Abstract
The human pathogen Campylobacter jejuni possesses a general N-linked glycosylation system that is known to play a role in pathogenicity; however, a detailed understanding of this role remains elusive. A considerable hindrance to studying bacterial N-glycosylation in vivo is the absence of small molecule inhibitors to reversibly control the process. This report describes a pathway-screening assay that targets the early enzymes of C. jejuni N-glycan biosynthesis that would enable identification of inhibitors to the first four steps in the pathway. The assay includes PglF, PglE, PglD, PglC and PglA; the enzymes involved in the biosynthesis of an undecaprenyl diphosphate-linked disaccharide and monitors the transfer [3H]GalNAc from the hydrophilic UDP-linked carrier to the lipophilic UndPP-diNAcBac (2,4-diacetamido-2,4,6-trideoxyglucose). The optimized assay has a Z′-factor calculated to be 0.77, indicating a robust assay suitable for screening. The diacylglycerol kinase from Streptococcus mutans, which provides a convenient method for phosphorylating undecaprenol, has been included in a modified version of the assay thereby allowing the screen to be conducted with entirely commercially available substrates.
1. Introduction
The general N-linked protein glycosylation (pgl) pathway of Campylobacter jejuni is the first discovered bacterial N-glycosylation system and has been demonstrated to play an important role in the pathogenicity of this organism.1, 2 The human pathogen C. jejuni is a major cause of gastroenteritis and is the most common antecedent to Guillain-Barré syndrome, a major cause of non-trauma-induced paralysis.3-5 N-linked glycosylation occurs in the periplasm of the Gram-negative pathogen, with the en bloc transfer of a heptasaccharide from an undecaprenyl diphosphate (UndPP) carrier to an asparagine in the acceptor protein. Over 40 proteins have been demonstrated to be N-glycosylated in C. jejuni, and it is predicted that more than 150 of the 340 secreted C. jejuni proteins may be glycosylated based on the presence of the recognition sequence D/E-X1-N-X2-S/T (where X1 and X2 may be any residue except proline).2, 6, 7 The role of this post-translational modification and how it affects pathogenicity are areas of active investigation. While a detailed understanding of how N-glycosylation enables pathogenicity is lacking, blocking N-glycosylation using genetic approaches renders C. jejuni dramatically less pathogenic, as has been determined in several studies employing chick colonization pathogenicity models.8-11 The genes mutated in these studies include the oligosaccharyl transferase PglB and those involved in the biosynthesis of the undecaprenyl diphosphate-heptasaccharide; which strongly suggests that an inhibitor of one of the biosynthetic enzymes along this pathway would block N-glycosylation and potentially modulate pathogenicity.
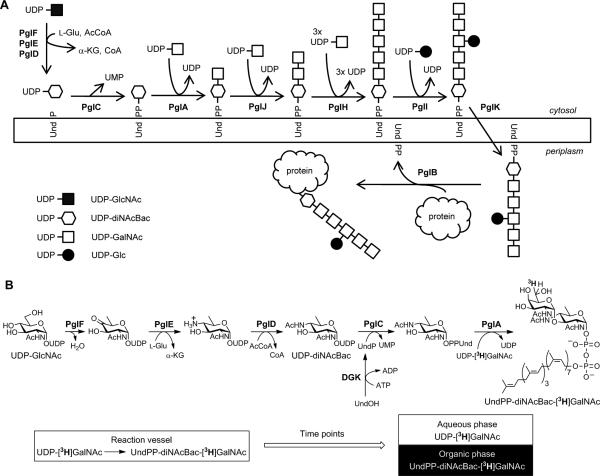
The biosynthesis of the C. jejuni heptasaccharide involves the sequential addition of sugars to generate a heptasaccharide linked to undecaprenyl diphosphate (Und-PP) anchored to the cytoplasmic face of the inner membrane (Figure 1A). The first sugar in the heptasaccharide is the unusual sugar N,N′-diacetylbacillosamine (2,4-diacetamido-2,4,6-trideoxyglucose, herein referred to as diNAcBac), which is transferred from UDP-diNAcBac and is only known to exist in certain strains of bacteria.12 In addition to the involvement in C. jejuni N-glycosylation, UDP-diNAcBac is also a known intermediate in the biosynthesis of legionaminic acid in Legionella pneumophilia (causative agent of Legionnaire's disease)13 and is putatively a component sugar in O-linked protein glycosylation in Neisseria gonorrhoeae (causative agent of gonorrhea).14 In C. jejuni, UDP-diNAcBac is derived from UDP-GlcNAc by the sequential action of the 4,6-dehydratase PglF, the 4-aminotransferase PglE and the 4-acetyltransferase PglD (Figure 1B).15, 16 The glycosyl-1-phosphate transferase PglC then uses UDP-diNAcBac and undecaprenyl phosphate (UndP) to generate diNAcBac-PP-Und.17 The glycosyltransferase PglA then adds GalNAc (via an α–1,3 linkage), then PglJ adds the second GalNAc (α1,4-linkage), and PglH adds three additional GalNAc sugars (α1,4-linkages, Figure 1A).18-20 PglI adds a branching glucose to the third GalNAc (β1,3-linkage) to afford the complete Und-PP-heptasaccharide.19 The undecaprenyl diphosphate-linked glycan is then flipped to the periplasmic face of the inner membrane by the flippase PglK,21 where the oligosaccharyltransferase PglB transfers the glycan to asparagine residues within the consensus sequence of proteins in the periplasm. A high-resolution magic angle spinning nuclear magnetic resonance study comparing C. jejuni mutants found that N-linked glycosylation was greatly reduced in pglF, pglE, pglD, pglA, pglJ, pglH, pglK and pglB mutants, while the pglI mutant is unaffected.11 A pglC mutant was unavailable as this mutation appears to be lethal.11, 22 The essential nature of the entire biosynthetic pathway (except PglI) strongly suggests one could target any one of the enzymes with inhibitors to reduce the flux of substrates through the N-glycosylation pathway of C. jejuni.
Figure 1.
A) The C. jejuni biosynthesis of UndPP-heptasaccharide and the en bloc transfer of the glycan to asparagine residues of target proteins in the periplasm. B) The lipophilicity-based pathway assay is composed of the first five enzymes of the C. jejuni UndPP-heptasaccharide biosynthesis pathway, which can be coupled with Streptococcus mutans diacylglycerol kinase (DGK).
In this report we describe a multicomponent kinetic assay for the early enzymes in the UndPP-heptasaccharide biosynthetic pathway (Figure 1B). PglF, PglE, PglD, PglC, and PglA are reconstituted in vitro and the conversion of UDP-GlcNAc into UndPP-diNAcBac-[3H]GalNAc is quantified by monitoring the transfer of [3H]GalNAc from the water soluble UDP carrier to the lipophilic UndPP-diNAcBac acceptor. This assay targets enzymes involved in the biosynthesis of the unusual bacterial sugar diNAcBac and the transfer of diNAcBac-phosphate to UndP. This multienzyme assay, together with the established assays for the individual enzymes,15-17, 23 is anticipated to be a useful screen for inhibitors, and may be used to evaluate substrate flux along the inhibited pathway. We also describe a modification of this assay, which incorporates the diacylglycerol kinase from Streptococcus mutans (DGK) which allows for use of the readily available undecaprenol in lieu of UndP,24 such that screening of five Pgl enzymes in a single assay can be carried out using entirely commercially available substrates.
2. Results
2.1 Reconstituted activities of PglF, PglE, PglD, PglC, PglA and DGK
The five pgl pathway enzymes with the appropriate cofactors and substrates, including UndP and UDP-[3H]GalNAc, were reconstituted for the quenched time-point assay. PglF and PglC are integral membrane proteins purified from cell envelope fractions, and therefore the assays are performed in the presence of the detergent Triton X-100. Samples collected at different time points were extracted with a chloroform-methanol-water system, allowing measurement of the ratio of water soluble substrate [3H]GalNAc, to organic soluble product UndPP-diNAcBac. As anticipated, a time-dependent transfer of radioactivity to the organic phase was observed (Figure 2A). This transfer of radioactivity to the organic phase was not observed in the absence of UndP (Figure S1). In a second experiment where DGK, ATP, and undecaprenol (UndOH) were used in place of UndP, similar results were observed (Figure 2B). In both of these experiments an initial lag period was observed, consistent with a coupled assay with several partially rate-limiting steps. This was followed by a linear phase representing the reaction progress from ~5 to 30% conversion. At higher conversion, the reaction rate decelerates due to substrate depletion (Figures 2A and 2B). These results are consistent with formation of the final PglA product UndPP-diNAcBac-[3H]GalNAc,19, 20, 24 as is the requirement of each enzyme for these observations (Figure 2C). The systematic removal of individual enzymes led to observation of only background levels of radioactivity in the organic phase from the quenched assay aliquot, confirming these observations measure the formation of the final product of these five enzymes, UndPP-diNAcBac-GalNAc.
Figure 2.
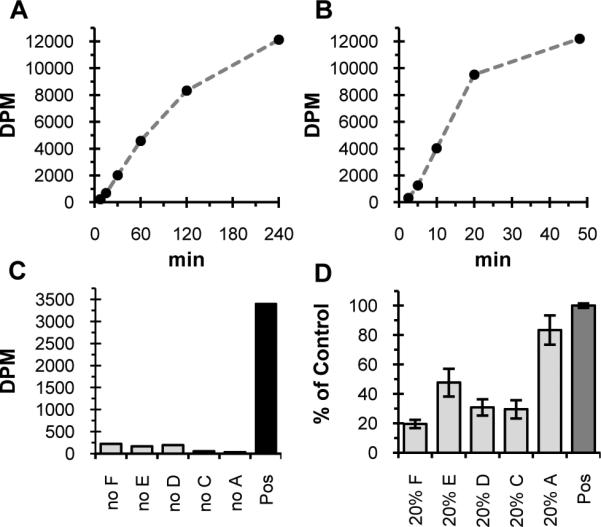
Transfer of radioactivity to an organic extract in the Pgl pathway screen (27000 DPM = theoretical yield). A) Initial activity of PglF, PglE, PglD, PglC, PglA assay with UndP measured with the lipophilicity-based assay (0.04% Triton X-100, 42 nM PglF, 146 μM PglE, 236 nM PglD, 52.5 nM PglC, 444 nM PglA). B) Activity of PglF, PglE, PglD, PglC, PglA, DGK assay with UndOH (0.04% Triton X-100, 84 nM PglF, 700 nM PglE, 675 nM PglD, 105 nM PglC, 222 nM PglA, 272 nM DGK). C) Transfer of radioactivity to organic phase is dependent on each enzyme. Radioactivity transferred to the organic phase at 30 min by PglF, PglE, PglD, PglC, PglA assay with UndP (control) compared to when individual enzymes are removed (0.04% Triton X-100, 42 nM PglF, 146 nM PglE, 1.2 nM PglD, 53 nM PglC, 444 nM PglA). Similar results were observed when UndOH and DGK used instead of UndP (not show). D) Assay is optimized for maximum sensitivity to inhibition of PglF, PglE, PglD, and PglC. Lipophilic radioactivity when each enzyme is individually lowered five-fold from control ([Triton X-100], [enzyme] as in C). Data normalized from three separate runs where lipophilic radioactivity in control was 2000 to 2500 DPM. Error bars represent the standard error of the mean (n = 3 for 20% enzyme data, n = 9 for control).
2.2 Balancing enzyme concentrations for kinetic sensitivity to PglF, PglE, PglD and PglC
We have chosen to include the first five enzymes in the UndPP-heptasaccharide pathway, as this represents several enzymes intimately involved with recognition of the unusual sugar diNAcBac or its precursor intermediates. Although PglA falls into this category, the primary role of this enzyme is to deliver the [3H] labeled GalNAc to UndPP-diNAcBac and drive the phosphoglycosyl transferase reaction forward and therefore, it is used in excess in the assay. In order to make the assay simultaneously sensitive to inhibitors of PglF, PglE, PglD, and PglC, the concentrations of these enzymes have been balanced such that the activity of each of these is partially rate determining. This was accomplished by systematically lowering the concentration of each enzyme until a significant effect on the overall rate was observed. A simulated inhibition of these enzymes demonstrates this balanced state has been achieved; when the concentration of PglF, PglE, PglD or PglC is lowered five-fold a lower overall rate results (Figure 2D).
2.3 Effects of Triton X-100 and DMSO
Variation of the detergent concentration has a significant effect on observed activity. Maximum activity was observed with 0.04% Triton X-100 (Figure S2), near the critical micelle concentration of this detergent (0.019%25). This activity 30-fold greater than that that measured at 1% Triton X-100, and no activity was observed below the critical micelle concentration of the detergent. These observations are consistent with the activity of known membrane-associated enzymes.26
Dimethyl sulfoxide (DMSO) is a common small molecule carrier in library screening and as such its compatibility with the assay was assessed. Up to 5% DMSO did not have a noticeable effect on the overall activity, and only a minor effect on background signal was observed. When PglF, PglE, or PglD was absent, slightly higher background radioactivity was observed than when PglC, PglA or DGK was absent (Figure 2C). In 5% DMSO the background radioactivity with PglF, PglE, or PglD removed was less than ~10% the radioactivity with all enzymes present, and the background radioactivity with PglC removed was less than ~3% the activity with all enzymes present (Figure S3). One possible explanation for these observations is that latent substrate promiscuity of PglC and PglA exists, and that this activity is enhanced with increasing DMSO concentration. Regardless of the origin of this effect, it is clear that up to 5% DMSO may be used with the assay while maintaining a signal to background ratio of ten to one or greater.
2.4 Volume-minimized assay for screening and inhibition with substrate analogues
For screening purposes the reaction volume has been minimized to allow for economical use of reagents and time. Reaction volumes of 20 μL, which allow for a single time point evaluation of reaction progress, have been deemed sufficient for screening purposes. Conditions are such that at 30 min ~10% conversion to final product is observed, which is in the linear phase of reaction progress. The assay was evaluated by running positive and negative controls on three different occasions. The average signal-to-background ratio was 56-fold and the standard deviation for the positive and negative controls was 7.3 and 0.27%, respectively. This data was used to calculate the Z′-factor,27 which is a statistical parameter used to evaluate assay quality. The Z′-factor takes into consideration the signal dynamic range and data reproducibility, and should be between 0.5 and 1 for an assay to be suitable for screening. The Z′-factor for the minimized assay was calculated to be 0.77.
To demonstrate inhibition in the context of the assay, the effects of several products and product analogues and the divalent metal ion chelator EDTA were investigated. UDP, α-ketoglutarate, CoASH, and UMP were all found to inhibit the assay (Figure 3). EDTA was also found to inhibit the assay, presumably by sequestering the divalent magnesium required for PglC and PglA, which are known to require a divalent cation for activity.17, 23
Figure 3.
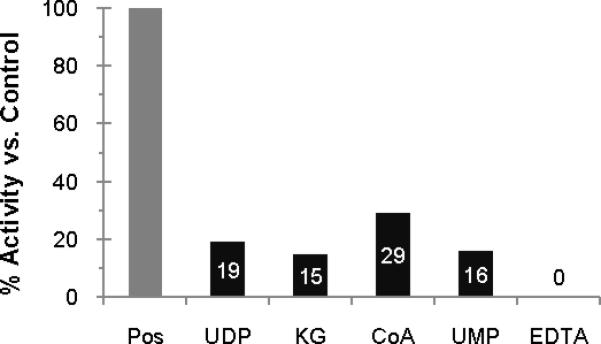
Inhibition of PglF, PglE, PglD, PglC, PglA assay with products, substrate analogs and EDTA. Numbers indicate percent remaining activity relative to the positive control. Concentrations used: 1 mM UDP, 20 mM α-ketoglutarate, 3.75 mM CoASH, 1 mM UMP, 18 mM EDTA. Background (no enzymes control) subtracted.
3. Discussion
A recurring theme in prokaryotic glycoconjugate assembly is the sequential transfer of sugars to a membrane-bound isoprene-linked carrier during glycan biosynthesis. Notable examples of this include the biosynthesis of GlcNAc-MurNAc-pentapetide subunits of peptidoglycan, the outer O-antigens of lipopolysaccharide (LPS), capsular polysaccharides and lipooligosaccharides. More recently discovered examples include bacterial glycosylation pathways, such as the O-glycosylation pathway of N. gonorrheae and the N-glycosylation pathway of C. jejuni. Having established an assay for C. jejuni, which takes advantage of the lipophilicity of the undecaprenyl diphosphate-linked glycan, it is anticipated that the assay described here could easily be adapted to these other pathways.
The development of this lipophilicity-based pathway screen is a useful tool to address the present need for inhibitors of C. jejuni N-glycosylation. The absence of such inhibitors is currently a hindrance for researchers studying the roles of this pathway in vivo. This is in contrast with eukaryotic N-glycosylation, which is inhibited by tunicamycin, the utility of which is demonstrated by more than 2500 peer-reviewed journal articles citing its use. Tunicamycin targets the GlcNAc phosphototransferase, which transfers GlcNAc-1-P from UDP-GlcNAc to dolichyl-phosphate. Tunicamycin is also known to inhibit the transfer of GlcNAc-1-P to UndP and the formation of the undecaprenyl-PP-MurNAc pentapeptide involved in bacterial peptidoglycan biosynthesis. However, tunicamycin does not inhibit C. jejuni PglC, as described previously in a study in which this lack of inhibition was contrasted with inhibition of Escherichia coli GlcNAc-1-phosphate transferase WecA.17 While it is unfortunate that tunicamycin may not be used to inhibit PglC in biological studies, the observed difference in tunicamycin sensitivity suggests the feasibility of targeting PglC of the C. jejuni pathway selectively over GlcNAc phosphototransferase of the eukaryotic pathway, which may be of therapeutic importance.
In the search for inhibitors of C. jejuni N-glycosylation, pathway screening offers several advantages over parallel screening of each individual enzyme. Chiefly, the efficiency of simultaneously screening multiple enzymes reduces the total number of assays required. It is estimated that one could easily screen 25 potential inhibitory interactions in a single tube with a cocktail of five compounds using the present assay. This would require additional experiments to identify the particular inhibitor/enzyme combination of interest, but does not necessarily require individual assays for each enzyme as was demonstrated in a similar pathway screen of the ADP-heptose biosynthetic pathway of LPS.28 In the case of PglF, PglE, PglD, PglC and PglA, however, individual assays are established.15-17, 23 A second benefit of the pathway screen is the use of commercially available substrates, which is particularly relevant here where every sugar intermediate beyond UDP-GlcNAc is rare and requires special preparation and handling. Finally, assaying the pathway also allows identification of potential modulators of enzyme complex formation.
4. Conclusion
In this report we have described how the lipophilic nature of the isoprene-linked glycan can be harnessed to screen multiple enzymes involved with unusual sugar biosynthesis. This assay has been optimized for maximum sensitivity to inhibition of PglF, PglE, PglD, and PglC by balancing the enzyme concentrations such that each is partially rate determining. The assay has been optimized for maximum activity, which was found at 0.04-0.06% Triton X-100, and the assay volume has been minimized to 20 μL to economize on reagent use. The robustness of the optimized assay has been demonstrated by the measured Z′-factor of 0.77, well within the desired range of 0.5 to 1 for reliable screening.
5. Experimental
5.1 Materials
All chemicals were purchased from Sigma-Aldrich unless otherwise noted. Undecaprenol (UndOH) and UDP-[3H]GalNAc were purchased from American Radiolabeled Chemicals Inc. Undecaprenyl phosphate (UndP) had been previously29 prepared from undecaprenol using phosphoramidite chemistry. The pure solvent upper phase (PSUP) used in the assay contains 49% MeOH, 48% 100 mM aqueous KCl, and 3% CHCl3.
5.2 Enzyme preparation and storage
Recombinant enzymes were purified by affinity chromatography as N-terminal GST-tagged (PglF) or N-terminal His-tagged/C-terminal T7-tagged (PglE, PglD, PglC, PglA, DGK) constructs as described previously.15, 17, 19, 24 His6-tagged enzymes were dialyzed into 50 mM triethanolamine pH 7.8 with 1 mM tris(2-carboxyethyl)phosphine (TCEP) added. GST-tagged enzymes were stored in the elution buffer containing 10 mM glutathione (and 0.1% Triton X-100 in the case of PglF). Purified enzyme aliquots were stored at -80 °C with 15% (PglE, PglD, PglC, PglA, DGK) or 30% (PglF) glycerol.
5.3 Optimized PglF, PglE, PglD, PglC, PglA assay protocol
Reactions were performed in parallel on a 20 μL scale in 1.65 mL Eppendorf tubes at 25 °C. Reactions were initiated by addition of an enzyme stock to a solution of reagents and substrate. For control reactions containing no enzyme an equivalent volume of buffer was added instead. Assay components were 42 nM PglF (78 ng), 146 nM PglE (134 ng), 1.2 nM PglD (0.56 ng), 53 nM PglC (27 ng), 444 nM PglA (400 ng), 500 μM UDP-GlcNAc, 10 μM UDP-[3H]GalNAc (specific activity of 225 nCi nmol-1), 3 μM undecaprenyl phosphate, 20 mM L-glutamate, 1.68 mM AcCoA, 450 μM NAD+, 300 μM PLP, 10 mM MgCl2, 0.04% Triton X-100, 2% DMSO, 50 mM pH 7.8 triethanolamine. Immediately after addition of enzyme the solution was gently vortexed, spun down, then incubated. After 30 min 18 μL was quenched by addition to a biphasic mixture containing 800 μL 2:1 CHCl3/MeOH and 200 μL of PSUP and vortexed vigorously. The lower organic layer was washed with 3 × 200 μL PSUP, then dried under a stream of nitrogen. The use of PSUP during the washing step is known to aid retention of lipids in the organic layer, and is superior to water in this respect.30 The dried organic layer residue was dissolved with 200 μL DMSO and 5 mL EcoLite scintillation fluid (MP Biomedicals) was then added. Each vial was subjected to a 5 min scintillation count (maximum theoretical conversion = 27 000 DPM [3H]).
5.4 Optimized PglF, PglE, PglD, PglC, PglA, DGK assay protocol
The assay including DGK was performed as described in section 5.3 with the following changes. UndP was replaced with 3 μM UndOH, 1 mM ATP and 106 nM DGK (38 ng).
5.5 Time-dependent assays
Time-dependent assays were performed as described in section 5.3 with the following modifications. Assays were scaled up according to how many time points were to be acquired, and 18 μL aliquots were removed at the appropriate times. Other changes to particular experiments are noted in figure captions.
5.6 Calculation of the Z′-factor
Data used for the calculation of the Z′-factor was gathered from three separate occasions in which three positive controls and one negative control was measured. These data were converted to percentages using the average positive control as 100%, and the mean of the positive (μ+) and negative (μ-) controls, and the standard deviation of the positive (σ+) and negative controls (σ-) were calculated. These data were used to calculate the Z′-factor with equation (1).27
| (1) |
Supplementary Material
Acknowledgements
We thank Dr. Nelson Oliver, Dr. Mark Chen, Meredith Hartley, and Michael Morrison for helpful discussions and Angelyn Larkin for helpful discussions and critical reading of the manuscript. This work was supported by NIH Grant GM39334 to B.I., a NIH postdoctoral fellowship to J.M.T. 5F32GM080794 and an NSERC postdoctoral fellowship to J.P.M.
Abbreviations
- DGK
Streptococcus mutans diacylglycerol kinase
- TCEP
tris(2-carboxyethyl)phosphine
- UndOH
undecaprenol
- UndP
undecaprenyl phosphate
- diNAcBac
2,4-diacetamido-2,4,6-trideoxyglucose
- DMSO
dimethyl sulfoxide
- DPM
disintegrations per minute
- PSUP
pure solvent upper phase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. Mol. Microbiol. 1999;32:1022. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 2.Nothaft H, Amber S, Aebi M, Szymanski C. In: Campylobacter. Nachamkin I, Szymanski CM, Baser MJ, editors. ASM Press; Washington, DC: 2008. p. 447.p. 469. [Google Scholar]
- 3.Zilbauer M, Dorrell N, Wren BW. T. Roy. Soc. Trop. Med. H. 2008;102:123. doi: 10.1016/j.trstmh.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Kaida K, Ariga T, Yu RK. Glycobiology. 2009;19:676. doi: 10.1093/glycob/cwp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas MR, Winer JB. Expert Rev. Neurotherapeutics. 2006;6:1569. doi: 10.1586/14737175.6.10.1569. [DOI] [PubMed] [Google Scholar]
- 6.Chen MM, Glover KJ, Imperiali B. Biochemistry. 2007;46:5579. doi: 10.1021/bi602633n. [DOI] [PubMed] [Google Scholar]
- 7.Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF, Wacker M, Aebi M. EMBO J. 2006;25:1957. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrixson DR, DiRita VJ. Mol. Microbiol. 2004;52:471. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- 9.Karlyshev AV, Everest P, Linton D, Cawthraw S, Newell DG, Wren BW. Microbiology. 2004;150:1957. doi: 10.1099/mic.0.26721-0. [DOI] [PubMed] [Google Scholar]
- 10.Jones MA, Marston KL, Woodall CA, Maskell DJ, Linton D, Karlyshev AV, Dorrell N, Wren BW, Barrow PA. Infect. Immun. 2004;72:3769. doi: 10.1128/IAI.72.7.3769-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly J, Jarrell H, Millar L, Tessier T, Fiori LM, Lau PC, Allan B, Szymanski C,M. J. Bacteriol. 2006;188:2427. doi: 10.1128/JB.188.7.2427-2434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharon N. Glycobiology. 2007;17:1150. doi: 10.1093/glycob/cwm089. [DOI] [PubMed] [Google Scholar]
- 13.Glaze PA, Watson DC, Young NM, Tanner ME. Biochemistry. 2008;47:3272. doi: 10.1021/bi702364s. [DOI] [PubMed] [Google Scholar]
- 14.Vik Å, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobson W, Koomey M. Proc. Nat. Acad. Sci. U.S.A. 2009;106:4447. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivier NB, Chen MM, Behr JR, Imperiali B. Biochemistry. 2006;45:13659. doi: 10.1021/bi061456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson J-R, Logan SM. J. Biol. Chem. 2006;281:723. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- 17.Glover KJ, Weerapana E, Chen MM, Imperiali B. Biochemistry. 2006;45:5343. doi: 10.1021/bi0602056. [DOI] [PubMed] [Google Scholar]
- 18.Linton D, Dorrell N, Hitchen PG, Amber S, Karlyshev AV, Morrison HR, Dell A, Valvano MA, Aebi M, Wren BW. Mol. Microbiol. 2005;55:1695. doi: 10.1111/j.1365-2958.2005.04519.x. [DOI] [PubMed] [Google Scholar]
- 19.Glover KJ, Weerapana E, Imperiali B. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14255. doi: 10.1073/pnas.0507311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troutman JM, Imperiali B. Biochemistry. 2009;48:2807. doi: 10.1021/bi802284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alaimo C, Catrein I, Morf L, Marolda CL, Callewaert N, Valvano MA, Feldman MF, Aebi M. EMBO J. 2006;25:967. doi: 10.1038/sj.emboj.7601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood AC, Oldfield NJ, O'Dwyer CA, Ketley JM. Microbiology. 1999;145:379. doi: 10.1099/13500872-145-2-379. [DOI] [PubMed] [Google Scholar]
- 23.Weerapana E, Glover KJ, Chen MM, Imperiali B. J. Am. Chem. Soc. 2005;127:13766. doi: 10.1021/ja054265v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartley MD, Larkin A, Imperiali B. Bioorg. Med. Chem. 2008;16:5149. doi: 10.1016/j.bmc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neugebauer JM. Methods. Enzymol. 1990;182:239. doi: 10.1016/0076-6879(90)82020-3. [DOI] [PubMed] [Google Scholar]
- 26.Womack MD, Kendall DA, MacDonald RC. Biochim. Biophys. Acta, Biomembr. 1983;733:210. doi: 10.1016/0005-2736(83)90524-2. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J-H, Chung TD, Oldenburg KR. J. Biomol. Screen. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 28.De Leon GP, Elowe NH, Koteva KP, Valvano MA, Wright GD. Chem. Biol. 2006;13:437. doi: 10.1016/j.chembiol.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Chen MM, Weerapana E, Ciepichal E, Stupak J, Reid CW, Swiezewska E, Imperiali B. Biochemistry. 2007;46:14342. doi: 10.1021/bi701956x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folch J, Lees M, Sloane Stanley GH. J. Biol. Chem. 1957;226:497. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.