Abstract
ASICs (acid-sensing ion channels) are proton-gated channels that are important for pain sensation. New work by Xu and coworkers in this issue of Neuron identifies synthetic ligands and related small molecules found in the inflammatory soup that activate ASICs. These new findings highlight the power of small molecule screening to find new compounds that can control channel function. They also demonstrate how the discovery and characterization of such molecules can lead to new insights regarding channel mechanism and natural ligands.
Acidification of extracellular fluid hurts and is a common consequence of injury, inflammation, and ischemia. Even though protons are a general component of aqueous solutions and all proteins carry elements that are titratable, the prominence of extracellular acidosis in injury has driven the idea that organisms might have a dedicated ‘receptor’ for detecting extracellular pH changes (Krishtal, 2003). The leading candidate molecules for such a role are a set of transmembrane proteins known as acid-sensing ion channels (ASICs). These molecules are voltage-independent, sodium selective, proton-gated ion channels that belong to a larger family of epithelial sodium channel/degenerin (ENaC/DEG) channels (Wemmie et al., 2006).
ASICs are exceptionally sensitive to changes in extracellular proton concentration, having a Hill coefficient of 6–8 (Jasti et al., 2007; Krishtal, 2003). Tissue acidosis accompanies pain, inflammation, and hyperalgesia and in the brain is associated with ischemia and neuronal injury. Because ASICs appear to be primed to respond to such changes, they are leading targets for the development of novel compounds that could be used to control pain or that might be effective to treat ischemic brain injury (Sluka et al., 2009; Xiong et al., 2008). Nevertheless, there has remained a lingering question regarding whether the pH changes to which the channel is so exquisitely sensitive happen in native settings (Wemmie et al., 2006). This question, together with the notably large extracellular domain present in each channel subunit, has fueled the hypothesis that ASICs might be sensitive to naturally occurring ligands other than protons (Wemmie et al., 2006).
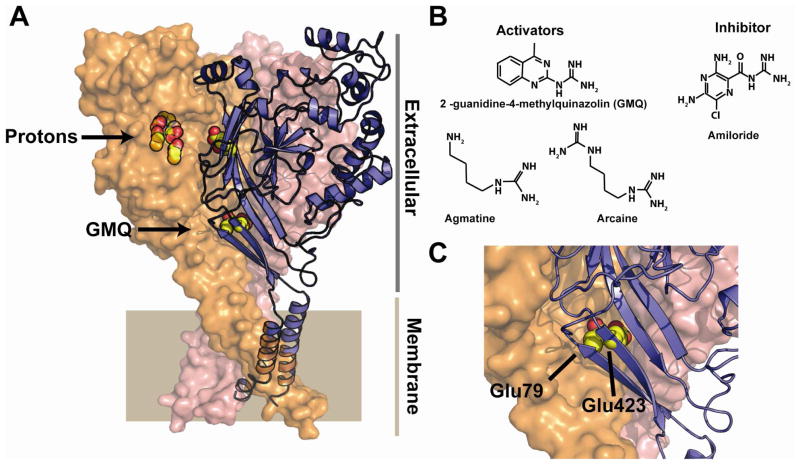
The notable functional signature of ASIC activation by protons is a rapid activation of the channel followed by a fast desensitization. The precise mechanistic details of how ASICs gate remain unknown. Recent, high-resolution crystallographic structural studies of what is likely to be a desensitized conformation of ASIC1 have revealed a pocket enriched in acidic residues that is part of the pH sensor (Gonzales et al., 2009; Jasti et al., 2007)(Figure 1A). It is in this context, that the new work by Xu and colleagues (Yu et al., 2010) makes an exciting new step in the development of ASIC chemical biology that bridges biophysical studies of the channel with functional studies of the discovered compounds in an authentic physiological setting.
Figure 1.
A, Structure of ASIC1 (3HGC) (Gonzales et al., 2009). Conserved acidic pairs in the putative proton sensing pocket and those that affect GMQ action are shown in yellow and red. Putative proton sensing pocket and the site of GMQ covalent modification are shown. Extracellular and membrane-spanning regions are indicated. B, Chemical structures of GMQ, agmatine, arcaine, and amiloride. C, Closeup of the positions of the conserved Glu79 and Glu423 residues. The structure is from ASIC1. The labels correspond to the ASIC3 numbering.
Similar to many other ion channel families, the pharmacological toolkit that can be used for specific manipulation of ASICs is limited (Deval et al., 2010; Krishtal, 2003; Xiong et al., 2008). There are only two high-affinity, selective ASIC inhibitors known. Both are venom-derived peptides, one from a tarantula, pslamotoxin 1, and the other, APETx2, from a sea anemone (Diochot et al., 2007). Pslamotoxin 1 acts as a potent analgesic (Mazzuca et al., 2007). This impressive physiological result highlights the power that selective ASIC modulators have to help dissect the underlying biology of the channel and suggests possible starting points for new pain-directed therapeutics (Deval et al., 2010; Sluka et al., 2009; Xiong et al., 2008). Although these peptides have been very useful tools other types of modulators are very much needed.
Xu and colleagues set out to find small molecules that could modify the activity of ASIC3, an ASIC that is particularly important for inflammatory pain (Deval et al., 2008). One of the major barriers to small molecule based exploration of ion channel chemical biology is that the available functional assays, such as standard whole cell electrophysiological recording, are not adept at processing the large numbers of candidate molecules (~10,000–1,000,000) that are generally used for screening campaigns (Dunlop et al., 2008). To circumvent this limitation, Xu and colleagues exploited the structural information and focused on screening a library of ~300 molecules bearing basic moieties motivated by the idea that such molecules might interact with the functionally-important acidic regions of the extracellular domains. This strategy was important as it limited the candidate molecules to a set that could be assessed by conventional electrophysiology. The authors identified a molecule, GMQ (2-guanidine-4-methylquinazolin), that evokes large ASIC3 currents and that unlike proton-evoked currents show little or no desensitization. GMQ contains a guanidinium moiety attached to an aromatic scaffold (Figure 1B). By following a classic ‘structure-activity relationship’ (SAR) approach that assayed the functional effects of changes in specific regions of the GMQ scaffold, the authors show that even though the potency of GMQ is not exceptionally high (EC50~350 μM), the series of GMQ related derivatives show characteristics that suggest specific interactions with the channel.
The initial expectation was that GMQ, which bears a guanido group, would target the proton-sensing, acidic residue rich pocket. However, mutational analysis suggests that this is not the case as mutations in the acidic-rich pocket that affect pH sensing spare the GMQ response. Further tests for residues affecting GMQ action identified two key acidic residues that lie in the subunit-subunit interface (Figure 1C) at a position that is nearer to the membrane than the proton-sensing site (Figure 1A). Previous studies had shown that substitution of one of these positions with cysteine, E79C, resulted in ASIC3 channels that could be modified in a state dependent manner by a variety of thiol reactive agents (Cushman et al., 2007). Further, E79C modification slowed desensitization substantially. Making use of these observations, the authors examined the classic thiol modifying Ellman’s reagent (5, 5′-dithiobis(2-nitrobenzoic acid), DTNB) as well as a DTNB-like GMQ derivative that could also act as a thiol exchange reagent. Strikingly, both activated the channel by covalent modification of E79C. This action, in the context of the structural data, is consistent with the idea that there is some type of conformational change in the subunit-subunit interfaces in the region of E79, as the conserved E79 equivalent in ASIC1 is not accessible in the desensitized structures and buried in the subunit-subunit interface.
Although the data point to the importance of E79 for activation by GMQ, the question of whether the interaction is direct remains unresolved. The simplest explanation in which the effect would involve a direct interaction between the E79 carboxylate and the GMQ guanido group does not appear to be satisfactory. Replacement of the E79 acidic sidechain with a 5-thio-2-nitrobenzoic acid (TNB) activates the channel. This effect depends on the presence of the carboxylate and happens in the absence of an obvious exogenous positive countercharge. Additionally, the portion of the disulfide linked GMQ that reacts with E79C is at the opposite end of the molecule from the guanido group. Thus, the mode of activation of the covalent GMQ may be different from that of GMQ alone. Nevertheless, the data support the idea that the essential factor for activation is a structural rearrangement at the subunit-subunit interface. Regardless of the actual mechanism, several lines of evidence, such as non-overlapping sensors for H+ and GMQ and differences in the activation kinetics support the idea that GMQ activation is different from proton-based activation. Further, the two mechanisms do not appear to be independent as changes in pH dramatically alter the response of the channel to GMQ. Together, these results hint at the possibility that the two interactive modes of activation may have physiological relevance.
The activity of GMQ prompted Yu et al. to look for natural molecules having similar chemical structures that might act as endogenous ligands. This search showed that two guanido-containing compounds, agmatine and arcaine, that are by-products of arginine metabolism (Regunathan, 2006), also cause persistent ASIC3 activation. Although these compounds are less effective than GMQ, the observation that naturally occurring molecules that may be present at sites of inflammation affect ASIC function raises the possibility that natural ligands such as these may be involved in the normal functioning of the channel. In support of these ideas, both GMQ and agmatine injection elicit pain-induced paw licking that is diminished in ASIC3 knockout mice and that is antagonized by the non-specific ASIC blocker amiloride (Deval et al., 2010; Xiong et al., 2008). Importantly, the pain-induced behavior caused by GMQ follows the biophysical SAR studies and provides further support that the measured effects are mediated via ASICs.
Together, the new Yu et al. studies provide an elegant example of the power of small molecule based ion channel screens to identify novel channel modulators. They further demonstrate that such molecules can have great utility for probing channel mechanisms of action and for developing ideas about the effects of naturally occurring ligands. Many questions remain. How many GMQ molecules does it take to activate the channel? What are the determinants that make GMQ, arcaine, and agmatine specific for ASIC3 over the other ASICs? Are agmatine and arcaine, which are both better known for suppressing, not causing pain responses (Regunathan, 2006), the natural ligands? Are there other endogenous amines released by inflamed tissue that will activate ASICs and act with a much greater potency? Amiloride, which as has a structure that has features similar to the activators (Figure 1B), is an inhibitor. Whether its site of action is the same as GMQ is not known.
The presence of a reactive cysteine that can be modified to affect channel behavior may provide a very useful handle for further advances towards each of these questions. Thiol-capture strategies have proven very effective for identifying and improving novel modulators of soluble proteins (Erlanson et al., 2004). The clear sensitivity of the E79C mutant to thiol exchange compounds suggests that such strategies could provide a facile way forward for further elaboration of ASIC modulators. Additionally, the dramatic effect of E79C covalent modification may provide a means for trapping an ASIC channel open state for structural studies. The development of potent pharmacological tools for ion channels remains an arduous task (Dunlop et al., 2008). The work by Xu and colleagues nicely demonstrates how the interplay between structure-directed approaches, functional studies, and classic SAR can discover new compounds and uncover unexpected facets of ion channel mechanism and functional biology.
Acknowledgments
This work was supported by grants to D.L.M. from NIH (DC007664, HL080050, NS065448) and the American Heart Association (0740019N) and to S.N.B. from the Life Sciences Research Foundation. D.L.M. is an AHA Established Investigator. S.N.B. is a Genentech Fellow of the Life Sciences Research Foundation.
Footnotes
Author contributions
S.N.B. and D.L.M. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Selected reading
- Cushman KA, Marsh-Haffner J, Adelman JP, McCleskey EW. A conformation change in the extracellular domain that accompanies desensitization of acid-sensing ion channel (ASIC) 3. J Gen Physiol. 2007;129:345–350. doi: 10.1085/jgp.200709757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval E, Gasull X, Noel J, Salinas M, Baron A, Diochot S, Lingueglia E. Acid-Sensing Ion Channels (ASICs): Pharmacology and implication in pain. Pharmacol Ther. 2010 doi: 10.1016/j.pharmthera.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Deval E, Noel J, Lay N, Alloui A, Diochot S, Friend V, Jodar M, Lazdunski M, Lingueglia E. ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 2008;27:3047–3055. doi: 10.1038/emboj.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diochot S, Salinas M, Baron A, Escoubas P, Lazdunski M. Peptides inhibitors of acid-sensing ion channels. Toxicon. 2007;49:271–284. doi: 10.1016/j.toxicon.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R. High-throughput electrophysiology: an emerging paradigm for ion-channel screening and physiology. Nat Rev Drug Discov. 2008;7:358–368. doi: 10.1038/nrd2552. [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- Krishtal O. The ASICs: signaling molecules? Modulators? Trends Neurosci. 2003;26:477–483. doi: 10.1016/S0166-2236(03)00210-8. [DOI] [PubMed] [Google Scholar]
- Mazzuca M, Heurteaux C, Alloui A, Diochot S, Baron A, Voilley N, Blondeau N, Escoubas P, Gelot A, Cupo A, et al. A tarantula peptide against pain via ASIC1a channels and opioid mechanisms. Nat Neurosci. 2007;10:943–945. doi: 10.1038/nn1940. [DOI] [PubMed] [Google Scholar]
- Regunathan S. Agmatine: biological role and therapeutic potentials in morphine analgesia and dependence. AAPS J. 2006;8:E479–484. doi: 10.1208/aapsj080356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluka KA, Winter OC, Wemmie JA. Acid-sensing ion channels: A new target for pain and CNS diseases. Curr Opin Drug Discov Devel. 2009;12:693–704. [PMC free article] [PubMed] [Google Scholar]
- Wemmie JA, Price MP, Welsh MJ. Acid-sensing ion channels: advances, questions and therapeutic opportunities. Trends Neurosci. 2006;29:578–586. doi: 10.1016/j.tins.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Xiong ZG, Pignataro G, Li M, Chang SY, Simon RP. Acid-sensing ion channels (ASICs) as pharmacological targets for neurodegenerative diseases. Curr Opin Pharmacol. 2008;8:25–32. doi: 10.1016/j.coph.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Z, Li W-G, Cao H, Feng E-G, Yu F, Liu H, Jiang H, Xu T-L. A non-proton ligand sensor in the Acid-sensing ion channel. Neuron. 2010;(This issue) doi: 10.1016/j.neuron.2010.09.001. [DOI] [PubMed] [Google Scholar]