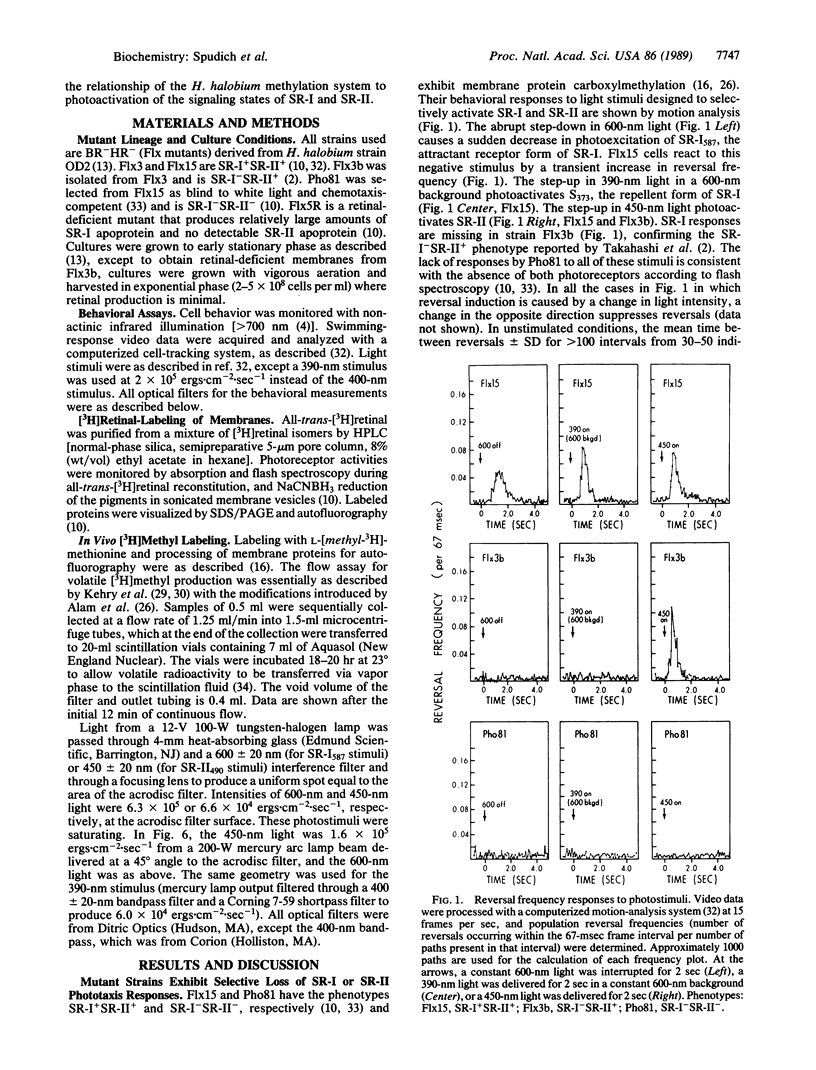
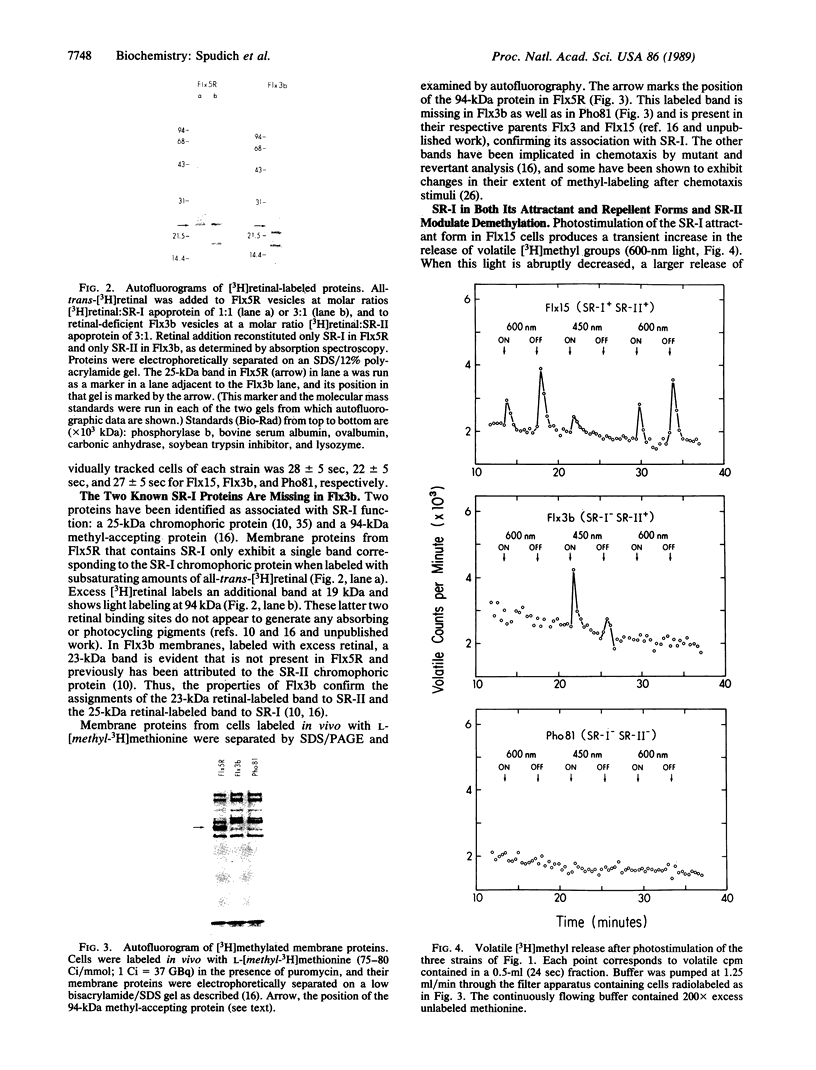
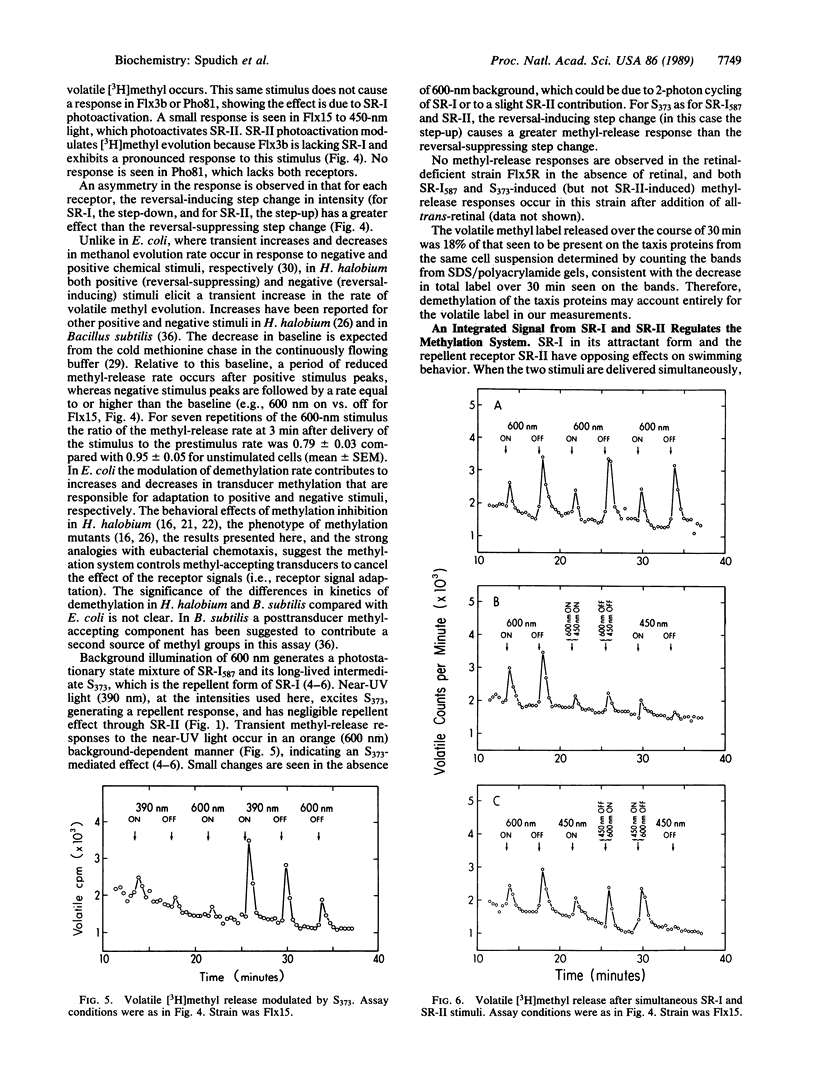
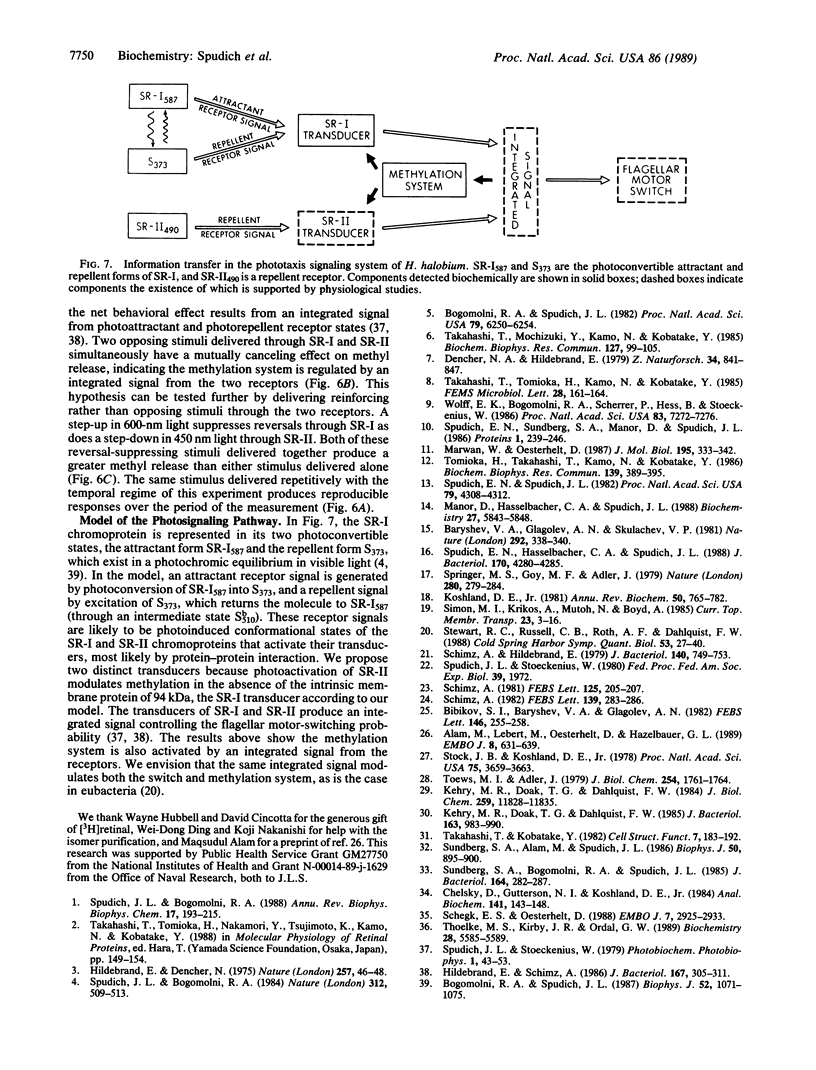
Abstract
This work demonstrates that phototaxis stimuli in the archaebacterium Halobacterium halobium control a methylation/demethylation system in vivo through photoactivation of sensory rhodopsin I (SR-I) in either its attractant or repellent signaling form as well as through the repellent receptor sensory rhodopsin II (SR-II, also called phoborhodopsin). The effects of positive stimuli that suppress swimming reversals (i.e., an increase in attractant or decrease in repellent light) and negative stimuli that induce swimming reversals (i.e., a decrease in attractant or increase in repellent light) through each photoreceptor were monitored by assaying release of volatile [3H]methyl groups. This assay has been used to measure [3H]methanol produced during the process of adaptation to chemotactic stimuli in eubacteria. In H. halobium positive photostimuli produce a transient increase in the rate of demethylation followed by a decrease below the unstimulated value, whereas negative photostimuli cause an increase followed by a rate similar to that of the unstimulated value. Photoactivation of the SR-I attractant and simultaneous photoactivation of the SR-II repellent receptors cancel in their effects on demethylation, demonstrating the methylation system is regulated by an integrated signal. Analysis of mutants indicates that the source for the volatile methyl groups is intrinsic membrane proteins distinct from the chromoproteins that share the membrane. A methyl-accepting protein (94 kDa) previously correlated in amount with the SR-I chromoprotein (25 kDa) is shown here to be missing in a recently isolated SR-I-SR-II+ mutant (Flx3b), thus confirming the association of this protein with SR-I. Photoactivated SR-II in mutant Flx3b controls demethylation, predicting the existence of a photomodulated methyl-accepting component distinct from the 94-kDa protein of SR-I. We present a model in which the three known phototaxis signaling receptor states (the attractant receptor SR-I587, its repellent form S373, and the repellent receptor SR-II490) are coupled to two distinct transducers the demethylation of which is controlled by one integrated signal.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam M., Lebert M., Oesterhelt D., Hazelbauer G. L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989 Feb;8(2):631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni R. A., Spudich J. L. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni R. A., Spudich J. L. The photochemical reactions of bacterial sensory rhodopsin-I. Flash photolysis study in the one microsecond to eight second time window. Biophys J. 1987 Dec;52(6):1071–1075. doi: 10.1016/S0006-3495(87)83301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D., Gutterson N. I., Koshland D. E., Jr A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal Biochem. 1984 Aug 15;141(1):143–148. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- Dencher N. A., Hildebrand E. Sensory transduction in Halobacterium halobium: retinal protein pigment controls UV-induced behavioral response. Z Naturforsch C. 1979 Sep-Oct;34(9-10):841–847. doi: 10.1515/znc-1979-9-1030. [DOI] [PubMed] [Google Scholar]
- Hildebrand E., Dencher N. Two photosystems controlling behavioural responses of Halobacterium halobium. Nature. 1975 Sep 4;257(5521):46–48. doi: 10.1038/257046a0. [DOI] [PubMed] [Google Scholar]
- Hildebrand E., Schimz A. Integration of photosensory signals in Halobacterium halobium. J Bacteriol. 1986 Jul;167(1):305–311. doi: 10.1128/jb.167.1.305-311.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Sensory adaptation in bacterial chemotaxis: regulation of demethylation. J Bacteriol. 1985 Sep;163(3):983–990. doi: 10.1128/jb.163.3.983-990.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem. 1984 Oct 10;259(19):11828–11835. [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Manor D., Hasselbacher C. A., Spudich J. L. Membrane potential modulates photocycling rates of bacterial rhodopsins. Biochemistry. 1988 Aug 9;27(16):5843–5848. doi: 10.1021/bi00416a004. [DOI] [PubMed] [Google Scholar]
- Marwan W., Oesterhelt D. Signal formation in the halobacterial photophobic response mediated by a fourth retinal protein (P480). J Mol Biol. 1987 May 20;195(2):333–342. doi: 10.1016/0022-2836(87)90654-1. [DOI] [PubMed] [Google Scholar]
- Schegk E. S., Oesterhelt D. Isolation of a prokaryotic photoreceptor: sensory rhodopsin from halobacteria. EMBO J. 1988 Sep;7(9):2925–2933. doi: 10.1002/j.1460-2075.1988.tb03151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimz A., Hildebrand E. Chemosensory responses of Halobacterium halobium. J Bacteriol. 1979 Dec;140(3):749–753. doi: 10.1128/jb.140.3.749-753.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimz A. Methylation of membrane proteins is involved in chemosensory and photosensory behavior of Halobacterium halobium. FEBS Lett. 1981 Mar 23;125(2):205–207. doi: 10.1016/0014-5793(81)80719-3. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Hasselbacher C. A., Spudich J. L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988 Sep;170(9):4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-transduction mutants of Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4308–4312. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Sundberg S. A., Manor D., Spudich J. L. Properties of a second sensory receptor protein in Halobacterium halobium phototaxis. Proteins. 1986 Nov;1(3):239–246. doi: 10.1002/prot.340010306. [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984 Dec 6;312(5994):509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Sensory rhodopsins of halobacteria. Annu Rev Biophys Biophys Chem. 1988;17:193–215. doi: 10.1146/annurev.bb.17.060188.001205. [DOI] [PubMed] [Google Scholar]
- Stewart R. C., Russell C. B., Roth A. F., Dahlquist F. W. Interaction of CheB with chemotaxis signal transduction components in Escherichia coli: modulation of the methylesterase activity and effects on cell swimming behavior. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 1):27–40. doi: 10.1101/sqb.1988.053.01.007. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Alam M., Spudich J. L. Excitation signal processing times in Halobacterium halobium phototaxis. Biophys J. 1986 Nov;50(5):895–900. doi: 10.1016/S0006-3495(86)83530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Bogomolni R. A., Spudich J. L. Selection and properties of phototaxis-deficient mutants of Halobacterium halobium. J Bacteriol. 1985 Oct;164(1):282–287. doi: 10.1128/jb.164.1.282-287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Mochizuki Y., Kamo N., Kobatake Y. Evidence that the long-lifetime photointermediate of s-rhodopsin is a receptor for negative phototaxis in Halobacterium halobium. Biochem Biophys Res Commun. 1985 Feb 28;127(1):99–105. doi: 10.1016/s0006-291x(85)80131-5. [DOI] [PubMed] [Google Scholar]
- Thoelke M. S., Kirby J. R., Ordal G. W. Novel methyl transfer during chemotaxis in Bacillus subtilis. Biochemistry. 1989 Jun 27;28(13):5585–5589. doi: 10.1021/bi00439a037. [DOI] [PubMed] [Google Scholar]
- Toews M. L., Adler J. Methanol formation in vivo from methylated chemotaxis proteins in Escherichia coli. J Biol Chem. 1979 Mar 25;254(6):1761–1764. [PubMed] [Google Scholar]
- Tomioka H., Takahashi T., Kamo N., Kobatake Y. Flash spectrophotometric identification of a fourth rhodopsin-like pigment in Halobacterium halobium. Biochem Biophys Res Commun. 1986 Sep 14;139(2):389–395. doi: 10.1016/s0006-291x(86)80003-1. [DOI] [PubMed] [Google Scholar]
- Wolff E. K., Bogomolni R. A., Scherrer P., Hess B., Stoeckenius W. Color discrimination in halobacteria: spectroscopic characterization of a second sensory receptor covering the blue-green region of the spectrum. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7272–7276. doi: 10.1073/pnas.83.19.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]






