fli1 on gene expression, we carried out DNA microarray analysis after specific knockdown of EWS-Fli1 through transfection of synthetic siRNAs. EWS-Fli1 knockdown increased expression of genes such as DKK1 and p57 that are known to be repressed by EWS-Fli1 fusion protein. Among other potential EWS-Fli1 targets identified by our microarray analysis, we have focused on the FOXO1 gene since it encodes a potential tumor suppressor and has not been previously reported in Ewing’s cells. To better understand how EWS-Fli1 affects FOXO1 expression, we have established a doxycycline-inducible siRNA system to achieve stable and reversible knockdown of EWS-Fli1 in Ewing’s sarcoma cells. Here we show that FOXO1 expression in Ewing’s cells has an inverse relationship with EWS-Fli1 protein level, and FOXO1 promoter activity is increased after doxycycline-induced EWS-Fli1 knockdown. In addition, we have found that direct binding of EWS-Fli1 to FOXO1 promoter is attenuated after doxycycline-induced siRNA knockdown of the fusion protein. Together, these results suggest that suppression of FOXO1 function by EWS-Fli1 fusion protein may contribute to cellular transformation in Ewing’s family tumors.
INTRODUCTION
Ewing's family tumors are malignancies that share histological features as well as a recurrent and specific t(11;22) chromosomal translocation [1]. This translocation results in a chimeric transcript encoding the N-terminal domain of the RNA-binding protein EWS and the DNA-binding domain of the ETS family transcription factor Fli1 [2]. The resultant EWS-Fli1 chimeric fusion protein is known to affect both gene transcription and RNA splicing [3, 4].
EWS-Fli11 has been proposed to be an oncogenic fusion protein based on its ability to transform cells. While a number of EWS-Fli1 target genes have been identified by ectopic expression of EWS-Fli1 in non-Ewing’s cells [5–8], several studies have indicated that EWS-FLi1 target genes identified in non-Ewing’s cells do not overlap with those in Ewing’s cells [9, 10]. To study how EWS-Fli1 influences gene expression in the genetic background of Ewing’s sarcoma, we and others have examined the effects of EWS-Fli1 knockdown by siRNA in actual Ewing’s cell lines [4, 11, 12]. We previously found that knockdown of EWS-Fli1 in Ewing’s cells leads to growth arrest and reduced invasiveness [4]. More recently we have shown that EWS-Fli1 abolishes cellular senescence in Ewing’s sarcoma cells by suppressing the functions of retinoblastoma protein [13].
To gain further insight into the oncogenic mechanisms of EWS-Fli1 fusion protein, in this manuscript we carried out microarray analysis of RNA samples from Ewing’s sarcoma cells transfected with synthetic siRNAs against EWS-Fli1. Analysis of our microarray data revealed that EWS-Fli1 affects many genes including repression of the FOXO1 gene in Ewing’s sarcoma cells. While synthetic siRNAs represent a convenient approach in the knockdown of EWS-Fli1, they also have severe limitations such as a relatively short duration of robust siRNA knockdown and a need to transfect fresh cells for each experiment. To achieve sustained and reversible siRNA knockdown of EWS-Fli1, we utilized the pSLIK (single lentivector for inducible knockdown) platform in Ewing’s sarcoma cells to conditionally turn on production of siRNA against EWS-Fli1. Using this inducible siRNA system in Ewing’s sarcoma cells, we have found that doxycycline-induced siRNA knockdown of EWS-Fli1 in Ewing’s sarcoma cells is accompanied by an increase in FOXO1 expression and a decrease in proliferation. Through chromatin immunoprecipitation (CHIP) assay, we showed that EWS-Fli1 binds directly to the promoter region of FOXO1. Taken together, these findings suggest that FOXO1 is a downstream target of EWS-Fli1, and that the tumor suppressor activity of FOXO1 is likely silenced by the oncogenic EWS-Fli1 fusion protein during tumorigenesis.
MATERIALS AND METHODS
Cell Culture
Human Ewing’s sarcoma cell lines A673, SK-ES and RD-ES were obtained from ATCC and maintained in RPMI, McCoy’s 5A and Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal bovine serum, respectively.
siRNA transfection and DNA microarray analysis
2 × 106 SK-ES cells in 0.1 ml PBS were electroporated with 4 µl of siRNA and plated onto 4 wells in a collagen-coated 6-well plate. 48 hrs later, total RNA was prepared for DNA microarray at the University of Washington Center for Expression Array. Target labeling and hybridization to Affymetrix Gene Chips (Human Genome U133 plus 2.0) were carried out with minor modifications from procedures recommended by the manufacturer. Probe sets with a “call” of (P) and a signal log ratio (SLR) of −1.0 (i.e. 2 fold decrease) and lower or a SLR of +1.0 (i.e. 2 fold increase) and higher were selected to obtain gene annotations via Affymetrix’s NetAffx Analysis Center (http://www.affymetrix.com/analysis/index.affx).
Lentivirus-mediated siRNA knockdown
Lentiviral siRNA constructs were obtained by annealing the pre-designed primers targeting EWS-Fli1 AGTACCCTTCTGACATCTCCT, firefly luficerase ACGTACGCGGAATACTTCGAA, or a scrambled sequence GGAAACATACTGTTACAAGAA, and subsequent cloning into the pEN_TGmiRC3 shuttle plasmid for transfer into the pSLIK-Neo lentiviral vector [14]. For generation of lentivirus, subconfluent 293FT packaging cells in a 10 cm plate were transfected with 3 µg of pSLIK-Neo-siRNA construct plus 9 µg ViraPower DNA Mixture (Invitrogen) by the Lipofectamine 2000 method. After 48 h, supernatants containing lentivirus were collected for infection of Ewing cells. Infected cells were then selected by their resistance to neomycin (G418). Induction of GFP and siRNA against EWS-Fli1 from the lenttiviral vector is controlled by addition of doxycycline to 3 µg/ml in the culture medium.
RT-PCR
Total RNA was extracted from 5 × 105 cells using the RNeasy Mini Kit (Qiagen, Valencia, CA) and eluted in 40 µl H2O. For RT-PCR, 2 µl RNA was analyzed in a total volume of 25 µl using the SuperScript One-Step RT-PCR Kit (Invitrogen). The primers for EWS-Fli1 were ACCGAGCAGCTATGGACA (EWS-U3) and CTTCAGCTAGAAGGCCACTGA (Fli-D1), the primers for FOXO1 were GCAGATCTACGAGTGGATGGTC (FOXO1-F) and AAACTGTGATCCAGGGCTGTC (FOXO1-R), and the primers for GAPDH were TGGTATCGTGGAAGGACTCAT and GTGGGTGTCGCTGTTGAAGTC.
Western blot analysis
To examine the effect of siRNA on protein expression, 2 × 106 cells were lysed with 0.1 ml Buffer X (50mM Tris, pH 7.4, 270mM NaCl, 0.5% Triton X-100) supplemented with protease and phosphatase inhibitors (Sigma-Aldrich Co). After separation on an 8% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane, the blot was incubated with a rabbit polyclonal anti-Fli1 or a rabbit anti-FOXO1 (clone C-19 for Fli1 and clone H-129 for FOXO1, Santa Cruz Biotechnology, Santa Cruz, CA), followed by horse radish peroxidase-conjugated goat anti-rabbit IgG. The protein bands were visualized using the ECL Plus Western Blotting Detection Reagents (Amersham Biosciences Corp., Piscataway, NJ).
Reporter plasmid construction and luciferase assay
Firefly luciferase reporter pGL3-hFOXO1-952 and pGL3-hFOXO1-286 were generated by cloning the PCR-amplified human FOXO1 promoter fragments into the Mlu I-Hind III sites of the pGL3-basic vector. G418-resistant Ewing’s sarcoma A673 cells in a 3.5-cm well were transfected with 2 µg pGL3-hFOXO1 luciferase reporter and 50 ng pRL-SV40 Renilla luciferase control using FuGENE 6 (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. 24 hrs after transfection, doxycycline was added to half of the wells for induction of siRNA, and cells were lysed after 3 days and assayed using the Promega dual-luciferase assay system. Transfection was repeated at least three times, and the luciferase activity was normalized to the internal Renilla luciferase control.
Chromatin immunoprecipitation assay
To cross-link DNA and protein, 1 × 108 of A673 cells were treated with 1% formaldehyde for 5 min at room temperature. Chromatin solution was then prepared as described previously [15]. For immunoprecipitation, 10 µl of antibody was incubated with chromatin solutions overnight at 4 °C on a rotating wheel. The immunocomplexes were collected with salmon sperm DNA/Protein A-Sepharose beads and washed extensively. Formaldehyde cross-linking was reversed by overnight incubation at 65 °C in 0.2 M NaCl plus 200 µg/ml proteinase K (Sigma). The mixture was then extracted with phenol/chloroform, and the DNA was precipitated with ethanol and resuspended in 20 µl of H2O. 20 ng of the DNA template was then used for PCR amplification of FOX1A and GAPDH promoter regions. Primers for FOX1A promoter were GGAAGAGGTTCCCACGGAGGGCAT and CCGGCGACACTTTGTTTA CT. Primers for GAPDH promoter were TCCTCCTGTTTCATCCAAGC and TAGTAGCCGGGCCCTACTTT.
RESULTS
Transient knockdown of EWS-Fli1 by synthetic siRNA affects gene expression in Ewing’s sarcoma cells
In earlier studies, we and others have found that genes affected by EWS-Fli1 and its related fusion proteins vary greatly in cells that were not derived from the tumors [9, 10]. Therefore, bona fide EWS-Fli1 target genes must be identified in the genetic background of Ewing’s sarcoma cells but not in surrogate cells such as NIH3T3. To this end, we were able to achieve a specific siRNA knockdown of EWS-Fli1 in SK-ES Ewing’s sarcoma cells, whereas a non-specific siRNA against the firefly luciferase gene had no effect on EWS-Fli1 expression (Fig. 1A). To efficiently identify EWS-Fli1 target genes in Ewing’s cells, we carried out DNA microarray analysis of RNA samples isolated from SK-ES cells transfected with a specific siRNA against EWS-Fli1 and a non-specific siRNA against luciferase. After comparison of the microarray data, we identified a set of genes that are significantly affected by EWS-Fli1 knockdown in these transfected Ewing’s sarcoma cells (Fig. 1B).
Fig. 1. siRNA knockdown of EWS-Fli1 affects gene expression in Ewing cells.
(A) SK-ES cells were transfected with a synthetic siRNA against EWS-Fli1 or a control non-specific siRNA against firefly luciferase (lanes 1–4). 3 days after transfection, cells lystes were blotted with a rabbit polyclonal anti-Fli1 antibody or a mouse monoclonal anti-β-actin. (B) After transfection with a synthetic siRNA against EWS-Fli1, gene expression in these SK-ES cells was analyzed by DNA microarray (in duplicates) and normalized to SK-ES cells transfected with a control siRNA against firefly luciferase. Only genes significantly affected by EWS-Fli1 knockdown are listed.
Among these genes, Dickkopf-1 (DKK1) is known to be the most up-regulated gene after siRNA knockdown in an Ewing cell line [12]. DKK1 is a secreted protein modulator of Wnt/β–catenin signaling. In addition to DKK1, other known EWS-Fli1 target genes from our list include platelet-derived growth factor receptor [16], cyclin-dependent kinase inhibitor p57 [17], and insulin-like growth factor binding proteins [12].
One of the genes in our list that were found to be up-regulated by EWS-Fli1 knockdown but have not been reported previously is FOXO1, a member of the Forkhead box family of transcription factors. FOXO1 has been known to function as a tumor suppressor [18], and we speculate that direct repression of this tumor suppressor gene by EWS-Fli1 may be involved in the pathogenesis of Ewing’s sarcoma.
Conditional siRNA knockdown of EWS-Fli1 can be induced by doxycycline in Ewing’s sarcoma cells
In our attempt to further carry out the characterization of FOXO1 as a potential EWS-Fli1 target gene in other Ewing’s cell lines, we realized that the use of synthetic siRNAs has severe limitations: knockdown by synthetic siRNA is transient in nature, and the knockdown is totally dependent on transfection efficiency. To study FOXO1 gene in other Ewing cell lines that are difficult to transfect, we have to use an alternative approach to deliver siRNA into the cells.
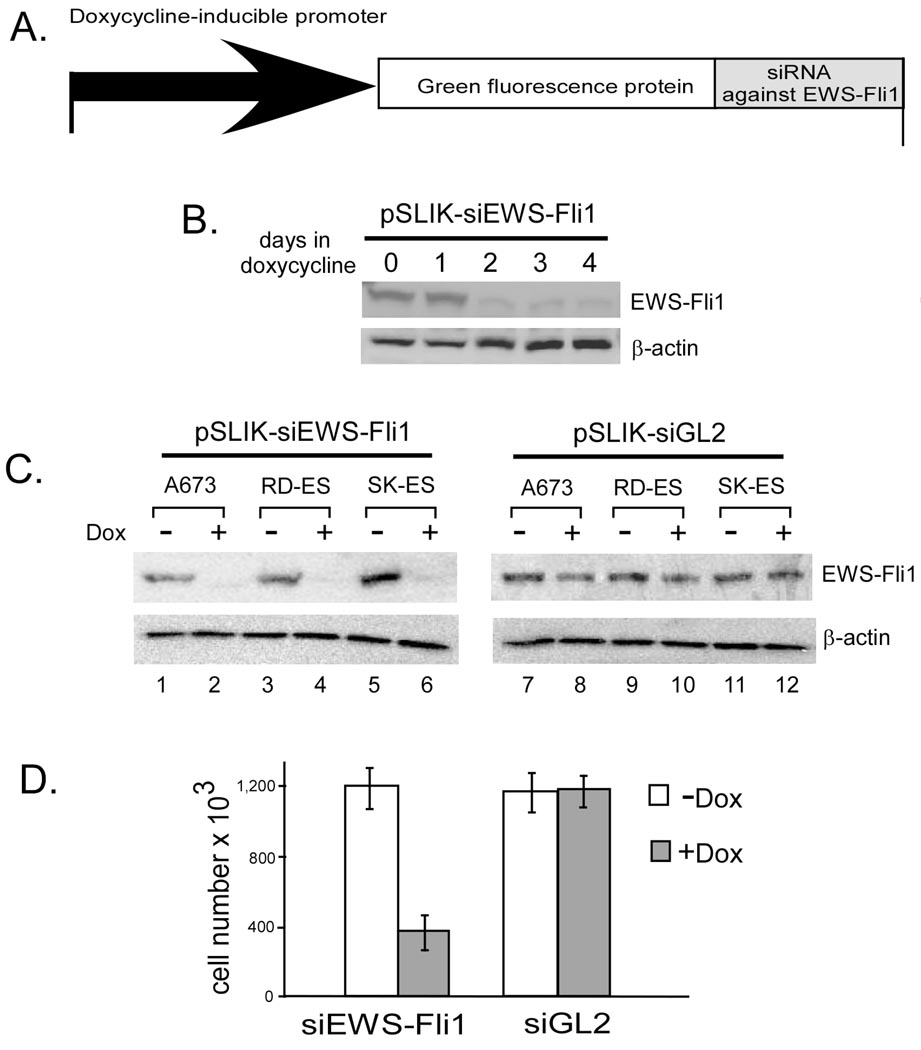
To achieve stable and efficient siRNA knockdown of EWS-Fli1 in various Ewing’s sarcoma cells, we engineered an siRNA construct based on the pSLIK-Neo lentiviral vector [14] for conditional expression of siRNA against EWS-Fli1. In experiments with the pSLIK-Neo-siEWS-Fli1 lentiviral vector, infected A673 Ewing’s cells were selected by their resistance to G418. Expression of GFP and siRNA against EWS-Fli1 from this vector is controlled by a doxycycline inducible promoter (Fig. 2A). In the absence of doxycycline, these G418-resistant cells do not express GFP or siRNA, and EWS-Fli1 level in Ewing’s sarcoma cells remained constant. When doxycycline was added to the culture medium, cells harboring the pSLIK-Neo-siEWS-Fli1 lentiviral construct express GFP (green under fluorescence microscope) and EWS-Fli1 protein level was significantly down-regulated by siRNA after doxycycline addition (Fig. 2B). In the absence of doxycycline, these cells could be easily expanded without a significant loss of their ability to silence EWS-Fli1 upon doxycycline addition. This pSLIK vector system has wide applicability since we were able to obtain efficient siRNA knockdown of EWS-Fli1 in other Ewing cell lines such as RD-ES and SK-ES cells (Fig. 2C, lanes 1–6). To confirm that doxycycline-induced siRNA knockdown of EWS-Fli1 is achieved by specific expression of siRNA against EWS-Fli1, we also constructed a control lentiviral vector pSLIK-Neo-siGL2 that expresses siRNA against the firefly luciferase gene not found in human cells. Doxycycline induction of this non-specific siRNA did not have an effect on EWS-Fli1 protein in any of the Ewing cell lines tested (Fig. 2C, lanes 7–12). Doxycycline-induced specific siRNA knockdown of EWS-Fli1 also inhibited proliferation of Ewing’s cells, whereas doxycyclin-induction of siRNA against luciferase did not have such an inhibitory effect (Fig. 2D).
Fig. 2. Conditional siRNA knockdown of EWS-Fli1 in Ewing’s cells.
(A) Diagram for the pSLIK-Neo-siEWS-Fli1 lentiviral siRNA vector. The doxycycline-inducible promoter is shown as an arrow, and miRNA cassette containing an shRNA against EWS-Fli1 is cloned downstream of the GFP gene. (B) A673 cells harboring the pSLIK-siEWS-Fli1 vector were treated with doxycycline and lysates were blotted with an anti-Fli1 antibody for detection of EWS-Fli1 protein. (C) Three Ewing cell lines expressing siRNA against EWS-Fli1 or luciferase were treated with doxycycline for 4 days and blotted with anti-Fli1. (D) 0.1 × 106 A673 cells harboring siEWS-Fli1 or siGL2 were cultured in medium with or without doxycycline, and cell numbers were counted 4 days later.
EWS-Fli1 knockdown induces FOXO1 expression in Ewing cell lines
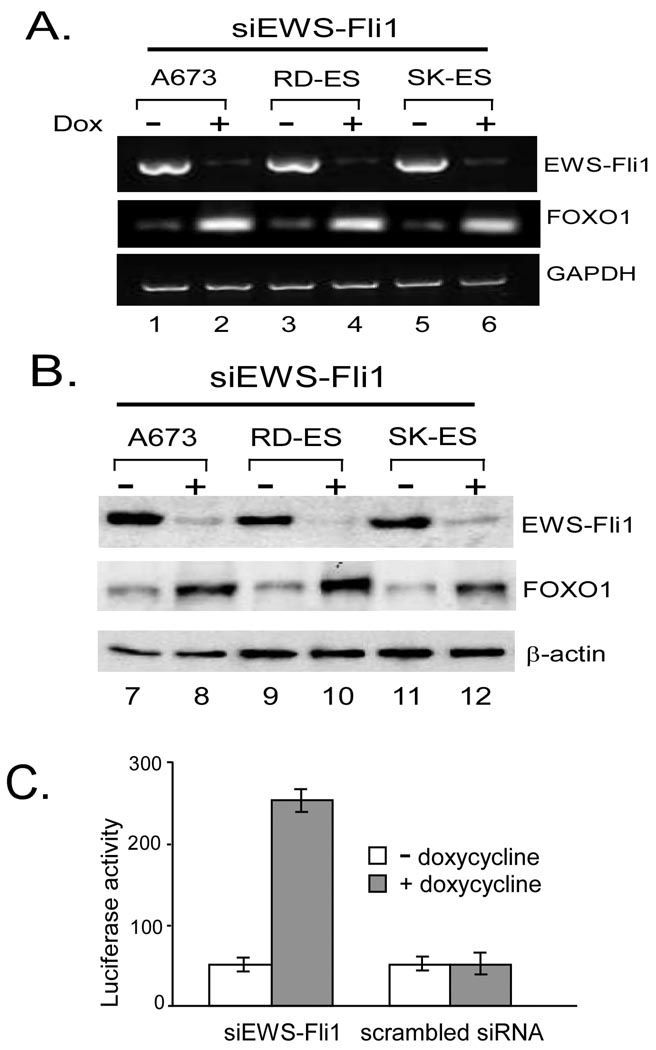
We focused our attention on FOXO1 since it is one of the genes that were shown to have an inverse relationship with EWS-Fli1 by our microarray experiment but have not been reported in the literature. FOXO1 is a member of the Forkhead box family of transcription factors that may function as a tumor repressor [18]. To confirm that the FOXO1 gene is indeed up-regulated after siRNA knockdown of EWS-Fli1, we carried out RT-PCR analysis of FOXO1 mRNA in three Ewing’s cell lines before and after doxycycline addition to the culture medium. As shown in Fig. 3A, doxycycline-induced siRNA knockdown of EWS-Fli1 in these Ewing cell lines led to an increase in FOXO1 mRNA. We then carried out western blotting and were able to show that this increase in FOXO1 mRNA was also reflected by an increase in FOXO1 protein (Fig. 3B). To confirm that the induced FOXO1 protein is functionally active, we transfected Ewing cells with the pFHRE-Luc reporter plasmid in which expression of the luciferase gene is driven by a FOXO1-responsive element. As shown in Fig. 3C, doxycycline induction of FOXO1 was associated with an increase in luciferase activity driven by the FOXO1-responsive element. This increase in luciferase activity was only observed in cells harboring specific siRNA against EWS-Fli1 but not in cells with a scrambled non-specific siRNA.
Fig. 3. Induction of FOXO1 by EWS-Fli1 knockdown.
(A) siRNA against EWS-Fli1 were turned on in three Ewing cell lines harboring the pSLIK-Neo-siEWS-Fli1 vector, and RNAs were collected for RT-PCR analysis. (B) Lysates from these three Ewing cell lines were blotted to show induction of FOXO1 protein after EWS-Fli1 knockdown. (C). pFHRE-Luc reporter was transfected into A673 cells harboring siEWS-Fli1 or a scrambled siRNA. The transfected cells were then cultured in medium with or without doxycycline for 4 days, and luciferase activities were measured and normalized to the pRL-SV40 controls.
EWS-Fli1 binds directly to the promoter region of the FOXO1 gene
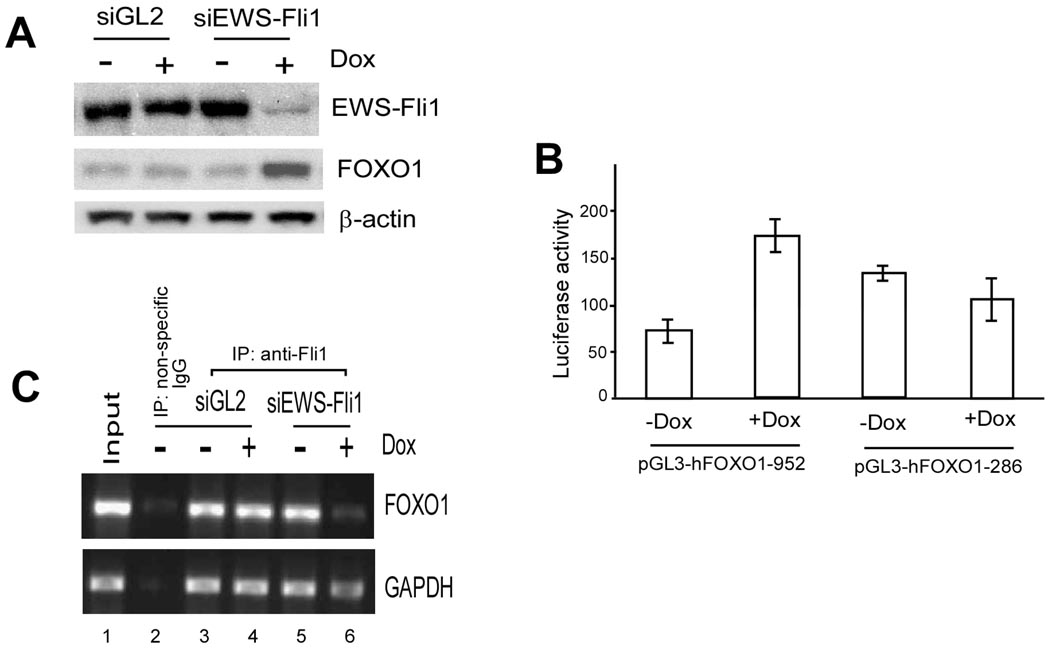
To further confirm that up-regulation of FOXO1 after EWS-Fli1 knockdown is a specific event related to changes in EWS-Fli1 but not an artifact due to the presence of doxycycline in the culture medium, we tested its expression in control A673 Ewing’s cells expressing an siRNA against luciferase. As shown in Fig. 4A, doxycycline addition to these control A673 cells did not have an effect on either EWS-Fli1 or FOXO1, while under the same conditions FOXO1 protein was increased in A673 cells expressing siRNA against EWS-Fli1.
Fig. 4. Direct binding of EWS-Fli1 to FOXO1 promoter.
(A) Western blotting was carried out using A673 cells after 3-day treatment with doxycycline to induce siRNA production. (B) The luciferase reporter plasmids were transfected into A673 cells harboring inducible siRNA against EWS-Fli1, treated with or without doxycycline for 3 days before luciferase assay. (C) CHIP assay and PCR were performed with fragmented chromatin from A673 cells following doxycycline-induced EWS-Fli1 knockdown.
To investigate whether EWS-Fli1 knockdown has an effect on FOXO1 transcription, we PCR amplified the human FOXO1 promoter and cloned the promoter fragment into the pGL3 luciferase reporter for transfection and luciferase assay. We found that doxycycline induction of EWS-Fli1 knockdown resulted in an increase in luciferase activity in A673 cells transfected with pGL3-hFOXO1-952 but not in A673 cells trasnfected with pGL3-hFOXO1-286, suggesting that EWS-Fli1 repression of the FOXO1 gene is mediated through an element between −952 to −286 of the FOXO1 promoter region (Fig. 4B). To show that EWS-Fli1 binds to this promoter region of the FOXO1 gene, we carried out chromatin immunoprecipitation (CHIP) experiments using an anti-Fli1 antibody. As shown in Fig. 4C, DNA fragments covering −952 to −286 region of the FOXO1 promoter was enriched by an anti-Fli1 antibody but not by the non-specific IgG (compare lanes 2 and 3). In addition, less DNA was recovered by the anti-Fli1 antibody from A673 cells with doxycycline-induced siRNA knockdown of EWS-Fli1 (compare lanes 5 and 6). Together, these findings demonstrate that the FOXO1 gene is indeed one of the target genes of EWS-Fli1 fusion protein in Ewing’s sarcoma cells.
DISCUSSION
In addition to synthetic siRNA, we previously also used an adenoviral vector to deliver siRNA and achieved near complete depletion of EWS-Fli1 for a sustained period of time in infected Ewing cells [13]. However, siRNA knockdown via adenoviral vector is irreversible and fresh infection has to be carried out for each new experiment. In this study, we have established a conditional lentiviral siRNA system to reversibly knockdown EWS-Fli1 in several Ewing cell lines. This doxycycline inducible siRNA system offers at least two advantages: first, those Ewing cell lines that are difficult to transfect can now be subjected to siRNA knockdown since the lentivirus will infect all Ewing cells, and the infected cells can be selected by their resistance to G418; second, siRNA knockdown of EWS-Fli1 in infected Ewing cells can now be turned on by addition of doxycycline to the culture medium and turned off by its withdrawal. Once G418-resistant Ewing cells are selected, they can be saved for all future experiments without the need of repeated infection.
In this study we have provided evidence that FOXO1 is a direct target of the EWS-Fli1 fusion protein in Ewing’s sarcoma cells. FOXO1 belongs to the forkhead O (FOXO) transcription factor subgroup based on the presence of a conserved DNA-binding forkhead box among its protein sequences. Genetic deletion of FOXO1 results in modest neoplastic phenotype in a lineage-restricted manner, suggesting its role as a potential tumor suppressor in certain cell types [18]. Loss of FOXO1 through chromosomal deletion was shown to promote androgen-independent prostate cancers [19]. Compared to normal endometrium, endometrioid endometrial cancer demonstrates a marked loss of FOXO1 expression [20]. In non-small cell lung cancer, FOXO1 expression is a favorable prognostic factor [21].
From our studies shown here and reported previously [13], EWS-Fli1 knockdown has been associated with activation of the S-phase repressor pRb, cell cycle arrest, decreased expression of cyclin D and increased expression of FOXO1. Since FOXO1 is known to down-regulate cyclin D at the transcriptional level [22], we speculate that tumorigenesis by EWS-Fli1 fusion protein may involve releasing the cyclin D promoter from repression by FOXO1 protein, thus providing an oversupply of cyclin D for partnering with CDK4 to inactivate the pRb protein.
Extensive studies have been carried out to show that the activities of FOXO1 protein are regulated by phosphorylation, acetylation, methylation and O-linked glycosylation [23]. FOXO1 protein can also associate with different cofactor complexes to regulate context-dependent programs of gene expression [24]. In comparison, not much is known about transcriptional regulation of the FOXO1 gene. It was reported that the transcription factor E2F-1 induces FOXO1 expression via binding to the E2F-1 sites located within 150 bp of the transcription initiation site of the FOXO1 gene [25]. FOXO1 can also be up-regulated by other forkhead transcription factors such as FOXC1 and FOXO3 through binding to the proximal promoter sequence [26, 27]. Since there has been no report on negative regulators of FOXO1 transcription, our study thus provides first evidence that the EWS-Fli1 oncogenic fusion protein can function as a repressor of the FOXO1 gene in Ewing’s sarcoma and related tumors.
ACKNOWLEDGEMENT
This work was supported by Public Health Service Grant RO1 AR051455 (to L. Y.) and by a Veterans Affairs Merit Review Award (to H. A. C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Desmaze C, Zucman J, Delattre O, Thomas G, Aurias A. Unicolor and bicolor in situ hybridization in the diagnosis of peripheral neuroepithelioma and related tumors. Genes Chromosomes Cancer. 1992;5:30–34. doi: 10.1002/gcc.2870050105. [DOI] [PubMed] [Google Scholar]
- 2.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Chansky HA, Hickstein DD. EWS.Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. J Biol Chem. 2000;275:37612–37618. doi: 10.1074/jbc.M005739200. [DOI] [PubMed] [Google Scholar]
- 4.Chansky HA, Barahmand-Pour F, Mei Q, Kahn-Farooqi W, Zielinska-Kwiatkowska A, Blackburn M, Chansky K, Conrad EU, 3rd, Bruckner JD, Greenlee TK, Yang L. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing's sarcoma cells in vitro. J Orthop Res. 2004;22:910–917. doi: 10.1016/j.orthres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Bailly RA, Bosselut R, Zucman J, Cormier F, Delattre O, Roussel M, Thomas G, Ghysdael J. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol. 1994;14:3230–3241. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahm KB, Cho K, Lee C, Im YH, Chang J, Choi SG, Sorensen PH, Thiele CJ, Kim SJ. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet. 1999;23:222–227. doi: 10.1038/13854. [DOI] [PubMed] [Google Scholar]
- 7.May WA, Arvand A, Thompson AD, Braun BS, Wright M, Denny CT. EWS/FLI1-induced manic fringe renders NIH 3T3 cells tumorigenic. Nat Genet. 1997;17:495–497. doi: 10.1038/ng1297-495. [DOI] [PubMed] [Google Scholar]
- 8.Nakatani F, Tanaka K, Sakimura R, Matsumoto Y, Matsunobu T, Li X, Hanada M, Okada T, Iwamoto Y. Identification of p21WAF1/CIP1 as a direct target of EWS-Fli1 oncogenic fusion protein. J Biol Chem. 2003;278:15105–15115. doi: 10.1074/jbc.M211470200. [DOI] [PubMed] [Google Scholar]
- 9.Zwerner JP, Guimbellot J, May WA. EWS/FLI function varies in different cellular backgrounds. Exp Cell Res. 2003;290:414–419. doi: 10.1016/s0014-4827(03)00371-9. [DOI] [PubMed] [Google Scholar]
- 10.Zou J, Ichikawa H, Blackburn ML, Hu HM, Zielinska-Kwiatkowska A, Mei Q, Roth GJ, Chansky HA, Yang L. The oncogenic TLS-ERG fusion protein exerts different effects in hematopoietic cells and fibroblasts. Mol Cell Biol. 2005;25:6235–6246. doi: 10.1128/MCB.25.14.6235-6246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu-Lieskovan S, Zhang J, Wu L, Shimada H, Schofield DE, Triche TJ. EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing's family of tumors. Cancer Res. 2005;65:4633–4644. doi: 10.1158/0008-5472.CAN-04-2857. [DOI] [PubMed] [Google Scholar]
- 12.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24:7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu HM, Zielinska-Kwiatkowska A, Munro K, Wilcox J, Wu DY, Yang L, Chansky HA. EWS/FLI1 suppresses retinoblastoma protein function and senescence in Ewing's sarcoma cells. J Orthop Res. 2008;26:886–893. doi: 10.1002/jor.20597. [DOI] [PubMed] [Google Scholar]
- 14.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, Fraser ID. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsui Y, Chansky HA, Barahmand-Pour F, Zielinska-Kwiatkowska A, Tsumaki N, Myoui A, Yoshikawa H, Yang L, Eyre DR. COL11A2 collagen gene transcription is differentially regulated by EWS/ERG sarcoma fusion protein and wild-type ERG. J Biol Chem. 2003;278:11369–11375. doi: 10.1074/jbc.M300164200. [DOI] [PubMed] [Google Scholar]
- 16.Uren A, Merchant MS, Sun CJ, Vitolo MI, Sun Y, Tsokos M, Illei PB, Ladanyi M, Passaniti A, Mackall C, Toretsky JA. Beta-platelet-derived growth factor receptor mediates motility and growth of Ewing's sarcoma cells. Oncogene. 2003;22:2334–2342. doi: 10.1038/sj.onc.1206330. [DOI] [PubMed] [Google Scholar]
- 17.Dauphinot L, De Oliveira C, Melot T, Sevenet N, Thomas V, Weissman BE, Delattre O. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression. Oncogene. 2001;20:3258–3265. doi: 10.1038/sj.onc.1204437. [DOI] [PubMed] [Google Scholar]
- 18.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong XY, Chen C, Sun X, Guo P, Vessella RL, Wang RX, Chung LW, Zhou W, Dong JT. FOXO1 is a candidate for the 13q14 tumor suppressor gene inhibiting androgen receptor signaling in prostate cancer. Cancer Res. 2006;66:6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- 20.Goto T, Takano M, Albergaria A, Briese J, Pomeranz KM, Cloke B, Fusi L, Feroze-Zaidi F, Maywald N, Sajin M, Dina RE, Ishihara O, Takeda S, Lam EW, Bamberger AM, Ghaem-Maghami S, Brosens JJ. Mechanism and functional consequences of loss of FOXO1 expression in endometrioid endometrial cancer cells. Oncogene. 2008;27:9–19. doi: 10.1038/sj.onc.1210626. [DOI] [PubMed] [Google Scholar]
- 21.Maekawa T, Maniwa Y, Doi T, Nishio W, Yoshimura M, Ohbayashi C, Hayashi Y, Okita Y. Expression and localization of FOXO1 in non-small cell lung cancer. Oncol Rep. 2009;22:57–64. [PubMed] [Google Scholar]
- 22.Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, Burgering BM, Medema RH. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22:7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang JY, Hung MC. A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res. 2009;15:752–757. doi: 10.1158/1078-0432.CCR-08-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Vos KE, Coffer PJ. FOXO-binding partners: it takes two to tango. Oncogene. 2008;27:2289–2299. doi: 10.1038/onc.2008.22. [DOI] [PubMed] [Google Scholar]
- 25.Nowak K, Killmer K, Gessner C, Lutz W. E2F-1 regulates expression of FOXO1 and FOXO3a. Biochim Biophys Acta. 2007;1769:244–252. doi: 10.1016/j.bbaexp.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Berry FB, Skarie JM, Mirzayans F, Fortin Y, Hudson TJ, Raymond V, Link BA, Walter MA. FOXC1 is required for cell viability and resistance to oxidative stress in the eye through the transcriptional regulation of FOXO1. Hum Mol Genet. 2008;17:490–505. doi: 10.1093/hmg/ddm326. [DOI] [PubMed] [Google Scholar]
- 27.Essaghir A, Dif N, Marbehant CY, Coffer PJ, Demoulin JB. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J Biol Chem. 2009;284:10334–10342. doi: 10.1074/jbc.M808848200. [DOI] [PMC free article] [PubMed] [Google Scholar]