Abstract
The massive amount of genomic sequence data that is now available for analyzing evolutionary relationships among 31 placental mammals reduces the stochastic error in phylogenetic analyses to virtually zero. One would expect that this would make it possible to finally resolve controversial branches in the placental mammalian tree. We analyzed a 2,863,797 nucleotide-long alignment (3,364 genes) from 31 placental mammals for reconstructing their evolution. Most placental mammalian relationships were resolved, and a consensus of their evolution is emerging. However, certain branches remain difficult or virtually impossible to resolve. These branches are characterized by short divergence times in the order of 1–4 million years. Computer simulations based on parameters from the real data show that as little as about 12,500 amino acid sites could be sufficient to confidently resolve short branches as old as about 90 million years ago (Ma). Thus, the amount of sequence data should no longer be a limiting factor in resolving the relationships among placental mammals. The timing of the early radiation of placental mammals coincides with a period of climate warming some 100–80 Ma and with continental fragmentation. These global processes may have triggered the rapid diversification of placental mammals. However, the rapid radiations of certain mammalian groups complicate phylogenetic analyses, possibly due to incomplete lineage sorting and introgression. These speciation-related processes led to a mosaic genome and conflicting phylogenetic signals. Split network methods are ideal for visualizing these problematic branches and can therefore depict data conflict and possibly the true evolutionary history better than strictly bifurcating trees. Given the timing of tectonics, of placental mammalian divergences, and the fossil record, a Laurasian rather than Gondwanan origin of placental mammals seems the most parsimonious explanation.
Keywords: continental drift, Cretaceous warming, genome analysis, hybridization, phylogenomics, split decomposition
Introduction
As genomic sequences are the ultimate source of molecular data for evolutionary studies, the wealth of information available from the whole-genome sequencing of metazoans has revolutionized evolutionary studies. After publication of the human genome (Lander et al. 2001; Venter et al. 2001), it was decided that low-coverage (2×) genome sequences from additional mammalian species would be beneficial for more accurate sequence annotation and for studying the evolution of disease genes. The data from these genome projects also enable us to study the evolution of mammalian lineages with a previously unimaginable amount of data, opening the filed of phylogenomics. The first phylogenomic study of placental mammals initially involved some 200,000 nucleotides (nt) of protein-coding sequences (Nikolaev et al. 2007). Improving the sequence and taxon coverage, phylogenomic analyses of mammals included 2.2 million nucleotides (Mnt) from about 2,840 protein-coding genes (Hallström et al. 2007) or some 1,700 conserved genome loci (Wildman et al. 2007). The largest and most complete data set so far analyzed more than 2.8 Mnt from 3,012 protein-coding genes (Hallström and Janke 2008). However, phylogenomics has failed to reliably resolve certain branches of the placental mammal tree, thereby posing new problems and questions in animal evolution (Hallström and Janke 2008).
Despite the amount of available data, some branches in the placental mammal tree are only weakly supported and phylogenomic analyses leave some branches as yet completely unresolved due to the variable and poor support for different evolutionary scenarios (Hallström and Janke 2008). This was unexpected because, theoretically, the sheer amount of genomic data should have easily overcome stochastic errors from single and multiple (≈20) gene analyses, a problem that vexed molecular phylogenetic studies before the wealth of information from the genomic age (Kullberg et al. 2008).
To date, three important but difficult to resolve branching points in the mammalian tree have been identified. The first is the primary branching among placental mammals, that is, between the superorders Xenarthra (in this study: sloth and armadillo), Afrotheria (elephant, tenrec, and hyrax), and Boreoplacentalia (all remaining species used in this study). Previous resolutions of their divergences depended on the choice of analytical methodology and type of data but did not enable the rejection of alternative hypotheses (Nishihara et al. 2007; Hallström and Janke 2008). Even the analyses of retroposon insertions, otherwise generally regarded as a solid phylogenetic marker system, failed to resolve a clear bifurcation in the most basal divergences among placental mammals (Churakov et al. 2009; Nishihara et al. 2009), thus, supporting the original sequence-based findings (Hallström and Janke 2008). Retroposon insertions are, with a few exceptions (Cantrell et al. 2001; van de Lagemaat et al. 2005), free of homoplasies (Steel and Penny 2000). Therefore, the apparently contradicting results from sequence data and retroposon insertion analyses require a natural explanation. Similar observations have also been made for the other two poorly or unresolved mammalian phylogenetic branches. The detection of apparently conflicting phylogenetic signals for the position of the Scandentia (tree shrews) relative to Primates and Glires (rodents and lagomorphs) within the Euarchontoglires clade, and the position of Chiroptera (bats) relative to Artiodactyla (even-toed ungulates), Carnivora, Lipothypla (hedghog, common shrew, and allies), and Perissodactyla (odd-toed ungulates) within the Laurasiaplacentalia, leave both controversial and poorly supported (Nishihara et al. 2006; Janecka et al. 2007; Kriegs et al. 2007; Hallström and Janke 2008).
A common feature of these three problematic divergences is the divergence of groups within 1–4 million years (My) of one another (Hallström and Janke 2008). Such time intervals may have been too short for the respective genomes to gather enough substitutions to resolve rapid divergences some 100 My later or limited taxon sampling may impede such phylogenetic analyses. Another possible reason for the problems involved in clearly resolving short central branches of the placental mammalian tree by sequence and retroposon data may be speciation-associated processes, such as species hybridization and incomplete lineage sorting (Nei 1987). Species hybridization leads to introgression, the incorporation of genes from one species into the gene pool of another species, whereas incomplete lineage sorting produces a pattern of allele fixations from ancestral polymorphisms that does not reflect the species history. Both processes generate mosaic genomes in which different loci support alternative mammalian relationships (Hallström and Janke 2008; Churakov et al. 2009). This type of reticulated evolution and incomplete lineage sorting can produce conflicting evolutionary signals. Split network methods can better illustrate and explore such conflicts in the data than can traditional analyses that seek a strictly bifurcating tree (Huson and Bryant 2006) and can be used to detect important processes in the evolution of the placental mammal genome.
Although the exact reasons for the problematic resolution of certain branches in the mammalian tree are currently unknown, they seem to be connected to the rapid radiation of early placental mammals in the mid-Cretaceous (Hallström and Janke 2008). Such a process could have been triggered by a swift mid-Cretaceous temperature increase that reached a maximum at the Cenomanian–Turonian boundary at 93.5 million years ago (Ma) (Retallack 2002) and fragmentation of the supercontinents Laurasia and Gondwana at about the time of the earliest divergences of placental mammals (Smith et al. 2004; Reeves 2009; www.reeves.nl).
We have built a new alignment, including recently released genome data, to further investigate problematic branches in the mammalian tree and to study the tree likeness of the mammalian genome data. The number of placental mammals included in this study has been increased to 31, 12 (63%) more than in a previous phylogenomic analysis (Hallström and Janke 2008), and the genome data from six outgroup species were included to ensure a solid root of the tree from a maximal taxon sampling. The previously unresolved branchings inside the Laurasiaplacentalia, which include Perissodactyla, Carnivora, Artiodactyla, Cetacea, Chiroptera, and Lipotyphla (Arnason et al. 2008), may be clarified by the new genome data of a perissodactyl (horse), a second chiropteran (mega bat), a cetacean (dolphin), and an artiodactylan (alpaca). The genome data from a hyracoid (hyrax) and a second xenarthran (sloth) are invaluable additions to the hitherto long and undivided branches of Afrotheria and Xenarthra, allowing the basal divergences among placental mammals to be examined in more detail. Computer simulations were made to estimate the amount of data needed to resolve short branches, and finally, split decomposition methods (Huson and Bryant 2006) were used for visualizing conflict in the tree.
Materials and Methods
Data
Predicted cDNA sequences from all tetrapods with assemblies and gene builds in release 54 of ENSEMBL were downloaded from ftp://ftp.ensembl.org/pub/current_fasta/. In total, 37 species (table 1) were included in the data build. The taxon sampling represents 16 of the 21 extant eutherian orders. The sequence data from metatherian (marsupials), prototherian (monotremes), avian (bird), reptile, and amphibian species were collected for rooting the placental mammal tree. For increasing the usable sequence length several outgroups were included in the analysis.
Table 1.
Scientific and Common Names of Species Included in the Phylogenomic Analyses.
| Species | Common Name |
| Homo sapiens | Human |
| Pan troglodytes | Chimpanzee |
| Gorilla gorilla | Gorilla |
| Pongo pygmaeus | Orangutan |
| Macaca mulatta | Macaque |
| Otolemur garnettii | Galago |
| Microcebus murinus | Mouse lemur |
| Tarsius syrichta | Tarsier |
| Tupaia belangeri | Treeshrew |
| Rattus norvegicus | Common rat |
| Mus musculus | House mouse |
| Dipodomys ordii | Kangaroo rat |
| Spermophilus tridecemlineatus | Ground squirrel |
| Cavia porcellus | Guinea pig |
| Oryctolagus cuniculus | Rabbit |
| Ochotona princeps | Pika |
| Felis catus | Cat |
| Canis familiaris | Dog |
| Equus caballus | Horse |
| Vicugna pacos | Alpaca |
| Bos taurus | Cow |
| Tursiops truncatus | Dolphin |
| Myotis lucifugus | Little brown bat |
| Pteropus vampyrus | Large flying fox |
| Sorex araneus | Common shrew |
| Erinaceus europaeus | Western European hedgehog |
| Loxodonta africana | African elephant |
| Echinops telfairi | Tenrec |
| Procavia capensis | Hyrax |
| Dasypus novemcinctus | Armadillo |
| Choloepus hoffmanni | Sloth |
| Monodelphis domestica | Opossum |
| Ornithorhynchus anatinus | Platypus |
| Gallus gallus | Chicken |
| Taeniopygia guttata | Zebra finch |
| Anolis carolinensis | Anole lizard |
| Xenopus tropicalis | Frog |
Assembly
Orthologous cDNA sequences were detected using the recursive Blast method (Hallström and Janke 2008) with a cutoff e-value of 10−12. This approach efficiently precludes paralogous sequences from gene families being included in the analysis without assuming a phylogenetic tree. In cases where several transcript variants were present for a gene, the longest one was used. The sequences were translated to amino acids (aa), and multiple sequence alignments from the identified orthologs were constructed with the program MUSCLE (Edgar 2004) for all ortholog groups containing at least 20 species. The alignments were trimmed by removing all columns containing gaps, and alignments with an observed (p) aa distance larger than 40% were excluded from the analysis. Finally, nt alignments were constructed with the assistance of the aa alignments. A customized Perl script produced and drew a presence/absence matrix for illustrating the data density. Each row in the density map refers to a species and each column to a gene. Any gene that is present in a species is marked with a black dash. The matrix is sorted in both directions, placing the species with the best coverage at the top of the graph and the genes represented by most species to the left.
Phylogenetic Analysis
Maximum likelihood (ML) analyses were performed as described in detail in Hallström and Janke (2008) and are therefore only briefly outlined here: Treefinder (TF) version October 2008 (Jobb et al. 2004) was used with the general time reversible (GTR) model (Lanave et al. 1984) assuming rate heterogeneity with eight classes of gamma-distributed rate categories (Yang 1994) and one class of invariable sites (8Γ + I) for nt sequences. For aa sequence analyses, the WAG2000 model, WAG, (Whelan and Goldman 2001) with a rate heterogeneity and invariable sites model (8Γ + I) was used for the ML analyses. Uncertain or controversial relationships were further analyzed by an extended ML analysis. Alternative topologies were statistically evaluated in TF using Shimodaira–Hasegawa probabilities (pSH; Shimodaira and Hasegawa 1999). Neighbor joining (NJ) analyses were done in the SplitsTree4 program (Huson and Bryant 2006) on the WAG2000 + Γ + I model using parameters estimated by TF. The complete alignment was analyzed according to these parameters. In addition, single gene alignments, selected for the 10% of the longest sequences, were evaluated for their phylogenetic content using for simplicity and speed of computation the GTR 8Γ + I model.
Dating
Divergence times were estimated from both aa and nt sequence data using the nonparametric rate smoothing method on a logarithmic scale (NPRS-LOG) implemented in TF (Jobb et al. 2004). The ten fossil-based age constraints that were used to calibrate the tree were taken from Benton et al. (2009) and are detailed in table 2. Mean values and their standard deviations were calculated from the branch lengths of 100 bootstrapped ML analyses of aa and nt sequences.
Table 2.
Calibration Points Used for Dating Mammalian Divergences.
| Split | Minimum Age (Ma) | Maximum Age (Ma) | |
| Eutheria | Metatheria | 124 | 138.4 |
| Boreoplacentalia | Exafrotheriaplacentalia | 61.5 | 113 |
| Euarchontoglires | Laurasiaplacentalia | 61.5 | 113 |
| Primates | Glires | 61.5 | 100.5 |
| Lagomorpha | Rodentia | 61.5 | 100.5 |
| Cetacea | Cow | 70 | 52.4 |
| Caniformia | Feliformia | 39.7 | 63.8 |
| Apes | Old World monkeys | 23.5 | 33.7 |
| Rat | Mouse | 10.4 | 12.3 |
| Human | Chimpanzee | 6.5 | 10 |
Network
The sequence data were analyzed by the SplitsTree4 program using the neighbor-net method from aa ML distances under the WAG2000 model of sequence evolution accounting for rate heterogeneity (gamma) and invariable sites as in the ML analyses. The retroposon data from Churakov et al. (2009) were recoded and simplified for the presence (1) and absence (0) of retroposons in human, elephant, armadillo, and opossum. The data matrix was then analyzed by SplitsTree4 and presented as a split network simply by plotting each retroposon insertion event onto the corresponding internal branch. A chi-square test was performed in Excel for evaluating the uniformity of distribution of retroposon insertions between the three possible topologies.
Simulation
The amount of sequence data needed to resolve a branch that would lead to a hypothetical bifurcating Exafroplacentalia clade was estimated by computer simulation using the Seq-Gen program (Rambaut and Grassley 1997). The simulations of aa sequences were performed on the tree topology, and branch lengths obtained from the actual data utilizing the observed average aa composition and rate heterogeneity. The time interval from the divergence of Afrotheria and Exafroplacentalia until Exafroplacentalia itself split into Boreoplacentalia and Xenarthra was varied by changing the branch length of this internal node to 1/2, 1/3, 1/4, and 1/5th of the observed length. The amount of sequence data required for providing pSH values below 0.05 for both the two alternative hypotheses was recorded and plotted on a line graph. In parallel, the effect of missing data on the tree reconstruction was investigated by removing characters from the simulated data sets. The removal was done according to the proportions of missing data in each individual species of the original data. The impact on the amount of data required for statistical significance was recorded.
Results
Data Set and Quality
The total length of the filtered alignment data was 2,863,797 nt, derived from 3,364 genes of 31 placental mammal species plus 6 outgroups. The average lengths of the individual sequences were 851 ± 667 nt and the average observed distance between human and platypus was 15.7 ± 9.9%.
Figure 1 illustrates the data density. Each row represents a species and a black dash denotes the presence of a gene in the alignment. The average sequence coverage for mammalian species was 79%. The poorest sequence coverage among the placentals was observed for the common shrew, which was represented in 62% of the alignment. The nonmammalian outgroups had a sequence coverage of 49–82%, which, as expected, was generally lower than the mammalian average. However, the marsupial species, opossum, had an above average coverage of 82%, because, as one of the better quality genomes, it has been sequenced with over 7-fold coverage. Also, compared with nontherian outgroups, the opossum's close relation to placental mammals facilitated ortholog detection.
FIG. 1.
Presence/absence matrix for illustrating sequence density. Each dash indicates the presence of a given gene (x axis) in a given species (y axis). The matrix is sorted for maximum sequence density along both axes, placing the species with the best coverage at the top of the plot and the genes represented by the most species at the left.
The nt and aa frequencies appeared to be very similar among the species, but due to the large size of the data set, a possibly overly strict chi-square test rejected compositional homogeneity for many species, even for R/Y-coded nt data. The data properties, such as distance distribution, character composition, evolutionary rates, and type of genes, resemble those of previous phylogenomic studies and are not described in detail here (Hallström et al. 2007; Hallström and Janke 2008).
Tree
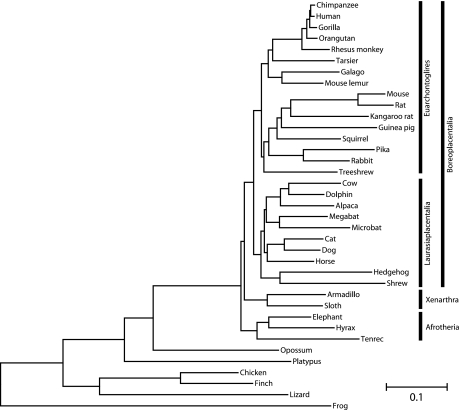
The ML tree based on aa sequence and a WAG + 8GI model is shown in figure 2. The tree generally conforms to that of previous phylogenomic studies (Hallström et al. 2007; Wildman et al. 2007; Hallström and Janke 2008). Most branches are supported by unit ML bootstrap and TF support values. Despite the increased taxon sampling, the same problematic branches found in previous studies were identified here. Thus, the earliest divergences of placental mammals, the Xenarthra, Afrotheria, and Boreoplacentalia, are only marginally supported. In this analysis of nt and aa sequences, Afrotheria represents the earliest split from the eutherians, but alternative topologies cannot be rejected by pSH tests based on different data types (supplementary table 1, Supplementary Material online). The Epitheria hypothesis, Afrotheria and Boreoplacentalia as sistergroups, receives the least support. The grouping of the tree shrew (Scandentia) differs from previous phylogenomic studies. In this study, Scandentia group with Glires (Rodentia plus Lagomorpha), but a grouping with primates or a position outside a primate/Glires clade cannot be rejected by a pSH analysis (supplementary table 2, Supplementary Material online). Finally, the phylogenetic position of the Chiroptera (bats) among the Laurasiaplacentalia receives only limited branch support from TF, 91% for NT12 and 95% for aa, and in pSH analyses, three alternative positions receive probabilities >0.05 (supplementary table 3, Supplementary Material online) and cannot be formally rejected by all data types. However, the Pegasoferae hypothesis, Chiroptera as sistergroup to Perissodactyla plus Carnivora, receives only low support and can be rejected on the basis of aa and NT12 ML analyses.
FIG. 2.
ML tree reconstructed from an alignment length of 954 ka aa from 37 species under the WAG2000 + 8GI of sequence evolution.
The pSH analysis remained ambiguous about the best supported tree when only one outgroup, the opossum, was used for these analysis (not shown) and the amount of usable data was reduced by 12%. Therefore, all analyses were done using multiple outgroups.
Dating
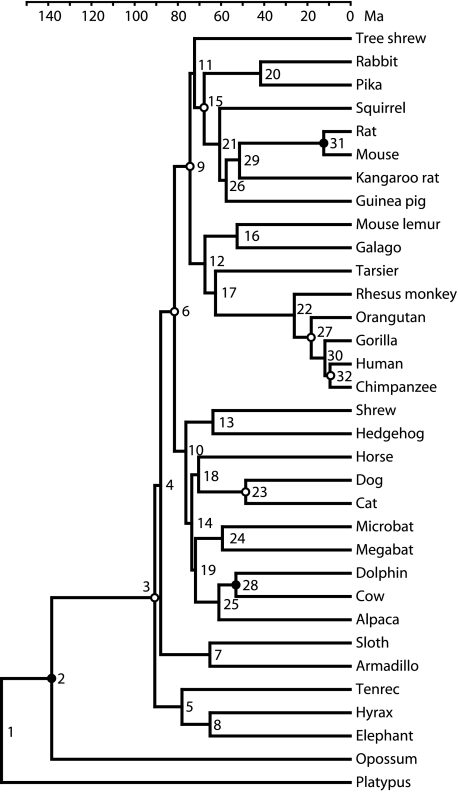
The estimations of divergence times shown in the chronogram of figure 3 and detailed in table 3 are based on the topology depicted in figure 2. The dating was performed solely by the NPRS-LOG method because previous studies showed virtually no differences relative to other algorithms (Roos et al. 2007; Nilsson et al. 2010). The numerous calibrating points are marked with circles. A circle is filled when the divergence time estimate reached either the upper or lower bound and open when it stayed anywhere between the boundaries. The most influential calibration point in this study was that between dolphin and cow (Cetacea and Artiodactyla). The lower bound (minimum) is given with 52.4 Ma as the latest possible divergence time between cetaceans from the remaining artiodactyls (Benton et al 2009). At this time, however, cetacean characters have already evolved and slightly older divergence times have been suggested by others (Bajpai and Gingerich 1998; Arnason et al. 2000; van Tuinen and Hadly 2004). Each million year, this lower bound is moved back in time almost equally affects all earlier divergences. None of the other calibration points exhibits an equally strong effect on the divergence time estimate, and most are estimated within their boundaries.
FIG. 3.
Chronogram of mammalian divergences. Open and filled circles indicate calibration points: Open circles represent those estimated divergence times anywhere between the boundaries, filled circles those reaching either the upper or lower boundary.
Table 3.
Divergence Times and Their Standard Deviation for Splits Shown in Figure 3.
| Split | Divergence Time (Ma) |
| 1 | 161.7 ± 0.35 |
| 2 | 138.4a |
| 3 | 90.60 ± 0.11 |
| 4 | 87.89 ± 0.12 |
| 5 | 78.01 ± 0.21 |
| 6 | 81.48 ± 0.32 |
| 7 | 65.08 ± 0.37 |
| 8 | 64.98 ± 0.38 |
| 9 | 74.28 ± 0.15 |
| 10 | 76.22 ± 0.30 |
| 11 | 72.18 ± 0.16 |
| 12 | 67.34 ± 0.27 |
| 13 | 63.64 ± 0.42 |
| 14 | 73.47 ± 0.21 |
| 15 | 67.72 ± 0.21 |
| 16 | 52.45 ± 0.33 |
| 17 | 62.47 ± 0.26 |
| 18 | 70.31 ± 0.26 |
| 19 | 71.75 ± 0.25 |
| 20 | 41.58 ± 0.17 |
| 21 | 60.54 ± 0.22 |
| 22 | 25.95 ± 0.17 |
| 23 | 48.48 ± 0.16 |
| 24 | 59.27 ± 0.26 |
| 25 | 60.90 ± 0.26 |
| 26 | 57.50 ± 0.21 |
| 27 | 18.10 ± 0.14 |
| 28 | 52.4a |
| 29 | 51.33 ± 0.20 |
| 30 | 11.76 ± 0.08 |
| 31 | 12.30a |
| 32 | 9.376a |
Estimates that have reached a limit set by the calibration point.
In the current phylogenomic ML tree the most basal divergence of placental mammals, between the Afrotheria and the remaining placental mammals, occurs at 90.6 Ma. Only 2.7 My later, at 87.9 Ma, the xenarthrans diverge from the Boreoplacentalia. Most other ordinal divergences occur between 80 and 65 Ma. The weakly supported position of the tree shrew and chiropterans are correlated with short divergence intervals of 2.1 and 2.0 My, respectively (table 3). Internal branches that have durations >4 My or are short but very recent are significantly supported despite compositional biases or other possible systematic errors in the data. Thus, the divergences among human, chimpanzee, and gorilla occur within 2 My but are significantly resolved. The slightly older age of the divergence of Afrotheria relative to our other phylogenomic studies is a consequence of the tree topology. The divergence times are similar or in some cases younger, except for the basal divergences, than those estimated before (Hallström and Janke 2008). This is probably due to the increased taxon sampling and larger number of calibration points. When the tree is constrained to the Xenafrotheria hypothesis (Afrotheria plus Xenarthra), a probable alternative (Hallström and Janke 2008), the early divergence times become younger, but other parts of the tree are left unaffected.
Network Analyses
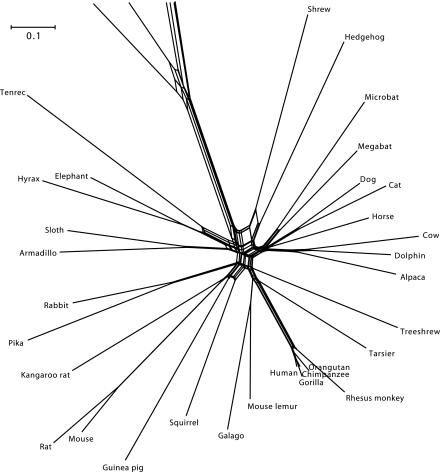
The neighbor-net based on aa sequence data is shown in figure 4 and a magnification of its central region with labels for the major splits in figure 5. The neighbor-net includes the tree in figure 1 but appears to favor the Xenafrotheria (Afrotheria plus Xenarthra) hypothesis. A NJ analysis conforms to the Xenafrotheria hypothesis, and NJ bootstrap supports this grouping with 90% and most others with unit support (Supplementary figure. 1, Supplementary Material online).
FIG. 4.
Neighbor-net based on the 954 ka aa alignment from 37 species. All intraordinal, ordinal, and most superordinal relationships are clearly defined in the neighbor-net by stretched boxes that are longer than they are wide, indicating limited conflict in the data. The clades that are poorly supported by ML aa sequence data analysis are characterized by boxed nodes that are nearly square or by negligibly short branch lengths.
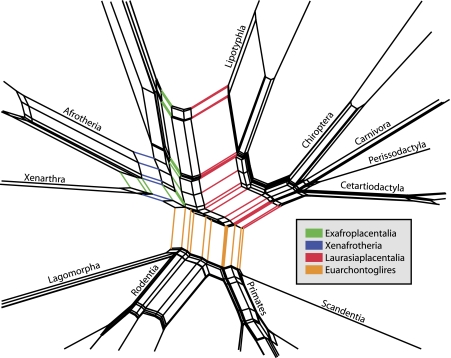
FIG. 5.
Close up of the neighbor-net highlighting major splits.
The data conflict is best exemplified for the splits of the Xenarthra, Afrotheria and Boreoplacentalia, certain splits within the Euarchontoglires, and particular relationships among the Laurasiaplacentalia. These branches are generally also those that were identified to be problematic by individual ML analyses. Thus, the placement of the Chiroptera and Lipotyphla relative to Carnivora, Perissodactyla, and Cetartiodactyla, as well as Scandentia relative to primates and Glires are uncertain by the neighbor-net analysis.
Differing from the ML tree, the neighbor-net shows a tendency to group the Chiroptera with a Carnivora, Perissodactyla, and Cetartiodactyla (Cetacea plus Artiodactyla) clade. Deep divergences of well-defined and supported groups, like Laurasiaplacentalia, Afrotheria, or Rodentia are separated by stretched boxes, illustrating a strong signal and limited conflict in the data, in agreement with the ML analysis. However, even among clearly resolved species, neighbor-net has the power to indicate possible conflict in the data, as exemplified in the cases of some primate, the rodent, or carnivore divergences. The reason for this conflict remains unknown, however.
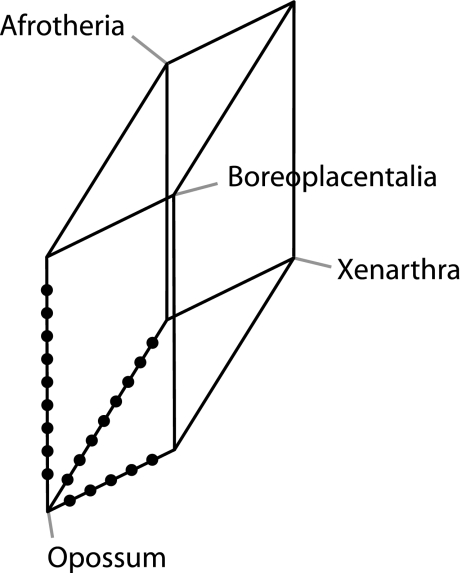
The tenth percentile of the longest genes (336 sequences) exceeded lengths of 1,850 nt. Even though phylogenetic analyses of some genes resolve particular branches as in the ML tree shown in figure 2, a majority-rule consensus tree of ML trees from individual genes resulted in a star-like tree at the ordinal level and above, resolving only relationships among the most closely related taxa. A consensus network using a 2% or 4% threshold value did not show more or other structures than the neighbor-net (Supplementary figure. 2a, and 2b, Supplementary Material online). Finally, a network analysis has been done for about 500 selected genes that are capable of rejecting two of the three possible hypotheses of a certain node, at pSH < 0.10. This analysis resulted in cube-like (i.e., unresolved) structures for the three critical nodes discussed above. The same analysis has been done for some selected and well-resolved nodes. Even these nodes showed a high degree of conflict, with about 20% of the sequences supporting either of the two alternative topologies (not shown). Thus, the analyses of single genes did not allow drawing further conclusions about the unresolved nature of certain branches that were encountered in the ML analysis of all sequence data. The split network from the retroposon insertion data (Churakov et al. 2009) illustrates the unresolved nature of the earliest placentalian divergences in an ideal way (fig. 6). There are nine retroposon insertions for Epitheria (all placentals except Xenarthra) and eight events that support the Xenafrotheria hypotheses. The Exafroplacentalia (Boreoplacentalia plus Xenarthra) hypothesis, which is supported by five retroposon insertions, cannot be excluded. Even though one may recognize a slight favor of the Epitheria and less support for the Exafroplacentalia hypothesis, a chi-square analysis yields a probability of P = 0.55, thus a uniform distribution of retroposon insertion cannot be rejected.
FIG. 6.
Retroposon split network.
Computer Simulation
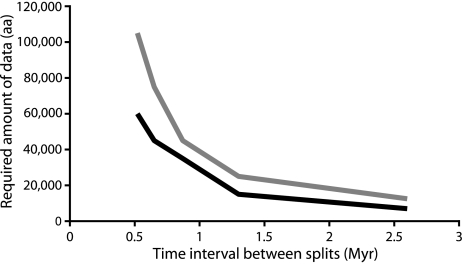
Computer simulations were performed to determine the theoretical minimum sequence length needed to significantly resolve the deepest divergence among placental mammals, based on pSH values. In this study, the aa ML analyses reconstruct Afrotheria as diverging first from the remaining placentals some 90 Ma. The following split between Xenarthra and Boreoplacentalia occurs only some 2.7 My later. Given this topology, the parameters used, and dating of the short branch, is surprising that as little as about 7,000 aa sites of evenly evolving sequence data are needed to significantly resolve this early divergence some 90 Ma. Even taking the, albeit limited, patchiness of the data into account by including the observed percentage of unknown data in the simulations, the amount of aa sequence data needed to resolve a very short early branch doubles. Thus, when including missing sites as a modeling parameter, the number of aa sites needed to significantly resolve such a branch increases to ca. 12,500. Under the same conditions, a time interval as short as 540 thousand years (ka) at 90 Ma could be significantly resolved with as little as 60,000 aa sites (supplementary table 4, Supplementary Material online). Figure 7 illustrates the results. It shows also that, as expected, the amount of data needed for resolving increasingly shorter branches increases approximately exponentially.
FIG. 7.
Simulation of aa sequence lengths required for resolving temporally tight divergences at 90 Ma. The black line is reconstructed from the simulation of a data set free of gaps, whereas the gray line represents a simulation where gaps were included according to their frequency in the original data.
Discussion
New genome data from mammalian species are becoming available at an accelerating rate. Within a year of a recent phylogenomic study (Hallström and Janke 2008) 11 new genomes were released in databases. These data enable more detailed analysis of the evolution of placental mammals. The new genome data alleviate concerns about reconstruction artifacts caused by limited taxon sampling and sequence length. The present phylogenomic analyses place the newly sequenced species into their expected positions on the mammalian tree and a consensus of their evolution emerges. Before the genomic era, comprehensive data sets and a dense taxon sampling for phylogenetic studies were available from mitochondrial protein-coding data, mitogenomics (Arnason et al. 2008), or from a few selected nuclear genes (Murphy et al. 2001). Mitogenomics clarified major parts of the mammalian tree, which are now being confirmed by phylogenomic analyses. For example, the unexpected sistergroup relationship between Carnivora and Perissodactyla was first identified by a mitogenomic study (Xu et al. 1996) and was reconstructed in later nuclear gene and phylogenomic analyses. Similarly, the grouping of the order Cetacea within Artiodactyla into the clade Cetartiodactyla was suggested by mitochondrial cytochrome b data (Irwin and Arnason 1994) and later strongly supported by nuclear gene (Gatesy et al. 1996), retroposon insertion (Nikaido et al. 1999), and mitogenomic (Arnason et al. 2000) analyses. Finally, the strong support for monophyly of the superorder Afrotheria was shown by nuclear gene analyses (Stanhope et al. 1998) and mitogenomics (Mouchaty et al. 2000) and is reconstructed from phylogenomic analyses.
However, in the current phylogenomic analyses, certain branches of the placental mammalian tree still received only limited support or remained—in effect—unresolved, despite the high data density, the authoritative amount of genome data, and the increased taxon sampling. Problematic branches involve the relationships among Xenarthra, Boreoplacentalia, and Afrotheria, the most ancient divergence of placental mammals. In addition, the position of the Chiroptera among Laurasiaplacentalia and the placement of the Scandentia (tree shrew) with the Glires or Euarchonta remain insufficiently supported or even unresolved. These branches were previously identified as problematic in early phylogenomic analyses (Nikolaev et al. 2007; Hallström and Janke 2008; Nishihara et al. 2007), and the resolution of their evolution remains vague even when new analytical approaches such as outgroup scoring are used (Schneider and Cannarozzi 2009).
Outgroup scoring offers an alternative approach to traditional phylogenetic reconstruction and may overcome some of its limitations. The method analyzes the topology and the support of a tree by extracting a signal utilizing outgroups at different distances. In this way, it overcomes, or at least reduces, the effect of model violations and long-branch attraction. Compared with standard ML analysis, simulations using outgroup scoring have demonstrated equal or better performances in resolving problematic relationships (Schneider and Cannarozzi 2009). This method, however, identifies the same problematic branches as the current analyses and provides no certain resolution. Although outgroup scoring favors the Xenafrotheria hypothesis (Xenarthra plus Afrotheria) as do some previous phylogenomic studies, a support value of 0.91 indicates that alternative topologies are possible. Inspections of alternative topologies by outgroup scoring unfortunately offer no more certainty than traditional ML analyses and provide no explanation for why some branches of the placental mammalian tree are refractory to resolution.
One might suspect that the lack of resolution in the placental mammalian tree is coupled to the amount of data available for each species. The common shrew (Lipothyphla) has the lowest sequence coverage (62%) in the alignment, yet it is still represented by 1.8 Mnt of sequence data and receives significant phylogenetic support. In contrast, the tree shrew (Scandentia) receives limited phylogenetic support despite having a similar or higher sequence coverage of 70% (2.0 Mnt). The phylogenetic position of the tree shrew is basically unresolved in ML and neighbor network analyses. As in the cases of other poorly resolved branches, the tree shrew joins other placental orders within Euarchontoglires with very short branches. This would explain the contradictory phylogenetic placements of this order and the limited number of synapomorphies from rare genomic changes that have been recovered in previous studies of scandentian evolution (Janecka et al. 2007; Kriegs et al. 2007). There simply may not have been enough time for genomes in temporally narrow divergences to accumulate sufficient numbers of informative sites. For this reason and the stochastic nature of short sequences, single gene analyses did not allow further conclusions about the nature of the conflicting signals.
Simulations
Computer simulations were made to investigate whether sequence length is a limiting factor in resolving short branches. For obvious reasons, such simulations are idealizations (simplifications) of natural processes and are not able to include all factors that shape a sequence during evolution. One potential pitfall of any simulation is that the same model is used for both simulating and analyzing the data. In our study, this probably leads to an underestimation of the required sequence length. However, the strength of simulations is their ability to isolate certain parameters intentionally, by computational or practical limitations. The aim was to identify the theoretical minimum amount of sequence data that enables significant reconstruction of the placentalian tree based on the general properties of the sequence data and their evolutionary history, that is, the tree. The present simulation includes a number of known sources of systematic errors, such as varying evolutionary rates between branches (varying branch lengths), varying rates along sequences (rate heterogeneity), multiple substitutions (absolute branch lengths), individual aa replacement probabilities (the aa replacement model), and isolated long branches from using the observed ML tree topology. In a separate simulation the fraction of missing data was added as a parameter in the simulation. Thus, the simulations are relatively realistic, even if all possible parameters were not included in the study. A model for simulating compositional bias has not yet been developed and was therefore not part of the simulation.
The analyses of the computer-simulated sequence evolution clearly show that under the modeled conditions and in the absence of introgression or lineage sorting a few ten thousand aa of random protein-coding data are capable of resolving even very short branches from lineages that existed for less than 1 My at 100 Ma. Yet, the problems encountered in resolving such branches, even with millions of nts, suggest that natural processes involved in speciation or significant systematic errors, other than those included in the current study, may mask the phylogenetic signal.
The simulation data show a strong inverse correlation between the data density and the amount of data needed for the resolution of short branches millions of years ago. When a parameter of missing data, represented by the same fraction missing in the original alignment, is included, the amount of data needed for significant resolution increases by more than 60%. A recent phylogenomic analysis of fundamental metazoan divergences that are at least six times as old as the mammalian radiation, utilized expressed sequence tags (EST) from several species as data sources. Consequently, the resulting data density in this study was only about 50% from 150 genes due to the lack of overlap between EST libraries (Dunn et al. 2008). The degree to which the reliability of the tree resolution is affected by increasing amounts of missing data and taxon sampling remains to be studied.
Networks
The limited resolution of certain branches may indicate that the evolution of placental mammals did not proceed in a strictly bifurcating way. Under these circumstances, current phylogenetic methods seeking a fully resolved two-dimensional tree are not suitable for reconstructing the history of placental mammals. Possibly, the phylogenetic signals of contradicting branches annihilate each other, which may lead to an apparently unresolved trifurcation. Collapsing problematic branches into trifurcations, however, hides invaluable evolutionary information. A case in point is best demonstrated from data of recent phylogenetic analyses of retroposon insertions on the early radiation of placental mammals (Churakov et al. 2009; Nishihara et al. 2009). The data of Churakov et al. (2009) are individually confirmed in other species by experimental sequence analysis, whereas the data of Nishihara et al. (2009) are based on database entries only.
A split network representation of the Churakov et al. (2009) data illustrates the complex radiation of the three early placental mammalian lineages. A traditional presentation of these data as a tree would require either a multifurcation, the presentation of three separate trees, or an extended tree-like diagram (see fig. 4A, Churakov et al. 2009), all of which blur the actual evolutionary message of a complex evolutionary process that led to the three clades of placental mammals. Other problematic divergences have not yet been analyzed by retroposon insertion data in the same detail, but sequence-based analyses are suggesting several such nonbifurcating radiations in the mammalian tree. The study of retroposon data for the evolution of the Chiroptera, Perissodactyla, and Carnivora (Nishihara et al. 2006), where only a single apparently contradicting signal has been described, may be such an example. The difficulty to resolve the placement of the Chiroptera via sequence analysis and conflict from the retroposon data suggest that a more detailed retroposon insertion analysis may also lead to a network-like picture of the radiation of these groups.
More complex split networks, like the phylogenomic neighbor-net of figures 4 and 5, depict the intricate evolutionary signals of the sequence data in a still decipherable way, whereas traditional bifurcating tree presentations hide alternative hypotheses or conflict in the data. Split networks have not been routinely used to study deep divergences among placental mammals because reticulate evolution events were not suspected to occur or be detectable over long timescales. Therefore, significantly conflicting data have rarely been observed in single gene analyses. Such conflict has only been sporadically observed in the phylogenetic analysis of single genes that support different phylogenies (Satta et al. 2000) or by testing individual hypotheses using pSH or similar statistical tests. In this way, many controversial branches are going unnoticed. This is especially the case when branches receive high support values, leading one not to suspect alternative hypotheses. Branches can receive high bootstrap and especially high Bayesian probabilities even in the presence of strong conflict in the data, which would leave them, in effect, unresolved by ML test statistics (Nishihara et al. 2007; Hallström and Janke 2008). The individual identification of such problematic branches occurs often only by chance or when they contradict preconceived hypotheses.
In contrast, network methods objectively present most inconsistencies in a tree in a single picture. This information can then be used to further investigate the nature and extent of conflict between alternative hypotheses. Unfortunately, the extreme age of basal mammalian divergences so far preclude the analysis of single genes for signs of conflict from reticulate evolution due to the limited amount of information each gene provides and its stochastic nature. Yet, the neighbor-net of the whole data set identifies all nodes where disagreement in the data is observed by other methods. From the network graphs in figures 4 and 5, it becomes immediately visible that the evolution and radiation of placental mammals has a more complex history than previously assumed.
Dating
The analysis of divergence time estimates may provide an explanation for the poor resolution of some branches. These branches are all characterized by short internal nodes that span intervals of 1–4 My. The computer simulations demonstrated that the limited resolution is not due to the lack of data. As little as 60,000 aa sites are sufficient to resolve branches that are much shorter. Thus, as discussed earlier (Hallström and Janke 2008), speciation-related processes, such as incomplete lineage sorting and hybridization, might explain the lack of resolution. These processes would explain the reticulate network from the reconstruction of retroposon data for the three fundamental mammalian lineages and the other contradictory results like the grouping of the Chiroptera with Perissodactyla and Carnivores in some analyses (Nishihara et al. 2006).
The molecular divergence time estimates indicate that the onset and rapid radiation of placental mammals at ca. 90 Ma coincides with an increase in global temperatures in the Albian (112–99.6 Ma) of the mid-Cretaceous (Puceat et al. 2003; Wilson and Norris 2001) that reached a maximum at the Cenomanian–Turonian boundary at 93.5 Ma (Retallack 2002). The climate change and a generally high biological turnover (Wilson and Norris 2001) are likely to have triggered the radiation of placental mammals to a similar extent as did plate tectonics, by producing more favorable and diversified environments. However, these developments may have also caused stress and extinction events by replacing ecosystems. These “cryptic” extinctions could be interpreted as rapid radiations (Crisp and Cook 2009) and may further complicate the interpretation of mammalian evolution. The cause of the true rapid radiations cannot be determined by genomic studies alone; however, careful correlations of the occurrence/disappearance of mammalian fossils, divergence times, networks, and paleoclimates may soon provide a more detailed picture.
Origin of Placental Mammals
When placental mammals began their radiation in the mid-Cretaceous some 90 Ma, the supercontinents Laurasia and Gondwana were fragmenting. It has therefore been suggested that the early radiation of placental mammals was shaped by vicariance (Hedges et al. 1996). Recently, tectonic movements have also been used to explain the difficulties encountered in resolving the early placental mammalian radiation and distribution pattern by analyzing sequence and retroposon data (Wildman et al. 2007; Churakov et al. 2009; Nishihara et al. 2009). These studies assume a Gondwanan distribution of the earliest placental mammals and a later distribution of Boreoplacentalia to Laurasia and Xenarthra to South America, whereas the members of Afrotheria remained in what became Africa. However, this scenario requires a highly choreographed, rapid, and nearly simultaneous splitting or reconnection of the continents (Nishihara et al. 2009) and seems rather unlikely.
Using geological events to explain the distribution and rapid divergence of three placentalian clades may be problematic because geological events are not points in time. They have durations of several million years, periods that exceed the short-spanned divergences of the early mammalian radiation and speciation, which occurred over a period of 2–4 My (Curnoe et al. 2006; van Dam et al. 2006). It is probable that during continental breakups recurring land bridges caused by ridges and sea-level fluctuations frequently connected continental plates overextended period of times. As an example, the Grande Rise and the Walvis Ridge were still exposed into Maastrichtian/Palaeocene times at 70–60 Ma (Sclater et al. 1977; Reyment and Dingle 1987), providing a semicontinuous south Atlantic connection between South America and Africa. Thus, the speciation process and divergence of lineages can be approximately an order of magnitude more rapid than continental drift dynamics.
A complete separation of South America, Africa, and Laurasian continents at 120 Ma has been suggested to explain the difficulty in resolving the Xenarthra–Afrotheria–Boreoplacentalia split by retroposon and sequence data (Nishihara et al. 2009). This date, 120 Ma, is considerably older than phylogenomic divergence time estimates among placental mammals, making their distribution by vicariance improbable. Furthermore, the Laurasian–Gondwanan separation occurred considerably earlier than 120 Ma (Smith et al. 2004; Reeves 2009; see www.reeves.nl) and was not concordant with the separation of South America and Africa.
Correlating the divergence of placental mammals in this way with continental drift is problematic because it requires speculations about the origin and distribution of living groups based on their current biogeography and ignores the mammalian fossil record. Such correlations seem to be intuitively convincing, but they presuppose a Gondwanan origin of placental mammals to parsimoniously explain their current distribution by continental drift (Wildman et al. 2007; Churakov et al. 2009; Nishihara et al. 2009). However, the existing fossil record does not support such a scenario. The most parsimonious interpretation of the actual fossil record suggests that basal divergences among placental mammals took place in Laurasia and not Gondwana (Archibald 2003; Hunter and Janis 2006; Wible et al. 2007). The current phylogenomic analyses estimate the origin of the Xenarthra at about 90 Ma. Although some placentalian orders may have already been present in the Cretaceous (Asher 2005; Benton and Donoghue 2007), no xenarthran fossils have been identified in South America over a time span of about 20 My from about this period (Albian/Cenomanian) until about 70 Ma (Maastrichtian). However, South America has an otherwise rich fossil record from archaic mammals, such as multituberculates, gondwanatheres, and sudamericids, prior to the Cretaceous–Tertiary (K/T) boundary (Flynn and Wyss 1998; Pascual and Ortiz-Jaureguizar 2007), suggesting that the lack of placentalian fossils is real and not an artifact. Therian mammals populated South America and replaced the archaic ones close to the K/T boundary (Pascual and Ortiz-Jaureguizar 2007). Molecular estimates of the origin and diversification time of the two major South American mammalian groups, marsupials at 68.5 Ma and xenarthrans at 65 Ma (Delsuc et al. 2004; Nilsson et al. 2004), correlate well with the fossil-based findings. Thus, molecular- and fossil-based data support a colonization of South America by xenarthrans and other therian mammals from the north (Laurasia) in the late Cretaceous and not via Africa in the mid-Cretaceous as some schemes suggest (Wildman et al. 2007; Churakov et al. 2009; Nishihara et al. 2009; Murphy et al. 2001; Waddell et al. 1999).
Likewise, the depauperate fossil record of African mammals does not indicate the presence of Xenarthra or members of the Afrotheria on the African continent during a period of 95–80 Ma, when, according to molecular-based divergence time estimates, major afrotherian radiations should have occurred. In fact, the oldest members of the crown group, Afrotheria are of Laurasian origin (Asher et al. 2003; Zack et al. 2005; Tabuce et al. 2008). This makes the hypothesis of a Laurasian origin and diversification of placental mammals currently the best supported by the fossil record.
Taking the fossil, tectonic, and molecular data into account yields a simple scenario for the early radiation of placental mammals. The first divergences, possibly triggered by climate change rather than plate tectonics, occurred in Laurasia. These rapid splits left no time for fixation of polymorphisms, but allowed for introgression, and led to mosaic genomes. This radiation was followed immediately by dispersal to different geographic regions, with the Xenarthra reaching South America at a later point in time. This scenario is not unreasonable given that xenarthrans and marsupials reached South America at approximately the same time, with marsupials obviously coming via North America, a Laurasian continent (Nilsson et al. 2004). Thus, the dispersal routes of xenarthrans and marsupials contradict speculations of a Gondwanan origin of placental mammals.
Summary
The advancement of sequencing technology enabled the generation of large quantities of genome data from numerous placental mammals, and eventually the genomes of most species will be sequenced. This gargantuan amount of data already makes it possible to resolve mammalian evolution and phylogeny in a detail that was unimaginable not too long ago (Novacek 1992). Exact divergence time estimates enable detailed studies about whether and how historic events such as plate tectonics and climate changes correlate with the evolution of placental mammals. However, current phylogenomic analyses already show that the evolution of placentalian genomes may not have been strictly bifurcating. Instead, incomplete lineage sorting, species hybridization, and possibly other as yet unknown processes led to mosaic genomes, with different parts having different phylogenetic histories (Ebersberger et al. 2007; Hallström and Janke 2008; Churakov et al. 2009). The current study as well as previous phylogenomic and retroposon analyses clearly show that certain branches of the mammalian tree, and possibly that of other species, cannot be simply resolved as strictly bifurcating by genome data, and are in many cases best viewed and interpreted as network-like processes.
Supplementary Material
Supplementary tables 1–4 and figures 1–2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We are grateful to Drs. Maria Nilsson and Adrian Schneider for critical comments on the study and the manuscript and Collin Reeves for comments on the plate tectonics. The Carl-Trygger, Nilsson-Ehle Foundations, and LOEWE supported the work.
References
- Archibald JD. Timing and biogeography of the eutherian radiation: fossils and molecules compared. Mol Phylogenet Evol. 2003;28:350–359. doi: 10.1016/s1055-7903(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Arnason U, Adegoke JA, Gullberg A, Harley EH, Janke A, Kullberg M. Mitogenomic relationships of placental mammals and molecular estimates of their divergences. Gene. 2008;421:37–51. doi: 10.1016/j.gene.2008.05.024. [DOI] [PubMed] [Google Scholar]
- Arnason U, Gullberg A, Gretarsdottir S, Ursing B, Janke A. The mitochondrial genome of the sperm whale and a new molecular reference for estimating eutherian divergence dates. J Mol Evol. 2000;50:569–578. doi: 10.1007/s002390010060. [DOI] [PubMed] [Google Scholar]
- Asher RJ. Insectivoran-grade placental mammals: character evolution and fossil history. In: Rose KD, Archibald JD, editors. The rise of placental mammals. Baltimore (MD): The Johns Hopkins University Press; 2005. pp. 50–70. [Google Scholar]
- Asher RJ, Novacek MJ, Geisler JH. Relationships of endemic African mammals and their fossil relatives based on morphological and molecular evidence. J Mamm Evol. 2003;10:131–162. [Google Scholar]
- Bajpai S, Gingerich PD. A new Eocene archaeocete (Mammalia, Cetacea) from India and the time of origin of whales. Proc Natl Acad Sci U S A. 1998;95:15464–15468. doi: 10.1073/pnas.95.26.15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PCJ, Asher RJ. Calibration and constraining molecular clocks. In: Hedges SB, Kumar S, editors. The timetree of life. Oxford: Oxford University Press; 2009. pp. 35–86. [Google Scholar]
- Cantrell MA, Filanoski BJ, Ingermann AR, Olsson K, DiLuglio N, Lister Z, Wichman HA. An ancient retrovirus-like element contains hot spots for SINE insertion. Genetics. 2001;158:769–777. doi: 10.1093/genetics/158.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Kriegs JO, Baertsch R, Zemann A, Brosius J, Schmitz J. Mosaic retroposon insertion patterns in placental mammals. Genome Res. 2009;19:868–875. doi: 10.1101/gr.090647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp MD, Cook LG. Explosive radiation or cryptic mass extinction? Interpreting signatures in molecular phylogenies. Evolution. 2009;63:2257–2265. doi: 10.1111/j.1558-5646.2009.00728.x. [DOI] [PubMed] [Google Scholar]
- Curnoe D, Thorne A, Coate JA. Timing and tempo of primate speciation. J Evol Biol. 2006;19:59–65. doi: 10.1111/j.1420-9101.2005.00989.x. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Vizcaino SF, Douzery EJ. Influence of Tertiary paleoenvironmental changes on the diversification of South American mammals: a relaxed molecular clock study within xenarthrans. BMC Evol Biol. 2004;4:11. doi: 10.1186/1471-2148-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, et al. (18 co-authors) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Ebersberger I, Galgoczy P, Taudien S, Taenzer S, Platzer M, von Haeseler A. Mapping human genetic ancestry. Mol Biol Evol. 2007;24:2266–2276. doi: 10.1093/molbev/msm156. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JJ, Wyss AR. Recent advances in South American mammalian paleontology. Trends Ecol Evol. 1998;13:449–454. doi: 10.1016/s0169-5347(98)01457-8. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Hayashi C, Cronin MA, Arctander P. Evidence from milk casein genes that cetaceans are close relatives of hippopotamid artiodactyls. Mol Biol Evol. 1996;13:954–963. doi: 10.1093/oxfordjournals.molbev.a025663. [DOI] [PubMed] [Google Scholar]
- Hallström BM, Janke A. Resolution among major placental mammal interordinal relationships with genome data imply that speciation influenced their earliest radiations. BMC Evol Biol. 2008;8:162. doi: 10.1186/1471-2148-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström BM, Kullberg M, Nilsson MA, Janke A. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol Biol Evol. 2007;24:2059–2068. doi: 10.1093/molbev/msm136. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Parker PH, Sibley CG, Kumar S. Continental breakup and the ordinal diversification of birds and mammals. Nature. 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- Hunter PJ, Janis CM. Spiny Norman in the garden of eden? J Mamm Evol. 2006;13:89–123. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Irwin DM, Arnason U. Cytochrome b gene of marine mammals: phylogeny and evolution. J Mamm Evol. 1994;2:37–55. [Google Scholar]
- Janecka JE, Miller W, Pringle TH, Wiens F, Zitzmann A, Helgen KM, Springer MS, Murphy WJ. Molecular and genomic data identify the closest living relative of primates. Science. 2007;318:792–794. doi: 10.1126/science.1147555. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Kullberg M, Hallström BM, Arnason U, Janke A. Phylogenetic analysis of 1.5 Mbp and platypus EST data refute the Marsupionta hypothesis and unequivocally support Monotremata as sister group to Marsupialia/Placentalia. Zool Scr. 2008;37:115–127. [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (254 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Mouchaty SK, Gullberg A, Janke A, Arnason U. Phylogenetic position of the Tenrecs (Mammalia: Tenrecidae) of Madagascar based on analysis of the complete mitochondrial genome sequence of Echinops telfairi. Zool Scr. 2000;29:307–317. [Google Scholar]
- Murphy WJ, Eizirik E, O'Brien SJ, et al. (11 co-authors) Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nikaido M, Rooney AP, Okada N. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interspersed elements: hippopotamuses are the closest extant relatives of whales. Proc Natl Acad Sci U S A. 1999;96:10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev S, Montoya-Burgos JI, Margulies EH, Program NCS, Rougemont J, Nyffeler B, Antonarakis S. Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 2007;3:1. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson MA, Arnason U, Spencer PB, Janke A. Marsupial relationships and a timeline for marsupial radiation in South Gondwana. Gene. 2004;340:189–196. doi: 10.1016/j.gene.2004.07.040. [DOI] [PubMed] [Google Scholar]
- Nilsson MA, Härlid A, Kullberg M, Janke A. The impact of fossil calibrations, codon positions and relaxed clocks on the divergence time estimates of the native Australian rodents (Conilurini) Gene. 2010;455:22–31. doi: 10.1016/j.gene.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Nishihara H, Hasegawa M, Okada N. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci U S A. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Maruyama S, Okada N. Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc Natl Acad Sci U S A. 2009;106:5235–5240. doi: 10.1073/pnas.0809297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Okada N, Hasegawa M. Rooting the eutherian tree: the power and pitfalls of phylogenomics. Genome Biol. 2007;8:R199. doi: 10.1186/gb-2007-8-9-r199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novacek MJ. Mammalian phylogeny: shaking the tree. Nature. 1992;356:121–125. doi: 10.1038/356121a0. [DOI] [PubMed] [Google Scholar]
- Pascual R, Ortiz-Jaureguizar E. The Gondwanan and South American episodes: two major and unrelated moments in the history of the South American mammals. J Mamm Evol. 2007;14:75–137. [Google Scholar]
- Puceat E, Lecuyer C, Sheppard SMF, Dromart G, Reboulet S, Grandjean P. Thermal evolution of Cretaceous Tethyan marine waters inferred from oxygen isotope composition of fish tooth enamels. Paleoceanography. 2003;18:1029. [Google Scholar]
- Rambaut A, Grassley NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Bioinformatics. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Reeves C. Re-examining the evidence from plate-tectonics for the initiation of Africa's passive margins. London: Geological Society of Huston/Petroleum Exploration Society of Great Britain; 2009. [Google Scholar]
- Retallack GJ. Carbon dioxide and climate over the past 300 Myr. Philos Trans R Soc Lond A. 2002;360:659–673. doi: 10.1098/rsta.2001.0960. [DOI] [PubMed] [Google Scholar]
- Reyment R, Dingle RV. Palaeogeography of Africa during the Cretaceous period. Palaeogogr Palaeoclimatol Palaeoecol. 1987;59:93–116. [Google Scholar]
- Roos J, Aggarwal RK, Janke A. Extended mitogenomic phylogenetic analyses yield new insight into crocodylian evolution and their survival of the Cretaceous-Tertiary boundary. Mol Phylogenet Evol. 2007;45:663–673. doi: 10.1016/j.ympev.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Satta Y, Klein J, Takahata N. DNA archives and our nearest relative: the trichotomy problem revisited. Mol Phylogenet Evol. 2000;14:259–275. doi: 10.1006/mpev.2000.0704. [DOI] [PubMed] [Google Scholar]
- Schneider A, Cannarozzi GM. Support patterns from different outgroups provide a strong phylogenetic signal. Mol Biol Evol. 2009;26:1259–1272. doi: 10.1093/molbev/msp034. [DOI] [PubMed] [Google Scholar]
- Sclater JG, Hellinger S, Tapscott C. The paleobathymetry of the Atlantic Ocean from the Jurassic to the present. J Geol. 1977;85:509–552. [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Smith AG, Smith DG, Funnell BM. Atlas of Mesozoic and Cenozoic Coastlines. Cambridge: Cambridge University Press; 2004. [Google Scholar]
- Stanhope MJ, Waddell VG, Madsen O, de Jong W, Hedges SB, Cleven GC, Kao D, Springer MS. Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. Proc Natl Acad Sci U S A. 1998;95:9967–9972. doi: 10.1073/pnas.95.17.9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel M, Penny D. Parsimony, likelihood, and the role of models in molecular phylogenetics. Mol Biol Evol. 2000;17:839–850. doi: 10.1093/oxfordjournals.molbev.a026364. [DOI] [PubMed] [Google Scholar]
- Tabuce R, Asher RJ, Lehmann T. Afrotherian mammals: a review of current data. Mammalia. 2008;72:2–14. [Google Scholar]
- van Dam JA, Abdul Aziz H, Alvarez Sierra MA, Hilgen FJ, Hoek Ostende LW, Lourens LJ, Mein P, Meulen AJ, Pelaez-Campomanes P. Long-period astronomical forcing of mammal turnover. Nature. 2006;443:687–691. doi: 10.1038/nature05163. [DOI] [PubMed] [Google Scholar]
- van de Lagemaat LN, Gagnier L, Medstrand P, Mager DL. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 2005;15:1243–1249. doi: 10.1101/gr.3910705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tuinen M, Hadly EA. Calibration and error in placental molecular clocks: a conservative approach using the cetartiodactyl fossil record. J Hered. 2004;95:200–208. doi: 10.1093/jhered/esh045. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. (273 co-authors) The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Waddell PJ, Cao Y, Hasegawa M, Mindell DP. Assessing the cretaceous superordinal divergence times within birds and placental mammals by using whole mitochondrial protein sequences and an extended statistical framework. Syst Biol. 1999;48:119–137. doi: 10.1080/106351599260481. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wible RJ, Rougier GW, Novacek MJ, Asher RJ. Cretaceous eutherians and Laurasian origin for placental mammals near the K/T boundary. Nature. 2007;447:1003–1006. doi: 10.1038/nature05854. [DOI] [PubMed] [Google Scholar]
- Wildman DE, Uddin M, Opazo JC, Liu G, Lefort V, Guindon S, Gascuel O, Grossman LI, Romero R, Goodman M. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci U S A. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PA, Norris RD. Warm tropical ocean surface and global anoxia during the mid-Cretaceous period. Nature. 2001;412:425–429. doi: 10.1038/35086553. [DOI] [PubMed] [Google Scholar]
- Xu X, Janke A, Arnson U. The complete mitochondrial DNA sequence of the Greater Indian Rhinoceros, Rhinoceros unicornis, and the phylogenetic relationship among Carnivora, Perissodactyla and Artiodactyla (+Cetacea) Mol Biol Evol. 1996;13:1167–1173. doi: 10.1093/oxfordjournals.molbev.a025681. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Zack SP, Penkrot TA, Bloch JI, Rose KD. Affinities of ‘hyopsodontids’ to elephant shrews and a Holarctic origin of Afrotheria. Nature. 2005;434:497–501. doi: 10.1038/nature03351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.