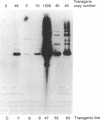
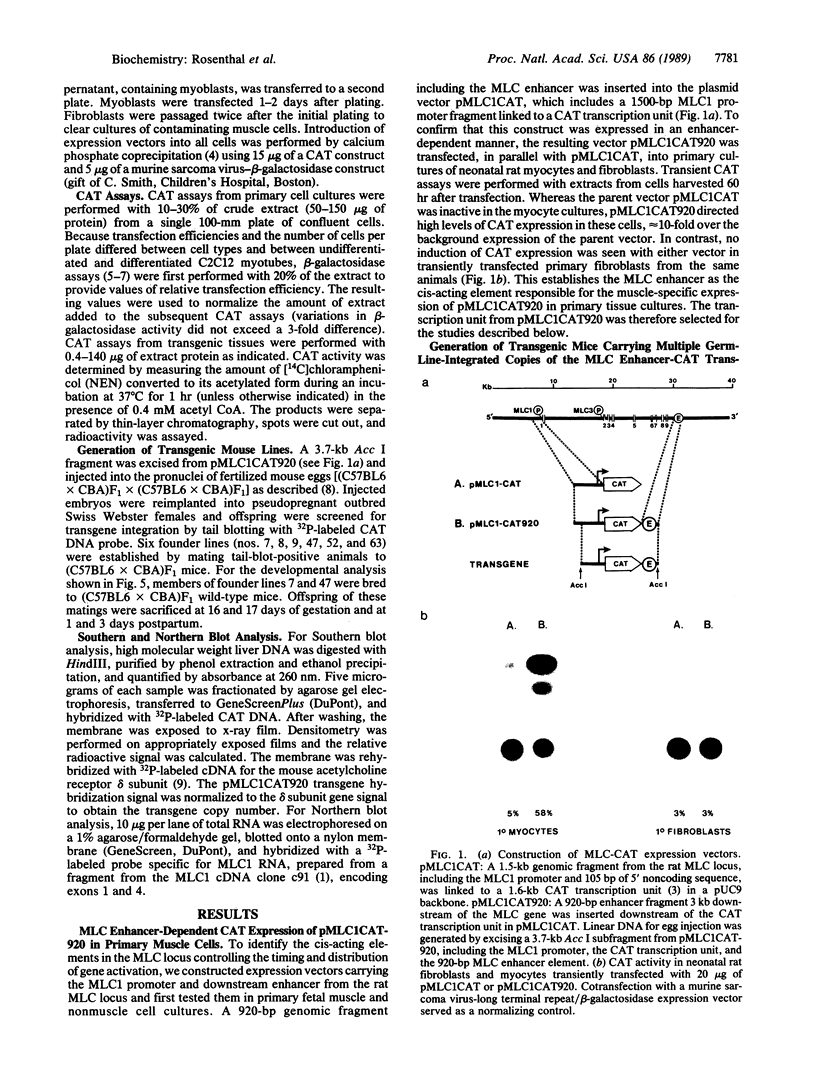
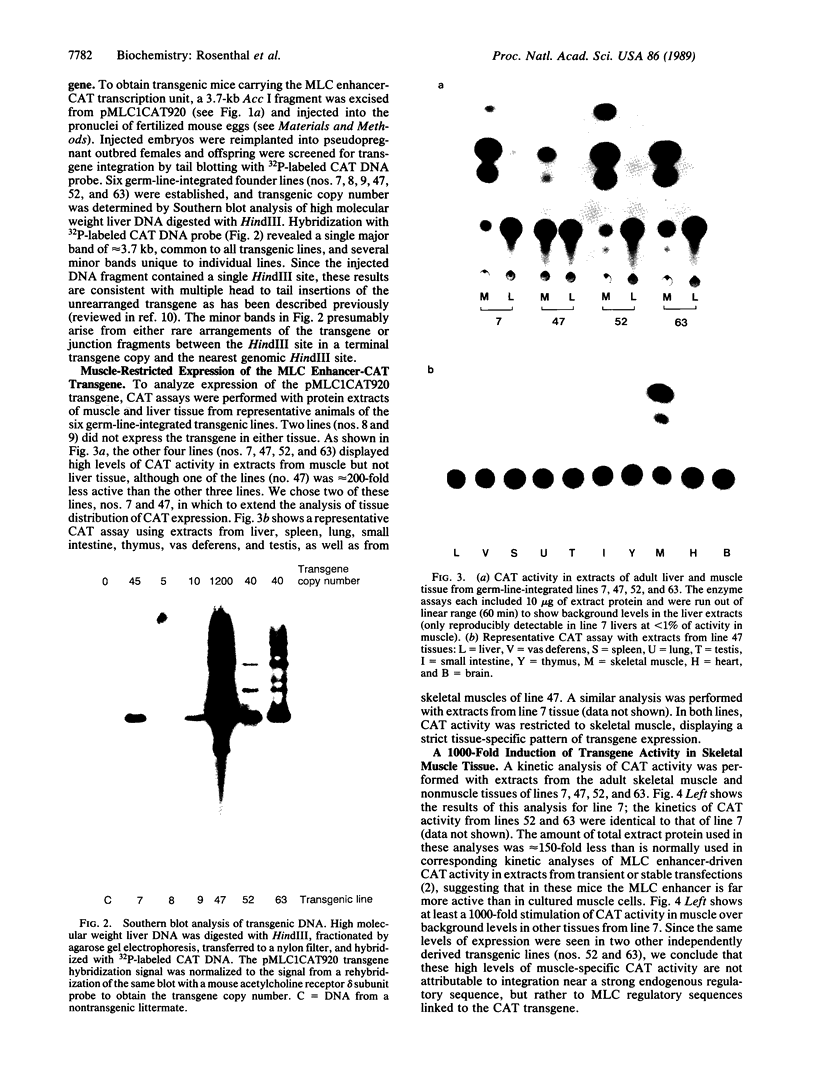
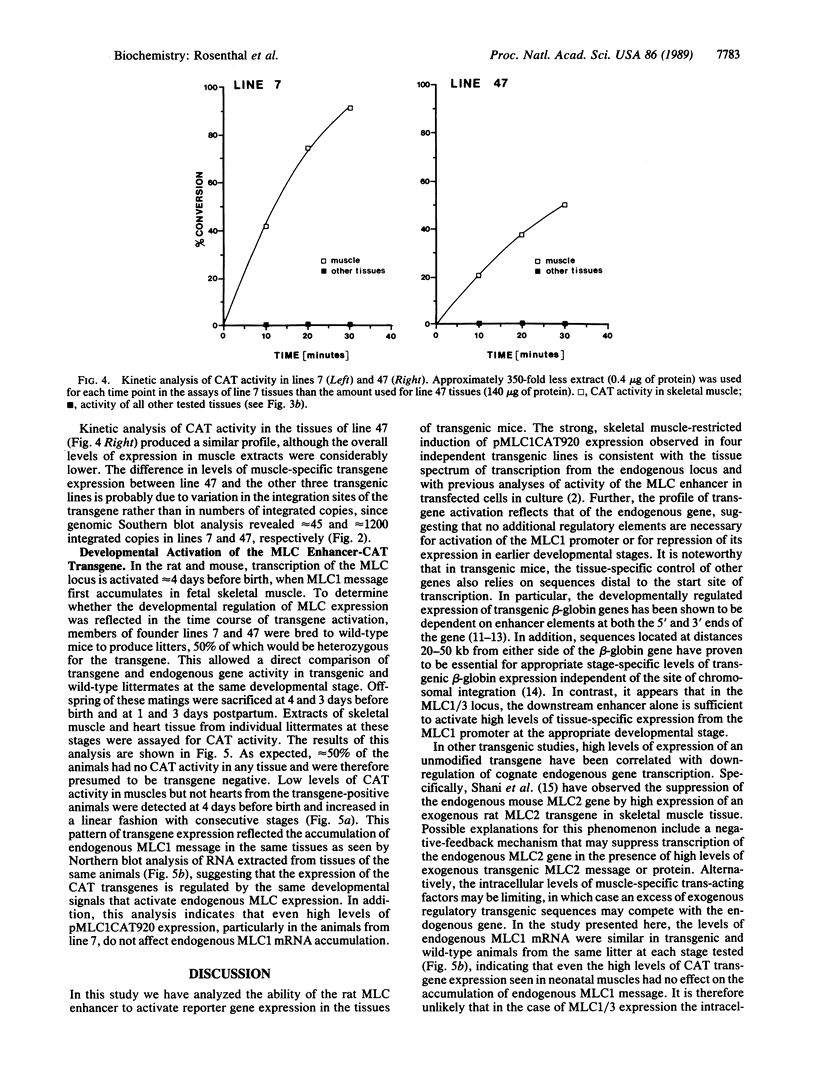
Abstract
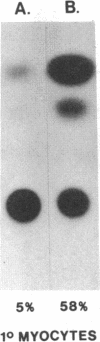
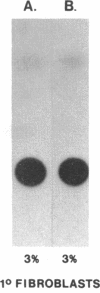
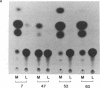
The rat myosin light chain (MLC)1/3 gene locus contains a potent muscle-specific enhancer, located downstream of the coding region, greater than 24 kilobases away from the MLC1 transcription start site. To assess the role of this enhancer in the activation of MLC expression during development, transgenic mice were generated carrying multiple copies of a MLC1 promoter-chloramphenicol acetyltransferase (CAT) transcription unit linked to a genomic fragment including the enhancer. CAT expression was detected in four mouse lines, up to 1000-fold higher in skeletal muscles than in other tissues. Activation of endogenous MLC1 transcription in these animals 4 days before birth was reflected in the onset of CAT transgene expression. This study identifies the transcriptional control elements necessary to activate the 21-kilobase MLC1/3 locus at the appropriate fetal stage and indicates that the MLC enhancer is sufficient to induce developmentally regulated expression from the MLC1 promoter exclusively in skeletal muscle cells.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behringer R. R., Hammer R. E., Brinster R. L., Palmiter R. D., Townes T. M. Two 3' sequences direct adult erythroid-specific expression of human beta-globin genes in transgenic mice. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7056–7060. doi: 10.1073/pnas.84.20.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow M. T., Olson P. S., Stockdale F. E. Myosin light-chain expression during avian muscle development. J Cell Biol. 1983 Mar;96(3):736–744. doi: 10.1083/jcb.96.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M., Ernst H., Wentworth B., Nadal-Ginard B., Rosenthal N. A muscle-specific enhancer is located at the 3' end of the myosin light-chain 1/3 gene locus. Genes Dev. 1988 Dec;2(12B):1779–1790. doi: 10.1101/gad.2.12b.1779. [DOI] [PubMed] [Google Scholar]
- Edlund T., Walker M. D., Barr P. J., Rutter W. J. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5' flanking elements. Science. 1985 Nov 22;230(4728):912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- Gauthier G. F., Lowey S., Benfield P. A., Hobbs A. W. Distribution and properties of myosin isozymes in developing avian and mammalian skeletal muscle fibers. J Cell Biol. 1982 Feb;92(2):471–484. doi: 10.1083/jcb.92.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987 Dec 24;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- LaPolla R. J., Mayne K. M., Davidson N. Isolation and characterization of a cDNA clone for the complete protein coding region of the delta subunit of the mouse acetylcholine receptor. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7970–7974. doi: 10.1073/pnas.81.24.7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F., Hall C. V., Ringold G. M., Dobson D. E., Luh J., Jacob P. E. Functional analysis of the steroid hormone control region of mouse mammary tumor virus. Nucleic Acids Res. 1984 May 25;12(10):4191–4206. doi: 10.1093/nar/12.10.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D., Brinster R. L. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy M., Strehler E. E., Garfinkel L. I., Gubits R. M., Ruiz-Opazo N., Nadal-Ginard B. Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J Biol Chem. 1984 Nov 10;259(21):13595–13604. [PubMed] [Google Scholar]
- Shani M., Dekel I., Yoffe O. Expression of the rat myosin light-chain 2 gene in transgenic mice: stage specificity, developmental regulation, and interrelation with the endogenous gene. Mol Cell Biol. 1988 Feb;8(2):1006–1009. doi: 10.1128/mcb.8.2.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudel M., Costantini F. A 3' enhancer contributes to the stage-specific expression of the human beta-globin gene. Genes Dev. 1987 Nov;1(9):954–961. doi: 10.1101/gad.1.9.954. [DOI] [PubMed] [Google Scholar]
- Trudel M., Magram J., Bruckner L., Costantini F. Upstream G gamma-globin and downstream beta-globin sequences required for stage-specific expression in transgenic mice. Mol Cell Biol. 1987 Nov;7(11):4024–4029. doi: 10.1128/mcb.7.11.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]