Abstract
Taste perception is an important component of an animal's fitness. The identification of vertebrate taste receptor genes in the last decade has enabled molecular genetic studies of the evolution of taste perception in the context of the ecology and dietary preferences of organisms. Although such analyses have been conducted in a number of species for bitter taste receptors, a similar analysis of sweet taste receptors is lacking. Here, we survey the sole sweet taste–specific receptor gene Tas1r2 in 42 bat species that represent all major lineages of the order Chiroptera, one of the most diverse groups of mammals in terms of diet. We found that Tas1r2 is under strong purifying selection in the majority of the bats studied, with no significant difference in the strength of the selection between insect eaters and fruit eaters. However, Tas1r2 is a pseudogene in all three vampire bat species and the functional relaxation likely started in their common ancestor, probably due to the exclusive feeding of vampire bats on blood and their reliance on infrared sensors rather than taste perception to locate blood sources. Our survey of available genome sequences, together with previous reports, revealed additional losses of Tas1r2 in horse, cat, chicken, zebra finch, and western clawed frog, indicating that sweet perception is not as conserved as previously thought. Nonetheless, we found no common dietary pattern among the Tas1r2-lacking vertebrates, suggesting different causes for the losses of Tas1r2 in different species. The complexity of the ecological factors that impact the evolution of Tas1r2 calls for a better understanding of the physiological roles of sweet perception in different species.
Keywords: bats, vampire bats, Tas1r2, sweet, taste, pseudogene
Introduction
The sense of taste provides important dietary information and is crucial for the survival of animals (Bachmanov and Beauchamp 2007). All tastes are combinations of five basic modalities: sweet, umami, bitter, salty, and sour (Kinnamon and Margolskee 1996; Lindemann 1996). Sweet and umami tastes allow the recognition of diets with nutritious carbohydrates and proteins, respectively, and often cause appetitive reactions; bitter and sour tastes warn against the ingestion of potentially harmful foods and often cause aversive reactions; the salty taste detects sodium and other minerals that are needed with varying concentrations in different species (Herness and Gilbertson 1999; Bachmanov and Beauchamp 2007).
Taste perception is conferred by taste receptor cells through the use of either ion channels (sour and salty) or G protein-coupled receptors (GPCRs) (sweet, umami, and bitter) (Roper 1989; Lindemann 1996; Bachmanov and Beauchamp 2007). Two families of GPCRs, Tas1rs and Tas2rs, function as sweet/umami and bitter taste receptors, respectively (Hoon et al. 1999; Adler et al. 2000; Chandrashekar et al. 2000; Matsunami et al. 2000; Kitagawa et al. 2001; Max et al. 2001; Montmayeur et al. 2001; Nelson et al. 2001; Sainz et al. 2001; Nelson et al. 2002; Zhao et al. 2003; Meyerhof 2005). Because the bitter taste is mainly responsible for the detection of toxins in food sources, which vary greatly among different animals, Tas2r genes have been hypothesized to differ substantially among species. This hypothesis is supported by multiple evolutionary studies (Conte et al. 2003; Shi et al. 2003; Wang et al. 2004; Fischer et al. 2005; Go et al. 2005; Shi and Zhang 2006). For example, there is a great variation in Tas2r gene number among mammals, from 21 in dog to 42 in rat (Shi and Zhang 2006).
By contrast, the Tas1r gene repertoire is small with only three genes in most mammals studied: Tas1r1, Tas1r2, and Tas1r3 (Hoon et al. 1999; Kitagawa et al. 2001; Max et al. 2001; Montmayeur et al. 2001; Nelson et al. 2001; Sainz et al. 2001; Zhao et al. 2003; Shi and Zhang 2006). Of these, Tas1r1 and Tas1r2 are expressed in separate taste receptor cells, although they are both coexpressed with Tas1r3. The Tas1r1 + Tas1r3 heterodimer functions as the umami taste receptor, whereas the Tas1r2 + Tas1r3 heterodimer functions as the sweet taste receptor (Nelson et al. 2001; Li et al. 2002; Mombaerts 2004). Thus, Tas1r2 is likely the sole sweet-specific taste receptor. Indeed, Tas1r2-knockout mice exhibit severely impaired responses to sweet but not other tastants and show human-like sweet taste preference after the subsequent knockin of human Tas1r2 (Zhao et al. 2003).
Compared with the evolution of Tas2rs, the evolution of Tas1rs is poorly characterized. Because the sweet and umami tastes are regarded as necessary for nutrient uptake, they are believed to be conserved among species (Shi and Zhang 2006) and therefore have not been well studied. Indeed, limited surveys conducted thus far found one Tas1r1, one Tas1r2, and one Tas1r3 in every mammal (Shi and Zhang 2006), with only two exceptions: Tas1r1 is pseudogenized in the giant panda (family Ursidae) (Li et al. 2010; Zhao et al. 2010) and Tas1r2 is inactivated in the cat family Felidae (Li et al. 2005). However, because the number of species that have been examined is small (Shi and Zhang 2006), it is unclear whether the observed conservation of Tas1rs is generally true. More importantly, the ecological causes of the two exceptions are unclear. For example, if the loss of Tas1r1 in the giant panda is due to its dietary shift from meat to bamboo, why do herbivores such as the cow and horse still have a functional Tas1r1 gene (Zhao et al. 2010)? Similarly, the inactivation of Tas1r2 in the cat family but not in other carnivores is puzzling. To examine the impact of diet on Tas1r evolution while avoiding confounding factors, it is preferable to examine relatively closely related species that nonetheless have diverse dietary preferences.
We choose to study the evolution of the sweet taste receptor gene Tas1r2 in 42 bat species because bats have a huge diversity in their diets, including insects, other arthropods, fish, reptiles, amphibians, mammals, birds, fruits, flowers, nectar, pollen, foliage, and blood (Altringham 1996). Around 70% of bat species are insectivorous, although some of these are also carnivorous or piscivorous. Only three species are sanguivorous, feeding exclusively on blood. Two groups of bats consume mainly plant products: Old World fruit bats (members of the suborder Yinpterochiroptera) exclusively eat fruits, flowers, pollen, and nectar, whereas New World (NW) fruit bats (members of the suborder Yangochiroptera) feed on some insects in addition to their main diets of fruits or nectar and pollen (Altringham 1996). For simplicity, we classify bats into three main dietary groups: insect eaters (species primarily feeding on insects and other arthropods), fruit eaters (species primarily feeding on plant products), and blood feeders (species feeding on blood) (fig. 1). We show that Tas1r2 is pseudogenized in blood feeders, whereas conserved in other bats. By surveying currently available genome sequences, we further report losses of Tas1r2 in horse and zebra finch and a possible loss in pig. We discuss possible ecological factors influencing the evolution of Tas1r2.
FIG. 1.
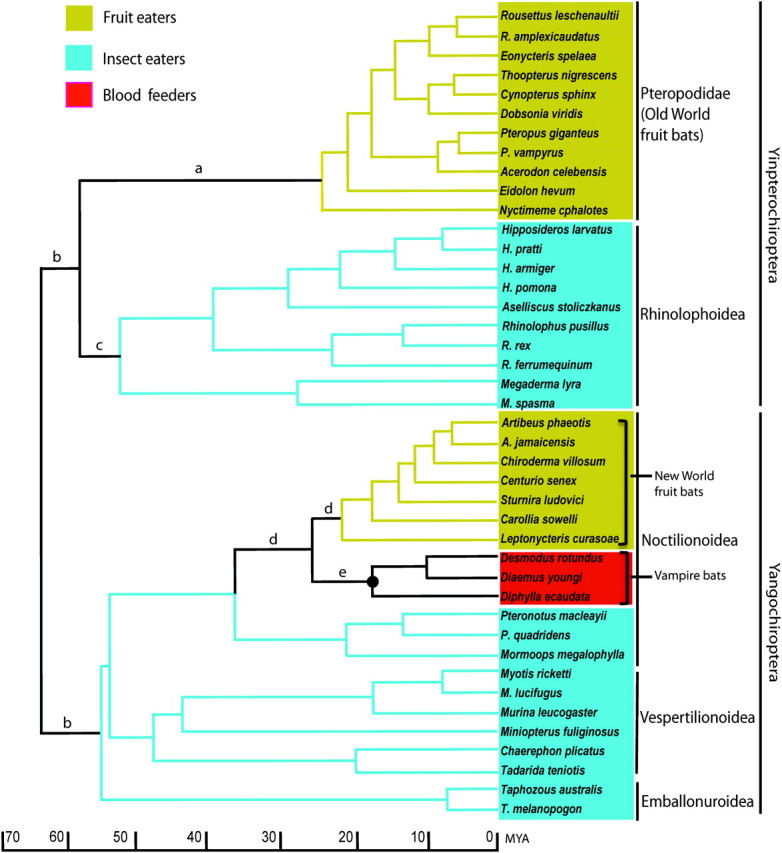
The species tree of the 42 bats studied, with the dietary preferences indicated by various colors. The tree topology and approximate divergence times follow Teeling et al. (2005). The common ancestor of vampire bats discussed in the paper is marked with a solid circle. Branch colors indicate sets of branches used in testing the selective pressures on Tas1r2 in Table 1.
Materials and Methods
Polymerase Chain Reaction and DNA Sequencing
The genomic DNAs of 40 bat species (supplementary table S1, Supplementary Material online) were isolated from wing membrane biopsies or liver tissue samples using the Qiagen DNeasy kit. Based on an alignment of currently available Tas1r2 sequences of human, mouse, dog, cow, flying fox, and little brown bat, we designed a pair of degenerate primers (SFF: 5′-GACRCYTCSTGCTTCAAGCGGC-3′ and SFR: 5′-GGTTGAGCACRGTGACCARGAG-3′) to amplify ∼720 nucleotides of the sixth exon of Tas1r2 from 39 of the 40 bats. For the remaining species (hairy-legged vampire bat), the above primers did not work. We thus redesigned a forward primer (SFF23: 5′-CGACTGGCCTTCCTCAAAT-3′) according to the Tas1r2 sequence acquired from the common vampire bat; SFF23 worked when coupled with SFR. The polymerase chain reaction (PCR) mixtures (10 μl) contained 1 μl (50 ng/μl) genomic DNA, 5 μl of 2× buffer, 1.5 μl (50 mM) MgCl2, 1 μl (10 μM) of each primer, and 1 U Taq DNA polymerase (Takara). PCRs were conducted on a DNA Engine Dyad Cycler (BioRad) under the following condition: 5 min of initial denaturation, 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 60 s; and a final extension at 72 °C for 5 min. PCR products were examined on agarose gels and were cloned into the pMD19-T vector (Takara). Positive clones were sequenced on an ABI DNA sequencer using the sequencing primer pair (M13−47: 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ and M13−48: 5′-GAGCGGATAACAATTTCACACAGG-3′). Three to five clones of each PCR product were sequenced from both directions to validate the results. In addition, we sequenced all six exons of the Tas1r2 gene from Myotis lucifugus and Pteropus vampyrus, using primers designed according to their low-coverage genome sequences. All newly acquired sequences have been deposited into the GenBank.
Evolutionary Analysis
Deduced amino acid sequences were aligned with CLUSTALX 1.81 (Thompson et al. 1997) and modified with BioEdit 7.0.4 (Hall 1999). The nucleotide sequence alignment was generated according to the protein sequence alignment. Bat phylogeny and divergence dates were taken from recently published phylogenetic studies (Teeling et al. 2005; Miller-Butterworth et al. 2007). Ancestral sequences were reconstructed using the Bayesian method (Yang et al. 1995) implemented in the BASEML program in PAML 4.1 (Yang 2007) and the parsimony method. Nonsynonymous (dN) and synonymous (dS) nucleotide substitution rates were estimated using the likelihood method implemented in the CODEML program in PAML 4.1 (Yang 2007).
The bat Cytb and RAG2 gene sequences were retrieved from the GenBank, with the accession numbers AY604444, AF382889, DQ077398, FJ155475, FJ155476, AF316447, AF316445, AF316444, AF316454, and AF316438.
Results
Equally Strong Purifying Selection on Tas1r2 of Fruit-Eating Bats and Insect-Eating Bats
Tas1r2 has six coding exons, encoding an approximately 800 amino acid peptide, which is composed of a long extracellular N-terminus, seven transmembrane domains, three extracellular and three intracellular loops, and an intracellular C-terminus (Li et al. 2002). From 40 bat species spanning the entire phylogeny of bats (fig. 1), we sequenced ∼720 nucleotides of the sixth exon of Tas1r2. The sequenced segment corresponds to the region from the end of the N-terminus to the beginning of the seventh transmembrane domain (fig. 2). We also obtained the DNA sequences of the same region from the draft genome sequences of two additional bat species (M. lucifugus and P. vampyrus). The maximum-likelihood tree reconstructed using the 42 bat Tas1r2 sequences is not significantly different from the established bat species tree (Teeling et al. 2005) (Shimodaria-Hasegawa test, P = 0.079), suggesting that the sequences acquired are orthologous. We thus used the established species tree in all subsequent analyses.
FIG. 2.
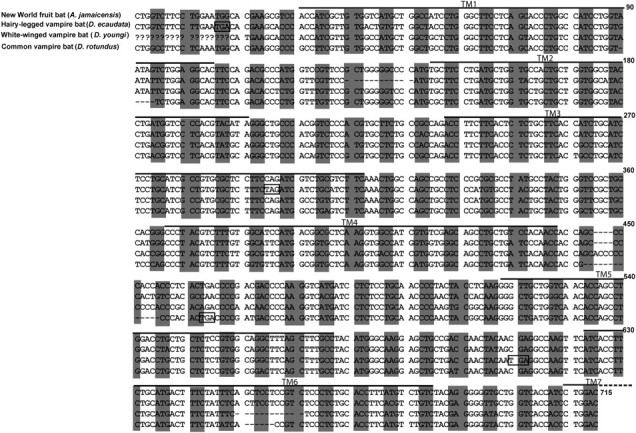
Alignment of Tas1r2 nucleotide sequences from the three vampire bats and one New World fruit bat for the regions examined in this work. Dashes indicate alignment gaps and question marks represent unamplified nucleotides. Codons in the correct reading frame are indicated by shading and premature stop codons are boxed. Regions corresponding to the seven transmembrane (TM) domains are indicated.
With the exception of vampire bats, the open reading frame (ORF) of Tas1r2 is maintained in the sixth exon in all bats surveyed. In addition, we sequenced all six exons of the Tas1r2 gene from M. lucifugus and P. vampyrus and confirmed that the entire Tas1r2 ORF is intact in these species (supplementary fig. S1, Supplementary Material online). To verify that Tas1r2 is under purifying selection and thus is putatively functional in non-vampire bats and to explore the possibility of differential selection on Tas1r2 in fruit eaters and insect eaters, we estimated the ratio (ω) of nonsyonymous to synonymous substitution rates by a likelihood method. We conducted three analyses on the dataset of 39 species, excluding the vampire bats. First, under the assumption of a uniform ω for all branches of the tree of the 39 bats (model A in Table 1), ω was estimated to be 0.197, which is significantly smaller than 1 (P < 10−100; see the comparison with model B in Table 1). This result suggests that Tas1r2 is under an overall strong purifying selection in these bats. Second, we tested whether a model that allows different ω values for fruit eaters and insect eaters (model C in Table 1) fits the data significantly better than a simpler model that does not allow the difference (model D in Table 1). In this analysis, each branch that cannot be classified into the clades of fruit eaters or insect eaters has its own ω (fig. 1). We did not find model C to fit the data significantly better than model D (P > 0.4), indicating similar levels of purifying selection on the Tas1r2 genes of fruit eaters and insect eaters. Third, we examined a model in which every branch has its own ω (model E). This model is not significantly better than the uniform ω model (model A), suggesting that the variation of ω among different lineages of nonvampire bats is minimal.
Table 1.
Likelihood Ratio Tests of Selective Pressures on Bat Tas1r2 Gene.
| Models | ω (dN/dS) | ln La | npb | Models Compared | 2Δ(ln L)c | P Valuesd |
| Dataset I: 39 sequences (all except vampire bats) | ||||||
| A. All branches have one ω | ω = 0.192 | −6223.77 | 78 | |||
| B. All branches have the same ω = 1 | ω = 1 | −6485.21 | 77 | B vs. A | 522.88 | 1.0 × 10−115 |
| C. Branches in yellow have ω1, branches in blue have ω2, and branches a, b, c, and d each has its own ω | ω1 = 0.183, ω2 = 0.205 | −6211.42 | 84 | |||
| D. Branches in yellow have ω1, branches in blue have ω2 = ω1, and branches a, b, c, and d each has its own ω | ω1 = ω2 = 0.196 | −6211.42 | 83 | D vs. C | 0.64 | 0.422 |
| E. Each branch has its own ω | Variable ω by branch | −6189.76 | 153 | A vs. E | 68.02 | 0.703 |
| Dataset II: 40 sequences (dataset I plus the ancestral sequence of vampire bats) | ||||||
| F. All branches have the same ω | ω = 0.211 | −6575.83 | 80 | |||
| G. Branch e (ancestral to vampire bats) has ω2 and other branches have ω1 | ω1 = 0.192, ω2 = 0.994 | −6559.21 | 81 | F vs. G | 33.23 | 8.2 × 10−9 |
| H. Branch e has ω2 = 1 and other branches have ω1 | ω1 = 0.192, ω2 = 1 | −6223.77 | 79 | H vs. G | 0 | 1 |
| Dataset III: 42 sequences (all bats, after removing gaps in pseudogenes) | ||||||
| I. Branch e and branches connecting the three vampire bats have ω2 whereas other branches have ω1 | ω1 = 0.193, ω2 = 0.424 | −6158.29 | 85 | |||
| J. Branch e has ω3, branches connecting the three vampire bats have ω2, and other branches have ω1 | ω1 = 0.193, ω2 = 0.393, ω3 = 911.06 | −6156.79 | 86 | I vs. J | 3.00 | 0.083 |
The natural logarithm of the likelihood value.
Number of parameters.
Twice the difference in ln L between the two models compared.
P values lower than 0.05 are shown in bold.
Pseudogenization of Tas1r2 in Vampire Bats
In striking contrast to the conservation of Tas1r2 in all fruit-eating and insect-eating bats, the ORF of Tas1r2 is disrupted in each of the three vampire bats, which feed exclusively on blood. The ORF-disrupting mutations include nonsense mutations, insertions, and deletions (fig. 2). Specifically, the hairy-legged vampire bat Diphylla ecaudata has a 12-nucleotide deletion and two premature stop codons that would result in the loss of all transmembrane domains in its Tas1r2 protein. The white-winged vampire bat Diaemus youngi has a 12-nucleotide deletion and a 4-nucleotide insertion, which leads to a frame shift and a premature stop codon, resulting in the truncation of the protein in the third transmembrane domain. The common vampire bat Desmodus rotundus has a five-nucleotide deletion, a six-nucleotide deletion, a nine-nucleotide deletion, and a premature stop codon, resulting in the loss of the last two transmembrane domains in its Tas1r2.
Despite the observation of several ORF-disrupting mutations in each vampire bat, none of these mutations are shared among any pair of the three species. However, because the region we sequenced is relatively short, it remains possible that shared ORF-disrupting mutations exist in the regions of Tas1r2 that are not sequenced here. Even if no shared ORF-disrupting mutations exist, it is still possible that the purifying selection on Tas1r2 was completely removed before the divergence of the three vampire bats because it takes a substantial amount of time for an ORF-disrupting mutation to occur and be fixed in a species. Below, we evaluate whether the relaxation of the functional constraint on Tas1r2 started before or after the divergence of the three vampire bats.
We inferred the Tas1r2 sequence of the common ancestor of vampire bats (i.e., the black circle in fig. 1) using a combination of Bayesian and parsimony methods. Because the Bayesian method (Yang et al. 1995) cannot deal with indels, we used this method to infer the ancestral sequence from the alignment of all 42 Tas1r2 sequences for regions without indels. For indel regions, we used parsimony to infer the ancestral sequence. We then estimated ω using the inferred ancestral vampire bat Tas1r2, which has an ORF, with the Tas1r2 sequences from the 39 other bats. Assuming that all branches in the tree of the 39 extant sequences and one ancestral sequence have the same ω (model F in Table 1), we estimated ω = 0.211. If we assume that the branch leading to the ancestral vampire bats has ω2, whereas all other branches have ω1 (model G in Table 1), we found that ω2 = 0.994 and model G fits the data significantly better than model F (P < 10−8). We further found by comparing model G and model H (ω2 is fixed at 1; see Table 1) that ω2 is not significantly different from 1. In addition to this analysis, we removed premature stop codons in the three vampire bat sequences and analyzed them together with the 39 other bat sequences. We found that a model that allows a variation in ω between the stem vampire bat branch and the four branches connecting the three vampire bats (model J in Table 1) is not significant better than a simpler model that assumes the same ω for these five branches (model I in Table 1) (P = 0.083). Together, these results support the hypothesis that a relaxation of functional constraint occurred in the branch leading to the common ancestor of vampire bats and that Tas1r2 may have been evolving neutrally since the divergence of vampire bats from NW fruit bats.
If the above conclusion is correct, what is the probability of the absence of any shared ORF-disrupting mutation in the Tas1r2 region sequenced in the three vampire bats? To answer this question, it is necessary to date the evolutionary events under investigation. We searched GenBank for DNA sequences available in all three vampire bats and found two suitable genes for this analysis: cytochrome b (Cytb) and recombination activating gene 2 (RAG2). The DNA sequences of these two genes are also available in two NW fruit bats, which we use as outgroups (fig. 3a). We concatenated the two genes and reconstructed a neighbor-joining (NJ) tree (fig. 3a), based on Tamura-Nei distances estimated by the maximum composite likelihood method implemented in MEGA4 (Tamura et al. 2007). We could not reject the molecular clock hypothesis using Tajima's test (Tajima 1993) and therefore converted the NJ tree to a linearized tree (Takezaki et al. 1995). Using the calibration time of 26 My since the divergence of NW fruit bats and vampire bats (Teeling et al. 2005), we estimated the divergence times of the three vampire bats (fig. 3b). One can see that the time between the divergence of vampire bats from NW fruit bats and the first speciation within vampire bats is relative short (1.1 My), whereas the time between the two speciation events within vampire bats is relatively long (7.2 My). We then estimated the probability that a Tas1r2 sequence contains no fixed ORF-disrupting mutations after neutral evolution for a given amount of time, using the PSEUDOGENE program (Zhang and Webb 2003). This estimation requires the neutral rates of point mutations and indel mutations. We estimated from a genomic comparison between mouse and rat (Gibbs et al. 2004) that the neutral point mutation rate is 4.83 × 10−9 per site per year and the indel mutation rate is 3.13 × 10−10 per site per year. Furthermore, previous studies estimated that about 83% of indel mutations are not of multiples of three nucleotides and therefore are ORF disrupting (Podlaha and Zhang 2003; Zhang and Webb 2003). Using these parameters and a NW fruit bat Tas1r2 sequence, we estimated that the half-life of Tas1r2 is 2.02 My, meaning that the probability that the ORF is still present in the sequenced Tas1r2 region is 50% after 2.02 My of neutral evolution. Using this half-life, we estimated that the probability of no fixation of ORF-disrupting mutations is P1 = 0.085 after 7.2 My of neutral evolution and is P2 = 0.058 after 8.3 (7.2 + 1.1) My of neutral evolution. Thus, it is possible that no shared ORF-disrupting mutations are observed in Tas1r2 even if the purifying selection on the gene has been completely removed since the separation of vampire bats from NW fruit bats. In the above calculation, we assumed that the mutation rate per site per year is the same in bats and murids. Because the generation time of vampire bats is longer than that of murids, the mutation rate per year may be lower in bats than in murids. Thus, the actual probabilities may be somewhat higher than the above estimates of P1 and P2.
FIG. 3.
Evolutionary trees of combined Cytb and RAG2 genes from the three vampire bats and two New World fruit bats. (a) The NJ tree. Bootstrap percentages from 500 replications are shown on interior branches. (b) The linearized tree. Estimated divergence dates are shown with open arrow heads, whereas the calibration time is indicated with a solid arrow head. The scale shows the Tamura-Nei distance.
As a comparison, we also estimated the selective pressures on Cytb and RAG2 in vampire bats and NW fruit bats. We designated the four branches connecting the three vampire bats as vampire bat branches and the two branches connecting the two NW fruit bats as NW fruit bat branches (fig. 3a). For RAG2, there is no significant difference in ω between the two groups of branches (P > 0.7). For Cytb, ω is significantly higher for vampire bats than for NW fruit bats (P < 0.01). Nonetheless, for both genes, vampire bat ω is significantly lower than 1 (0.158 for RAG2 and 0.028 for Cytb). Thus, the pseudogenization of vampire bat Tas1r2 is not part of any genome-wide relaxation of purifying selection but rather a gene-specific event.
Our multiple analyses of the selective pressure and pseudogenization process are consistent with each other. Together, they suggest that, although no shared ORF-disrupting mutations are observed in the Tas1r2 genes of the three vampire bats, the relaxation of functional constraint already happened in their common ancestor.
Survey of Tas1r2 in Mammalian Genome Sequences
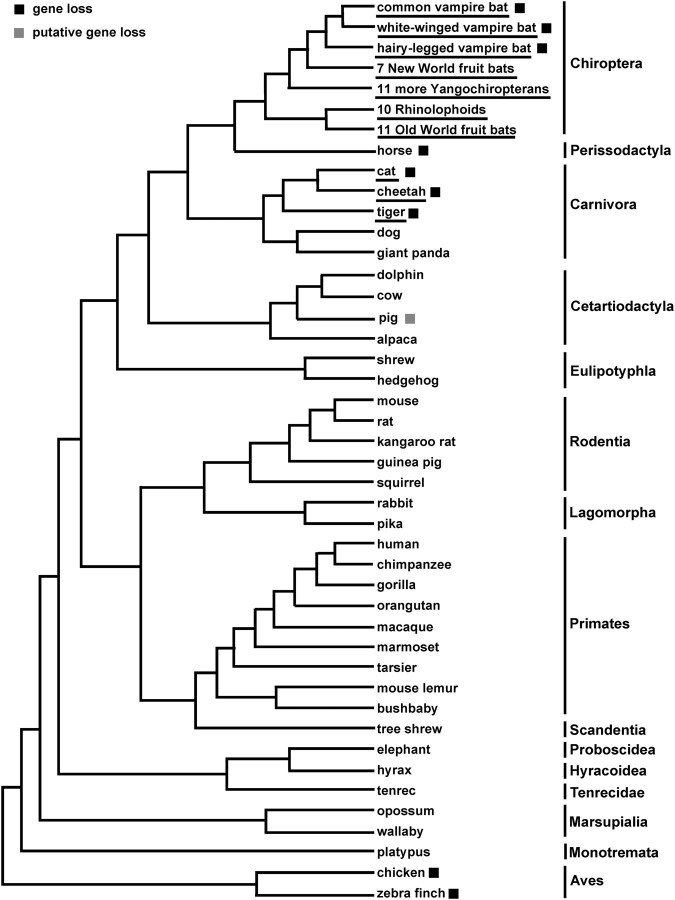
To gain a broader picture of Tas1r2 evolution, we used TBlastN and query sequences from closely related species to search for Tas1r2 in 32 currently available mammalian genome sequences (fig. 4; see the list of species names and gene accession numbers in supplementary table S2, Supplementary Material online). For the majority of the species surveyed, we were able to identify Tas1r2. Although in many low-coverage genome sequences, the identified sequences were incomplete, there were no premature stop codons. However, we were not able to find Tas1r2 from the horse (6.79× coverage) and pig (4×) genome sequences (fig. 4). In human and most species surveyed, PAX7 and ALDH4A1 are the two genes that are adjacent to TAS1R2 on the chromosome. We applied TBlastN to screen these two genes in horse and pig. In horse, PAX7 and ALDH4A1 were found to be next to each other, suggesting that Tas1r2 is truly lost. In pig, PAX7 but not ALDH4A1 was found, suggesting that the absence of Tas1r2 could be due to the incompleteness of the genome sequence, which may not be surprising given the low coverage (4×) of the sequence.
FIG. 4.
A species tree of mammals marked with multiple losses of functional Tas1r2. The Tas1r2 genes studied by amplification and sequencing of Tas1r2 are underlined, whereas those studied by genome sequence analysis are not. Birds are used as the outgroup of mammals. Note that Tas1r2 originated in the common ancestor of bony vertebrates (Grus and Zhang 2009), and therefore, the absence of Tas1r2 in individual mammalian and avian species are due to gene losses.
Consistent with earlier behavioral (Ganchrow et al. 1990) and electrophysiological (Halpern 1962) studies, Tas1r2 was reported to be absent in the chicken genome (Shi and Zhang 2006). Interestingly, we found that Tas1r2 is also missing from zebra finch, the only other bird with an available genome sequence (6× coverage), despite the presence of the two adjacent genes PAX7 and ALDH4A1. Because chicken and zebra finch belong to two distantly related avian orders (Hackett et al. 2008), if the absences of their Tas1r2 genes were due to the same evolutionary loss, Tas1r2 must be absent in a large number of birds including nectar feeders. It seems more likely that the absence of Tas1r2 in chicken and zebra finch were due to independent gene losses, but additional sampling of birds is required to test this hypothesis.
Discussion
In this study, we surveyed the sweet-specific receptor gene Tas1r2 from 42 species of bats to examine how different dietary preferences influence the evolution of Tas1r2. Because fruits and flowers contain glucose, fructose, and sucrose, one may expect Tas1r2 and the sweet taste to be much more important for fruit eaters than insect eaters. However, we found Tas1r2 to be under equally strong purifying selection in fruit eaters and insect eaters. Although the Tas1r2 region we surveyed does not include the putative ligand-binding domain (Cui et al. 2006), a relaxation of selective constraint is unlikely to be limited to that region. In fact, a previous study that documented a relaxation of selective constraint in human bitter taste receptors showed that the relaxation happens in all domains including those not involved in ligand binding (Wang et al. 2004). Furthermore, using the complete Tas1r2 sequences that include ligand-binding domains from P. vampyrus (fruit eater), M. lucifugus (insect eater), and dog, we confirmed that ω is not significantly different between the two bat lineages (P > 0.4). Based on the draft genome sequences, P. vampyrus and M. lucifugus have an intact gene for Tas1r3, which binds to Tas1r2 to form a functional sweet receptor. Thus, the sweet taste is likely under equally strong constraints in fruit eaters and insect eaters. It would be interesting to confirm our results by behavioral studies.
We found that Tas1r2 is a pseudogene in all three blood-feeding vampire bats. This finding appears consistent with previous behavioral studies. Specifically, the taste ability of common vampire bats is poorly developed compared with other mammals; they are able to discriminate salty, sour, and bitter tastants in high concentration but show indifference to sweet tastants even at the highest concentration tested (Thompson et al. 1982). Interestingly, a recent behavioral test found that common vampire bats lack taste-aversion learning, which is critical to poison avoidance (Ratcliffe et al. 2003). It would be interesting to study other taste receptors in vampire bats, especially bitter taste receptors, to understand the genetic basis of the lack of taste-aversion learning in vampire bats. Vampire bats are the only mammals that feed exclusively on blood. Because of the extreme narrowness in diet, their sense of taste is unlikely to be important. Furthermore, vampire bats use olfaction to find prey (Bahlman and Kelt 2006) and use infrared sensors to locate blood sources (Fenton 1992). These capabilities may have further rendered the taste sensitivity less important, as in several previously proposed cases of sensory tradeoffs (Zhang and Webb 2003; Zhao et al. 2009).
In addition to vampire bats, Tas1r2 is also known to be absent in the cat family Felidae (Li et al. 2005), chicken (Shi and Zhang 2006), and the tongueless western clawed frog (Shi and Zhang 2006) since its origin in the common ancestor of bony fishes (Grus and Zhang 2009). Furthermore, we found that Tas1r2 is missing in the genome sequences of horse, pig, and zebra finch, although its absence in pig may be an artifact of the low sequence coverage. Among the species that clearly lack a functional Tas1r2 gene, vampire bats are sanguivorous, cats are carnivorous, horse is herbivorous, chicken is herbivorous and insectivorous, zebra finch is herbivorous, and western clawed frog is insectivorous. The diverse diets of the Tas1r2-lacking vertebrates suggest no uniform dietary reason for the loss of the gene in these species. It is possible that the loss of Tas1r2 in each species has species-specific reasons, as explained earlier for vampire bats. The complexity of the potential ecological factors impacting Tas1r2 evolution calls for more extensive studies of the physiological roles of sweet perception, which apparently is not as conserved as previously thought.
Supplementary Material
Supplementary fig. S1 and table S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank Robert Baker for Desmodus and Dieamus tissues and laboratory support to C.M.P., Dong Xu for assistance with the lab work, Stephen Rossiter for helpful discussion, and Meg Bakewell for valuable comments on an earlier version of the manuscript. This work was supported by the Key Construction Program of the National “985” Project and the “211” Project of China to S.Z. and by a research grant from the U.S. National Institutes of Health to J.Z. and H.Z. was supported in part by the Fundamental Research Funds for the Central Universities.
References
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Altringham JD. Oxford University Press Oxford: 1996. Bats: Biology and Behavior. [Google Scholar]
- Bachmanov AA, Beauchamp GK. Taste receptor genes. Annu Rev Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahlman JW, Kelt DA. Use of olfaction during prey location by the common vampire bat. Biotropica. 2006;39:147–149. [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Conte C, Ebeling M, Marcuz A, Nef P, Andres-Barquin PJ. Evolutionary relationships of the Tas2r receptor gene families in mouse and human. Physiol Genomics. 2003;14:73–82. doi: 10.1152/physiolgenomics.00060.2003. [DOI] [PubMed] [Google Scholar]
- Cui M, Jiang P, Maillet E, Max M, Margolskee RF, Osman R. The heterodimeric sweet taste receptor has multiple potential ligand binding sites. Curr Pharm Des. 2006;12:4591–4600. doi: 10.2174/138161206779010350. [DOI] [PubMed] [Google Scholar]
- Fenton MB. New York: Facts on File; 1992. Bats. [Google Scholar]
- Fischer A, Gilad Y, Man O, Paabo S. Evolution of bitter taste receptors in humans and apes. Mol Biol Evol. 2005;22:432–436. doi: 10.1093/molbev/msi027. [DOI] [PubMed] [Google Scholar]
- Ganchrow JR, Steiner JE, Bartana A. Behavioral reactions to gustatory stimuli in young chicks (Gallus gallus domesticus) Dev Psychobiol. 1990;23:103–117. doi: 10.1002/dev.420230202. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, et al. (230 co-authors) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Go Y, Satta Y, Takenaka O, Takahata N. Lineage-specific loss of function of bitter taste receptor genes in humans and nonhuman primates. Genetics. 2005;170:313–326. doi: 10.1534/genetics.104.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grus WE, Zhang J. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol Biol Evol. 2009;26:407–419. doi: 10.1093/molbev/msn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, et al. (18 co-authors) A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Halpern BP. Gustatory nerve impulses in the chicken. Am J Physiol. 1962;203:541–544. doi: 10.1152/ajplegacy.1962.203.3.541. [DOI] [PubMed] [Google Scholar]
- Herness MS, Gilbertson TA. Cellular mechanisms of taste transduction. Annu Rev Physiol. 1999;61:873–900. doi: 10.1146/annurev.physiol.61.1.873. [DOI] [PubMed] [Google Scholar]
- Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJ, Zuker CS. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- Kinnamon SC, Margolskee RF. Mechanisms of taste transduction. Curr Opin Neurobiol. 1996;6:506–513. doi: 10.1016/s0959-4388(96)80057-2. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Kusakabe Y, Miura H, Ninomiya Y, Hino A. Molecular genetic identification of a candidate receptor gene for sweet taste. Biochem Biophys Res Commun. 2001;283:236–242. doi: 10.1006/bbrc.2001.4760. [DOI] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, et al. (123 co-authors) The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li W, Wang H, et al. (11 co-authors) Pseudogenization of a sweet-receptor gene accounts for cats' indifference toward sugar. PLoS Genet. 2005;1:27–35. doi: 10.1371/journal.pgen.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proc Natl Acad Sci U S A. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann B. Taste reception. Physiol Rev. 1996;76:718–766. doi: 10.1152/physrev.1996.76.3.719. [DOI] [PubMed] [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- Meyerhof W. Elucidation of mammalian bitter taste. Rev Physiol Biochem Pharmacol. 2005;154:37–72. doi: 10.1007/s10254-005-0041-0. [DOI] [PubMed] [Google Scholar]
- Miller-Butterworth CM, Murphy WJ, O'Brien SJ, Jacobs DS, Springer MS, Teeling EC. A family matter: conclusive resolution of the taxonomic position of the long-fingered bats, Miniopterus. Mol Biol Evol. 2007;24:1553–1561. doi: 10.1093/molbev/msm076. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Podlaha O, Zhang J. Positive selection on protein-length in the evolution of a primate sperm ion channel. Proc Natl Acad Sci U S A. 2003;100:12241–12246. doi: 10.1073/pnas.2033555100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe JM, Fenton MB, Galef BG., Jr An exception to the rule: common vampire bats do not learn taste aversions. Anim Behav. 2003;65:385–389. [Google Scholar]
- Roper SD. The cell biology of vertebrate taste receptors. Annu Rev Neurosci. 1989;12:329–353. doi: 10.1146/annurev.ne.12.030189.001553. [DOI] [PubMed] [Google Scholar]
- Sainz E, Korley JN, Battey JF, Sullivan SL. Identification of a novel member of the T1R family of putative taste receptors. J Neurochem. 2001;77:896–903. doi: 10.1046/j.1471-4159.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- Shi P, Zhang J. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol. 2006;23:292–300. doi: 10.1093/molbev/msj028. [DOI] [PubMed] [Google Scholar]
- Shi P, Zhang J, Yang H, Zhang YP. Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol. 2003;20:805–814. doi: 10.1093/molbev/msg083. [DOI] [PubMed] [Google Scholar]
- Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RD, Elias DJ, Shumake SA, Gaddis SE. Taste preferences of the common vampire bat (Desmodus-Rotundus) J Chem Ecol. 1982;8:715–721. doi: 10.1007/BF00988313. [DOI] [PubMed] [Google Scholar]
- Wang X, Thomas SD, Zhang J. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet. 2004;13:2671–2678. doi: 10.1093/hmg/ddh289. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci U S A. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- Zhao H, Rossiter SJ, Teeling EC, Li C, Cotton JA, Zhang S. The evolution of color vision in nocturnal mammals. Proc Natl Acad Sci U S A. 2009;106:8980–8985. doi: 10.1073/pnas.0813201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yang J-R, Xu H, Zhang J. Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol Biol Evol. 2010 doi: 10.1093/molbev/msq153. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.