Abstract
Summary: PedMerge allows users to accurately and efficiently merge separately ascertained pedigrees that belong to the same extended family. In addition to validation checks of pedigree structure, the software provides files in LINKAGE or PEDSYS format that easily allow to be used by a variety of genetic statistical software packages including LINKAGE, SOLAR, SLINK or can be further manipulated with Mega2.
Availability: http://sites.google.com/site/rosemarieplaetke/home/s/pedmerge
Contact: rplaetke@alaska.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
To investigate the genetic background of a disease or other traits, a variety of genetic statistical software has been developed (Stein and Elston, 2009). The development of this type of analytical software has been phenomenal. However, researchers continue to have to manage a variety of complex, time-consuming tasks to prepare data for these analyses, indicating that the development of data management software has not kept up with that of analytical tools. For example, data management of extended families remains still tedious, especially when pedigree ascertainment is ongoing, pedigree structure continues to change and interim analyses need to be performed. Additionally, fieldwork may demand a more flexible handling of pedigree data when, e.g. families are large and include multiple and/or consanguineous marriages/matings.
To date, this kind of merging can only be performed in a restricted way. For example, the pedigree drawing program Progeny allows merging of pedigrees in the drawings. However, pedigrees with multiple and/or consanguineous marriages/matings can have complex visual presentations. Merging them in this way may not be easy and error prone (e.g. linking false individuals). No summary report of the merge is provided, thus making it difficult to document and quality-control the development of a pedigree. The pedigree drawing programs Cyrillic and Pedigree-Draw cannot perform such a merge at all.
PedMerge fills in this gap. It supports pedigree management in any stage of fieldwork and data management without any restrictions on size, number of marriages/matings or consanguinity loops. It therefore can be used for pedigree management in (i) human genetics and (ii) animal breeding projects (see, examples below), as well as (iii) pedigree projects investigating, e.g. inbreeding loads and evolutionary potentials of (endangered) wild animal populations (Pemberton, 2008).
2 FEATURES
PedMerge provides a text-based, menu-driven interface. To invoke a merge the user needs to (i) specify two input files, (ii) select the format of the pedigree output file and (iii) specify whether to keep (default) additional information in the pedigree output file. Before merging it validates the pedigree structures and performs completeness and consistency checks.
2.1 Input files and options
PedMerge needs two input files. One file provides the pedigree structure of the sub-pedigrees and is in pre-Makeped LINKAGE format (Terwillger and Ott, 1994). Other data including phenotypes and genotypes can be included or excluded in the merge according to the user's preference.
The second input file contains information about the ‘key-individuals’, i.e. individuals that occur in two or more sub-pedigrees and are the components connecting them. A key-individual can be a spouse in one sub-pedigree and a child in another one and/or has multiple mates. The program allows an unlimited number of key-individuals in a family. The number and size of sub-pedigrees to be merged are also unlimited.
2.2 Output files
The user can choose to obtain a pedigree output file in pre-Makeped LINKAGE format or a Master file for PEDSYS, a database to manage pedigrees that is freely available from the Southwest Foundation for Biomedical Research (San Antonio, TX, USA). Optional, these two output files can contain additional information, e.g. phenotypic or genotypic data. If necessary, these output files can be further managed with Mega2 (Mukhopadhyay et al., 2005). A log file is provided that (i) reports the findings of the various validations checks, and (ii) summarizes the merge and assignment of new pedigree and individual IDs to allow the tracking of the entire merging process.
2.3 Implementation
Pedmerge is written in C# and interactively runs on versions of Windows but also on Linux and Mac OS X. We successfully ran it on the last two operating systems by using Mono, an open source implementation of Microsoft's .NET Framework that allows cross platform development (http://www.mono-project.com).
Detailed documentation of the software, how to set it up and example input files for testing are available on our website.
3 EXAMPLES OF APPLICATION
(I) CANHR Study: PedMerge simplifies and accelerates pedigree management for repeated analyses while ascertainment is ongoing. We performed variance component analyses and power evaluations with SOLAR (Almasy and Blangero, 1998) for a genetic obesity project at CANHR (Boyer et al., 2005; Plaetke et al., 2006). Family ascertainment was ongoing and performed with Progeny. After each field visit, several pedigree structures had changed because of recruitment of additional relatives and/or merging of separate pedigrees that had been found to be related. Additional merging with PedMerge was then performed. For the final analysis, 26 partially modified sub-pedigrees consisting of 507 participants were merged with PedMerge to one pedigree consisting of 986 individuals.
(II) Alaskan Husky Dog Study: Convenient and fast data preparation when pedigree structures are provided in different formats. Our current ascertainment of extended pedigrees for a behavior genetics study in Alaskan Husky dogs provides several challenges that can be managed more easily with PedMerge. (i) Pedigrees from kennels have been separately ascertained; ascertainment is ongoing. Controlled mating between dogs from different kennels can produce pedigrees that are related among kennels. (ii) Owners track their breeding efforts by collecting pedigree information themselves and using different approaches. In this case, we directly obtain from them their pedigree descriptions.
For example, one kennel provided basic information (i.e. name, sex, birth date) of each dog and its parents. Constructing these pedigrees was straight forward. We entered the data of these nuclear pedigrees, determined the key-individuals and applied PedMerge. In this case, data entry was done within 1.5 h. PedMerge needed less than a second to merge 19 pedigrees consisting of 89 individuals to finally obtain four merged pedigrees with a total of 70 dogs.
From another kennel we obtained four-generation ‘upside-down’ pedigrees, each showing the names of an individual currently in the kennel, its siblings belonging to the same litter, parents, two grandparents and four great-grand parents. For PedMerge, we generated the input files and checked data of nuclear two or three generation pedigrees within 3 h. The merging of these 27 pedigrees (Fig. 1A) consisting of 142 dogs to one large pedigree of 102 individuals lasted less than a second. We performed a second merge ‘by hand’ that needed 12 h. It included multiple checking of data but no data entry for further analysis. Because of multiple matings among dogs, two visual presentations were necessary to detect key-individuals, perform the manual merge and assign the new individual IDs (Fig. 1).
Fig. 1.
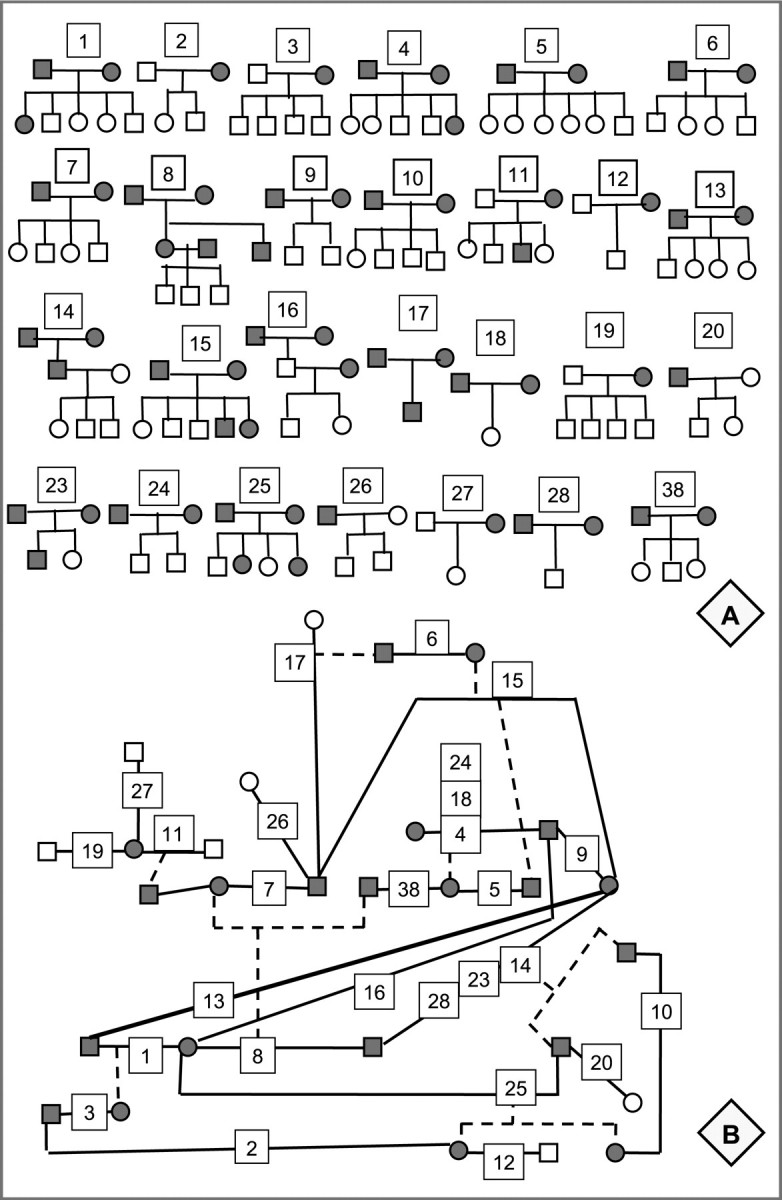
Demonstration of a ‘manual’ pedigree merge. (A) Nuclear pedigrees obtained from original data (key-individuals: dark symbols). (B) Structure of merged pedigree showing matings and key-individuals. If a child is a key-individual, it is connected to its parents by a dashed line. Numbers show which pedigree in (A) belongs to a mating. For example, mating 24–18–4 shows that these dogs mated three times and had 3 litters. New IDs for the individuals in the merged pedigree was given by simultaneously working through pedigrees in (A) and (B).
Funding: National Center for Research Resources at the National Institutes of Health (P20 RR16430). Software development: Federico Balbi. We thank the dog owners for providing the pedigree information of their dogs.
Conflict of Interest: none declared.
REFERENCES
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer BB, et al. Building a community-based participatory research center to investigate obesity and diabetes in Alaska Natives. I nt. J. Circumpolar Health. 2005;64:300–309. doi: 10.3402/ijch.v64i3.18002. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay N, et al. Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- Pemberton JM. Wild pedigrees: the way forward. Proc. R. Soc. B. 2008;275:613–621. doi: 10.1098/rspb.2007.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaetke R, et al. Linkage analysis of obesity related traits on chromosomes 3, 7, 10 and 17 in a Yup'ik Eskimo population. Annual Conference 2006 of the American Society of Human Genetics; 2006. available at http://www.ashg.org/genetics/ashg06s/index.shtml (last accessed date October 10, 2006) [Google Scholar]
- Stein CM, Elston RC. Finding genes underlying human disease. Clin. Genet. 2009;75:101–106. doi: 10.1111/j.1399-0004.2008.01083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Ott J. Handbook of Human Genetic Linkage. Baltimore: Johns Hopkins University Press; 1994. [Google Scholar]


