Abstract
The perciform suborder Notothenoidei provides a compelling opportunity to study the adaptive radiation of a marine species-flock in the cold Southern Ocean that surrounds Antarctica. To facilitate genome-level studies of the diversification of these fishes, we present estimates of the genome sizes of 11 Antarctic species and describe the production of high-quality bacterial artificial chromosome (BAC) libraries for two, the red-blooded notothen Notothenia coriiceps and the white-blooded icefish Chaenocephalus aceratus. Our results indicate that evolution of phylogenetically derived notothenioid families (e.g., the crown group Channichthyidae [icefishes]), was accompanied by genome expansion. Six species from the basal family Nototheniidae had C-values between 0.98 and 1.20 pg, a range that is consistent with the genome sizes of proposed outgroups (e.g., percids) of the notothenioid suborder. In contrast, four icefishes had C-values in the range 1.66–1.83 pg. The BAC libraries VMRC-19 (N. coriiceps) and VMRC-21 (C. aceratus) comprise 12× and 10× coverage of the respective genomes and have average insert sizes of 138 and 168 kb. Paired BAC-end reads representing ∼0.1% of each genome showed that the repetitive element landscapes of the two genomes (13.4% of the N. coriiceps genome and 14.5% for C. aceratus) were similar. The availability of these high-quality and well-characterized BAC libraries sets the stage for targeted genomic analyses of the unusual anatomical and physiological adaptations of the notothenioids, some of which mimic human diseases. Here we consider the evolution of secondary pelagicism by various taxa of the group and illustrate the utility of Antarctic icefishes as an evolutionary-mutant model of human osteopenia (low-mineral density of bones).
Introduction to the notothenioid radiation
The modern ichthyofauna of the Southern Ocean is severely restricted in species diversity due to dramatic paleoclimatic and paleogeographic changes in Antarctica (Eastman 1993) that occurred during the Cenozoic Era. The onset of widespread glaciation in Antarctica ∼34 million years ago (mya) (Zachos et al. 2001), attributable to (1) declining atmospheric CO2 (DeConto and Pollard 2003); (2) establishment of the Antarctic Circumpolar Current due to opening of the Drake Passage ∼40–34 mya (Kennett 1977; Livermore et al. 2005; Scher and Martin 2006); and (3) development of the Antarctic Polar Front (between ∼50 and 60°S), decoupled the Southern Ocean from warmer, subtropical waters to the north (Kennett 1977). As the Southern Ocean cooled to the freezing point of seawater (−1.9°C), the shallow-water, cosmopolitan, and temperate fish fauna of the late Eocene (38 mya) became largely extinct due to destruction of inshore habitat and changes in trophic structure caused by repeated ice-sheet scouring of the continental margin (Eastman 2005). Today, species of a single perciform suborder, the Notothenioidei, constitute 46% of the ∼300 fish species of the Southern Ocean and, at the highest latitudes, represent 77% of species diversity and 90% of biomass (Eastman 2005). Given their geographical restriction, high endemism, and rapid speciation (average time for speciation of 0.76–2.1 million years), the Notothenioidei are considered the best example of a marine species flock (Eastman 2000; Eastman and McCune 2000).
The ancestral notothenioid stock was a negatively buoyant, bottom-dwelling perciform species that arose ∼40–60 mya (DeWitt 1971; Eastman 1993; Eastman and Clarke 1998). As competing taxa became locally extinct, the notothenioids diversified to occupy the vacated ecological niches, many of which were present in the water column. Lacking a swim bladder, the Notothenioidei evolved pelagic, or partially pelagic, lifestyles by reduction of skeletal mineralization and enhancement of lipid deposition (Eastman 1993). Tailoring of morphology for life in the water column, termed secondary pelagicism, is the hallmark of the notothenioid radiation, has arisen independently several times in different clades (Eastman 1997, 1999; Near et al. 2007), and reflects the retention of larval characteristics in the adult (paedomorphism) (Eastman 1997).
The long residence of the notothenioids in a freezing marine environment has also driven the evolution of biochemical and physiological adaptations that resist or compensate for the effects of extreme cold. The evolution of the antifreeze glycoprotein genes from a pancreatic, trypsinogen-like gene (Chen et al. 1997; Cheng and Chen 1999) is a remarkable genetic “resistance” innovation that fostered survival of the group (Montgomery and Clements 2000). Major examples of “compensatory” adaptation in notothenioids include efficient microtubule assembly (Williams et al. 1985; Detrich et al. 1989, 1992, 2000; Paluh et al. 2004) and protein translocation at low temperatures (Römisch et al. 2003), homeoviscous adaptation of membrane lipids to preserve membrane fluidity (Logue et al. 2000), and cold-stable lens crystallins that prevent formation of cataracts (Kiss et al. 2004). Expansion of gene families involved in metabolic processes (e.g., biosynthesis, folding, and degradation of proteins; lipid metabolism) that are critical to the physiological fitness of these organisms has also been reported (Chen et al. 2008).
Less well appreciated are the regressive changes or losses of function (“disaptations” per Montgomery and Clements [2000]) that evolved in the notothenioids due to relaxed selection pressure in their stable, cold environment. Striking regressive changes include the loss of erythrocytes (Ruud 1954) and the respiratory transport protein hemoglobin (Cocca et al. 1995, 2000; Zhao et al. 1998; Near et al. 2006) by all species of the icefish family and independent losses of cardiac myoglobin in a subset of those species (Sidell et al. 1997; O’Brien and Sidell 2000; Sidell and O’Brien 2006). Antarctic notothenioids have also lost the inducible heat-shock response (Hofmann et al. 2000; Buckley et al. 2004), which appears to have been recruited to a constitutive status to deal with elevated denaturation of proteins caused by cold stress (Place et al. 2004; Place and Hofmann 2005; Todgham et al. 2007).
Enablement of the genomes of the Notothenioidei
The novel phenotypes evolved by the high-Antarctic notothenioids beg mechanistic explanation, which arguably may best be approached using comparative genomics. In 2003, we collected high-molecular-weight DNA from many species of these hard-to-obtain fishes with the aim of enabling genomic studies in the group. We then submitted a white paper to the Bacterial Artificial Chromosome (BAC) Resource Network at the National Human Genome Research Institute (NHGRI) to request construction of high-quality and high-representation BAC genomic libraries from two notothenioid species, the blackfin (Scotia Arc) icefish Chaenocephalus aceratus and the bullhead notothen (yellowbelly rock cod) Notothenia coriiceps. Our choice of these two species was driven strategically both by their hematological and skeletal differences and by their relative numerical dominance among large notothenioids that can be collected near Palmer Station. Chaenocephalus aceratus represents a highly derived lineage (icefishes) that is devoid of erythrocytes, lacks functional globin genes, and has poorly ossified skeletons, whereas N. coriiceps represents a more basal lineage whose members possess erythrocytes, synthesize functional hemoglobins, and are robustly ossified. Thus, we anticipate that targeted genomic comparisons of these two species will illuminate the physiological, anatomical, and gene-regulatory changes associated with diversification within the notothenioid suborder. We illustrate our approach by considering the evolution of reduced mineralization of bone in the secondarily pelagic icefish. We also emphasize that utility of this naturally selected trait as a model for a common, maladaptive human condition, osteopenia (low density of minerals in bone), which predisposes to the disease osteoporosis.
Genome expansion correlates with phyletic diversification in the suborder Notothenioidei
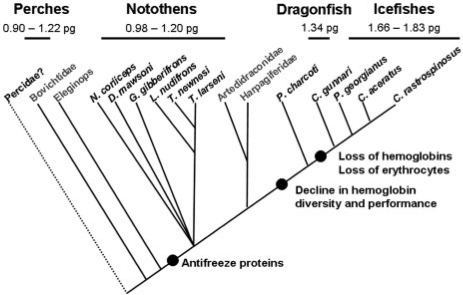
As a first step in developing genome resources for the Notothenioidei, we measured the sizes of the genomes of 11 Antarctic species by flow cytometry (Detrich et al. 2010). Figure 1 shows the phylogenetic relationships of the species relative to other notothenioid clades and indicates that values of C increase substantially as one moves from basal (Nototheniidae) to derived (Bathydraconidae, Channichthyidae) families. The basal Nototheniidae have genomes whose sizes are comparable to those of outgroups Percidae, Serranidae, and Gasterosteidae, which suggests that a C-value of ∼1 pg is ancestral for the notothenioid suborder. Our estimates of genome size for the 11 notothenioids fall in a range that is consistent with the limited number of prior measurements (Morescalchi et al. 1992, 1996; Hardie and Hebert 2003, 2004; Chen et al. 2008). Taken together, these results support the hypotheses: (1) that genome expansion accompanied the evolution of phylogenetically derived notothenioids; and (2) that genome expansion may have been driven in ways that offset reduction (Bathydraconidae) or loss (Channichthyidae) of hemoprotein function.
Fig. 1.
Phylogenetic relationships among the 11 notothenioid species examined in this study. Six notothens of the basal, red-blooded family Nototheniidae (Dissostichus mawsoni, Gobionotothen gibberifrons, Lepidonotothen nudifrons, N. coriiceps, Trematomus larseni, and T. newnesi) and four icefishes of the white-blooded notothenioid crown group Channichthyidae (C. aceratus, Champsocephalus gunnari, Chionodraco rastrospinosus, and Pseudochaenichthys georgianus) are shown with the size ranges of their genomes. One species (Parachaenichthys charcoti) of the red-blooded dragonfishes (Bathydraconidae), the sister group to the icefishes, is also presented. The perches (Percidae) are the probable sister group to the Notothenioidei (Dettai and Lecointre 2004), and the size range of their genomes is based on Perca flavescens (Hinegardner and Rosen 1972; Hardie and Hebert 2004), P. fluviatilis (Vialli 1957; Vinograd 1998), and Sander lucioperca (Vinograd 1998). The positions of the notothenioid families Bovichtidae, Eleginops, Artedidraconidae, and Harpagiferidae (gray font; not examined herein) are shown for comparison. The closed circles indicate key evolutionary events during the diversification of the Notothenioidei: the acquisition of antifreeze proteins; the decline in hemoglobin multiplicity and performance; and the losses of hemoglobins and of red blood cells by the icefishes. The tree is based on the studies of Lecointre et al. (1997), Balushkin (2000), Near et al. (2004), and Near and Cheng (2008). Reproduced from Detrich et al. (2010). Copyright (2010 Wiley-Liss, Inc. a subsidiary of John Wiley & Sons, Inc.); Reprinted with permission of John Wiley & Sons, Inc.
BAC libraries for N. coriiceps and C. aceratus
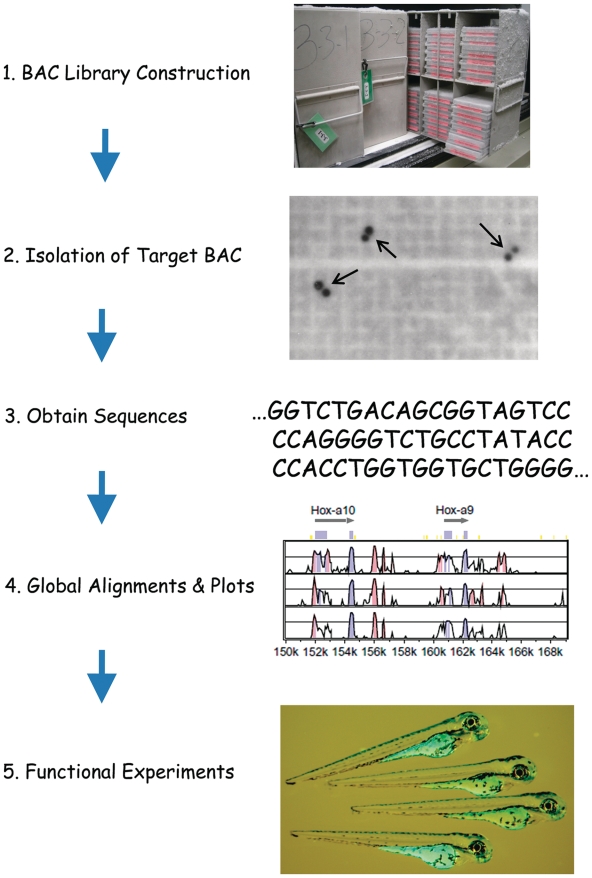
Although new technologies continue to reduce the costs of DNA sequencing, non-traditional vertebrate models, such as the Antarctic notothenioids, are unlikely to be chosen for complete genomic analysis in the foreseeable future. Nonetheless, one can envision targeted strategies (Fig. 2) whereby BAC libraries are generated from the genomes of two or more organisms with known phylogenetic relationships, BAC clones containing orthologous genomic regions of interest are isolated and their inserts sequenced; and computational tools are used to compare the regions to identify conserved and nonconserved DNA elements. The results could then be used to derive hypotheses amenable to empirical testing. One large-scale example of the targeted genomic sequencing strategy is the Encyclopedia of DNA Elements (ENCODE) initiative, which evaluated the regulatory logic of the human genome by comparison of selected regions encompassing 1% of the genome to orthologous regions from many other vertebrate species (Birney et al. 2007). Conversely, BAC libraries can be applied, at minimal cost, to quasi-global analyses, such as sequencing the ends of randomly selected BAC inserts to compare the compositions of repetitive elements of the genomes of two or more species (cf Shedlock et al. [2007]). Here we describe BAC libraries for N. coriiceps and C. aceratus and illustrate their use in both quasi-global and targeted genomic studies.
Fig. 2.
Flowchart for targeted comparisons of genomic sequences. BAC libraries are used to isolate orthologous regions of interest from the genomes of two species (1, 2). Sequences are obtained from the respective BAC clones (3) and aligned for comparison (4). Testable hypotheses derived from the sequence comparisons, such as putative regulation of gene expression by conserved DNA sequence elements, are then analyzed by functional experiments in vivo and in vitro. See Miyake and Amemiya (2004) and Amemiya and Gomez-Chiarri (2006) for in-depth reviews of the comparative genomics pipeline. Modified from Miyake and Amemiya (2004) with permission. Copyright (2004) Elsevier, Inc.
BAC libraries are based on plasmid vectors that contain an F-factor origin of replication (Shizuya et al. 1992). The F-factor replicon ensures that the plasmid replicates as a single-copy entity in Escherichia coli, a prerequisite for stable propagation of cloned inserts >100 kb. Large inserts are desirable for many genomic applications, including physical mapping of genomes, positional cloning of mutated genes, targeted genomic sequencing, and generation of transgenic animals (Yang et al. 1997; Amemiya et al. 1999; Miyake and Amemiya 2004).
Construction of the notothenioid BAC libraries was performed under the auspices of the NHGRI and followed rigid standards of quality control that mandated: (1) 10× genomic coverage; (2) average insert size of ∼150 kb; (3) assurance that the library contained a minimum of 95% insert-containing clones; and (4) little or no detectable contamination by bacterial DNA. The most critical challenge was to prepare high-molecular-weight DNA from blood cells of the two species, erythrocytes in the case of N. coriiceps and leukocytes for C. aceratus, using the laboratory facilities at Palmer Station, Antarctica. Because the laboratory lacks a pulsed field-gel apparatus, we operated “blind” and could not assess the quality of the DNAs until the samples were returned to our laboratories. Analyses of these DNAs through high-resolution, pulsed field-gel electrophoresis verified that the DNAs were suitable for BAC library construction, which subsequently followed slight modifications of published methods (Osoegawa et al. 1998; Danke et al. 2004); greater detail may be found in Detrich et al. (2010).
Table 1 provides summary statistics for the two libraries, VMRC-19 (N. coriiceps) and VMRC-21 (C. aceratus). The libraries comprise 12× and 10× coverage of the respective genomes based on the total number of clones in the library, the average insert sizes (138 kb for N. coriiceps, 168 kb for C. aceratus), and the genome sizes determined for the taxa. A survey of randomly selected BAC clones showed that non-recombinant and vector-artifact clones are rare (8% for VMRC-19, 5% for VMRC-21), and inserts of bacterial origin are not detectable. Thus, the two libraries meet nearly all of the criteria mandated by the NHGRI and are sufficiently robust to be used in targeted or global genomic studies. For inquiries regarding access to the BAC libraries, please contact Pieter de Jong (pdejong@chori.org).
Table 1.
Summary statistics for the notothenioid BAC libraries
| Chaenocephalus aceratus (VMRC-21) | Notothenia coriiceps (VMRC-19) | |
|---|---|---|
| Vector used | pCC1BAC | pCC1BAC |
| Restriction enzyme used | EcoRI | EcoRI |
| DNA source | Leucocytes | Erythrocytes |
| Number of 384-well plates | 336 | 288 |
| Eschericia coli strain used | DH10B T1 resistant | DH10B T1 resistant |
| Empty wells | 0 | 0 |
| Estimated non-recombinant clones and vector artifact clones, % | 6451 (5) | 8847 (8) |
| Estimated insert-containing clones, % | 122 573 (95) | 101 745 (92) |
| Average insert size | 168 kb | 138 kb |
| Median insert size | 168 kb | 155 kb |
| Estimated genome coverage | 10X | 12X |
| Restriction enzyme used | EcoRI | EcoRI |
Reproduced from Detrich et al. (2010). Copyright (2010 Wiley-Liss, Inc. a subsidiary of John Wiley & Sons, Inc.); Reprinted with permission of John Wiley & Sons, Inc.
Repetitive-element landscapes of the N. coriiceps and C. aceratus genomes
Large-scale BAC-end sequencing of C. aceratus and of N. coriiceps (samples represent ∼0.1% of each genome) indicated that the GC contents of the two species are identical (42%) and agree with prior estimates of notothenioid genomes (41–43%) obtained by analytical ultracentrifugation (Bucciarelli et al. 2002). The proportion of repetitive elements in the two samples (14.5% for C. aceratus and 13.4% for N. coriiceps) is similar, as are the percentages of most individual elements (Table 2). However, genomic representation of the Mariner/Tc1 transposon, of the Gypsy and Copia LTR retrotransposons, and of SINE elements appears to differ between C. aceratus and N. coriiceps. Whether these differences in retrotransposon composition contributed to the differential expansion of the icefish genomes and of individual protein-coding gene families, as postulated for LINEs by Chen et al. (2008), remains to be tested by large-scale, whole-genome sequencing and by functional approaches.
Table 2.
Repetitive elements in a sample of BAC end sequencesa
| Repetitive element class |
n (%) |
|
|---|---|---|
| Chaenocephalus aceratus | Notothenia coriiceps | |
| Transposable elements | 1390 | 618 |
| DNA transposons | 566 (40.9) | 253 (41.1) |
| Mariner/Tc1 | 27 (1.9) | 32 (5.2) |
| Kolobok | 32 (2.3) | 17 (2.8) |
| Harbinger | 30 (2.2) | 7 (1.1) |
| hAT | 211 (15.2) | 91 (15.0) |
| Other | 266 (19.1) | 106 (17.0) |
| LTR retrotransposons | 258 (18.3) | 80 (13.0) |
| Gypsy | 156 (11.0) | 49 (7.9) |
| Copia | 25 (1.8) | 1 (0.2) |
| DIRS | 46 (3.3) | 17 (2.8) |
| Other | 31 (2.2) | 13 (2.1) |
| Non-LTR retrotransposons | 468 (33.4) | 241 (38.6) |
| CR1 | 337 (24.0) | 157 (25.0) |
| L1 | 46 (3.3) | 31 (5.0) |
| SINE | 52 (3.7) | 37 (6.0) |
| Other | 33 (2.4) | 13 (2.6) |
| Endogenous Retrovirus | 98 (7.0) | 44 (7.0) |
| SSRs and satellites | 474 | 246 |
aRepetitive elements in paired-end reads of the BAC inserts were evaluated by use of RepeatMasker v3.0 (Smit et al. 2004), CENSOR (Kohany et al. 2006), and the Repbase Update (Jurka et al. 2005). Reproduced from Detrich et al. (2010). Copyright (2010 Wiley-Liss, Inc. a subsidiary of John Wiley & Sons, Inc.); Reprinted with permission of John Wiley & Sons, Inc.
CR1, chicken repeat 1; DIRS1, Dictyostelium intermediate sequence repeat 1; ERV, endogenous retroviral sequence; hAT, hobo, Ac and Tam3 family; L1, long-interspersed element 1; LTR, long-terminal repeat; SINE, short-interspersed element; SSR, simple sequence repeat; TE, transposable element.
An evolutionary-mutant model for human osteopenia: paedomorphism in notothenioid skeletal development
Albertson et al. (2009) have proposed that the analysis of non-traditional “evolutionary-mutant models” that mimic human disorders provides a complementary approach to standard forward genetics (induction of mutations in traditional laboratory animals [e.g., mice, zebrafish, flies, and worms]) for discovering genes and mechanisms that contribute to human disease. The severity, early onset, and coding-sequence bias of induced mutations often mask developmental pleiotropy and preclude phenotypic examination at the later life stages relevant to many human diseases. In contrast, many naturally occurring mutations in humans that either cause or predispose to disease result from alterations to the cis-regulatory regions of genes, whose combinatorial complexity and tissue specificity can yield normal gene activity early in development but abrogate gene regulation in adults. Given that most genetic pathways are functionally conserved in animals, one may reasonably anticipate that a molecular understanding of atypical phenotypes favored by natural selection in wild populations will contribute to identifying novel genetic factors and environmental interactions that affect human health. Here we consider the evolution of secondary pelagicism by Antarctic notothenioids and its relationship to maladaptive osteopenia in humans.
Notothenioids evolved secondary pelagicism by paedomorphism, the retention of ancestrally juvenile traits by adults of a descendant taxon (Eastman 1997). Paedomorphism results from heterochronic processes that change the schedule of developmental events (Gould 1992). When compared to benthic notothenioids, pelagic and benthopelagic species of the suborder show several paedomorphic skeletal characters: (1) delayed and reduced skeletal ossification; (2) partial or complete retention of the notochord; (3) reduction of the pterygoid process of the palatoquadrate; and (4) reduced numbers of teeth and of tooth rows.
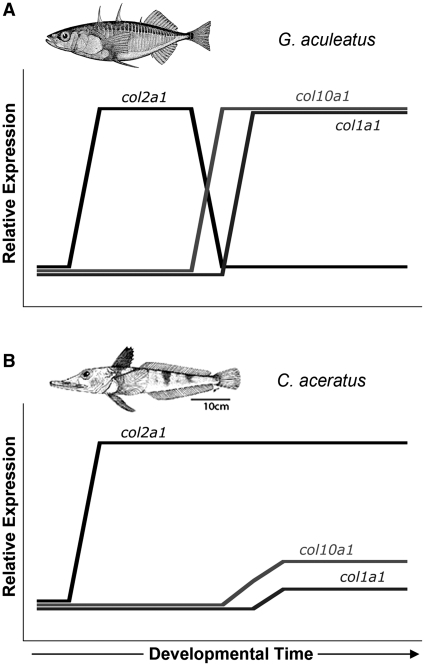
To address the molecular mechanisms that cause skeletal reduction and morphological change in notothenioids, Albertson et al. (2010) examined craniofacial development in a true pelagic species, Pleuragramma antarcticum, a benthopelagic species, C. aceratus, and a benthic species, N. coriiceps. The closely related percomorph, Gasterosteus aculeatus (three-spine stickleback), and the distantly related cyprinid, Danio rerio (zebrafish), were selected as outgroups. Using collagen genes as markers of cartilage development (col2a1) and bone formation (col1a1 and col10a1), they found that pelagic and benthopelagic notothenioid larvae exhibit delays in osteogenic development. Figure 3 shows schematically that expression of these genes in the stickleback and zebrafish follows the typical vertebrate pattern; col2a1 is expressed early in development (in differentiating chondrocytes) and then is down-regulated as expression switches to the bone marker genes col1a1 and col10a1, which are up-regulated and maintained at high levels (panel A). The pattern for pelagic/benthopelagic notothenioids differs dramatically, with early and sustained expression of col2a1 throughout larval development (panel B). Low levels of col10a1 were expressed in a subset of craniofacial elements, and col1a1 was expressed weakly in the pharyngeal skeleton (morphological features not shown). Thus, the evolution of bone loss in Antarctic notothenioids can be explained in part by prolongation of the early chondrogenic developmental program through extended periods of larval development. These data provide an initial molecular view of the signaling cascades that modulate bone density and suggest that changes in the regulation of suites of genes are linked to the adaptive radiation of Antarctic notothenioids into pelagic habitats. [The strong conservation of collagen protein sequences between the osteopenic C. aceratus and the strongly mineralized N. coriiceps reinforces the deduction that the critical evolutionary changes occurred in gene regulatory regions (Albertson et al. (2010)]. They are also likely to provide insights into the development of human osteopenia, which in turn may suggest new therapeutic approaches to prevent or treat this condition.
Fig. 3.
Schematic illustration of heterochronic alteration of collagen gene expression in the notothenioid larval skeleton relative to “conventional” teleost outgroups. (A) Expression in the three-spine stickleback G. aculeatus and in the zebrafish D. rerio follows the typical vertebrate pattern, with col2a1 expressed first in differentiating chondrocytes, followed by down-regulation of col2a1 and subsequent up-regulation of the bone markers, col10a1, and col1a1. (B) The notothenioid pattern is quite different, with sustained high levels of col2a1 expression throughout later periods of larval development, expression of col10a1 limited to a small subset of skeletal elements, and weak col1a1 expression throughout the pharyngeal skeleton. The x-axis denotes developmental time starting just before chondrocyte differentiation, and the y-axis represents relative (not quantitative) levels of collagen gene expression. Adapted from Albertson et al. (2010) with permission of the authors.
We are now ready to use the targeted genomic strategy to test the hypothesis that alteration of the regulation of collagen gene expression is involved in adaptive modulation of bone density. Using our BAC libraries for the benthic notothenioid N. coriiceps and the benthopelagic C. aceratus, we will isolate orthologous BAC clones whose inserts contain the col1a1, col2a1, and col10a1 genes. Sequence analysis of these clones and bioinformatic comparison of non-coding regions should reveal whether or not the regulatory motifs governing the expression of orthologous collagen genes in the two species have diverged. Simultaneous comparison of the notothenioid regulatory elements to those of collagen genes from stickleback and zebrafish is necessary to ensure that sound, phylogenetically controlled, inferences are drawn. Functional testing by generation of transgenic stickleback or zebrafish whose collagen gene regulatory motifs have been swapped for those of the two notothenioids would complete the formal chain of logic.
Recently, Chan et al. (2010) showed that the pelvic reduction in freshwater populations of the three-spine stickleback has evolved repeatedly through deletion of a tissue-specific enhancer for the Pituitary homeobox transcription factor 1 (Pitx1) gene. Restoration of Pitx1 expression by transgenic insertion of the enhancer into a pelvic-reduced laboratory line rescued pelvic girdle and spine formation. We are tempted to speculate that a similar mechanism for deletion may underlie the evolutionary transition between the switching of collagen gene expression observed in robustly mineralized notothenioids and the loss of switching observed in osteopenic species.
Funding
National Institutes of Health (grant AG031922 to H.W.D. and others); National Institutes of Health (grant HG02526 to C.T.A.); National Science Foundation (grants OPP-0336932, ANT-0635470 to H.W.D.) National Science Foundation (grant ANT-0632527 to C.T.A.).
Acknowledgments
We thank the personnel of the Office of Polar Programs of the National Science Foundation, of Raytheon Polar Services Company, and of the ARSV Laurence M. Gould, without whose support this work could not have been accomplished. Guillaume Lecointre (Muséum National d’Histoire Naturelle, Paris) and Andrew Shedlock (Museum of Comparative Zoology, Harvard University) provided valuable advice on the phylogeny of the notothenioids and on analysis of the repetitive sequence data, respectively.
References
- Albertson RC, Cresko W, Detrich HW, III, Postlethwait JH. Evolutionary mutant models for human disease. Trends Genet. 2009;25:74–81. doi: 10.1016/j.tig.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Yan YL, Titus TA, Pisano E, Vacchi M, Yelick PC, Detrich HW, III, Postlethwait JH. Molecular pedomorphism underlies craniofacial skeletal evolution in Antarctic notothenioid fishes. BMC Evol Biol. 2010;10:4. doi: 10.1186/1471-2148-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya CT, Gomez-Chiarri M. Comparative genomics in vertebrate evolution and development. J Exp Zool A Comp Exp Biol. 2006;305:672–82. doi: 10.1002/jez.a.308. [DOI] [PubMed] [Google Scholar]
- Amemiya CT, Zhong TP, Silverman GA, Fishman MC, Zon LI. Zebrafish YAC, BAC, and PAC genomic libraries. Methods Cell Biol. 1999;60:235–58. doi: 10.1016/s0091-679x(08)61904-4. [DOI] [PubMed] [Google Scholar]
- Balushkin AV. Morphology, classification, and evolution of notothenioid fishes of the Southern Ocean (Notothenioidei, Perciformes) J Ichthyol. 2000;40:S74–109. [Google Scholar]
- Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciarelli G, Bernardi G, Bernardi G. An ultracentrifugation analysis of two hundred fish genomes. Gene. 2002;295:153–62. doi: 10.1016/s0378-1119(02)00733-3. [DOI] [PubMed] [Google Scholar]
- Buckley BA, Place SP, Hofmann GE. Regulation of heat shock genes in isolated hepatocytes from an Antarctic fish, Trematomus bernacchii. J Exp Biol. 2004;207:3649–56. doi: 10.1242/jeb.01219. [DOI] [PubMed] [Google Scholar]
- Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–5. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-HC, Chen L. Evolution of an antifreeze glycoprotein: a blood protein that keeps Antarctic fish from freezing arose from a digestive enzyme. Nature. 1999;401:443–4. doi: 10.1038/46721. [DOI] [PubMed] [Google Scholar]
- Chen Z, et al. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc Natl Acad Sci USA. 2008;105:12944–49. doi: 10.1073/pnas.0802432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, DeVries AL, Cheng C-HC. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notohthenioid fish. Proc Natl Acad Sci USA. 1997;94:3811–6. doi: 10.1073/pnas.94.8.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocca E, Detrich HW, III, Parker SK, di Prisco G. A cluster of four globin genes from the Antarctic fish Notothenia coriiceps. J Fish Biol. 2000;57:33–50. [Google Scholar]
- Cocca E, Ratnayake-Lecamwasam M, Parker SK, Camardella L, Ciaramella M, di Prisco G, Detrich HW., III Genomic remnants of α-globin genes in the hemoglobinless antarctic icefishes. Proc Natl Acad Sci USA. 1995;92:1817–21. doi: 10.1073/pnas.92.6.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danke J, Miyake T, Powers T, Schein J, Shin H, Bosdet I, Erdmann M, Caldwell R, Amemiya CT. Genome resource for the Indonesian coelacanth, Latimeria menadoensis. J Exp Zool A Comp Exp Biol. 2004;301:228–34. doi: 10.1002/jez.a.20024. [DOI] [PubMed] [Google Scholar]
- DeConto RM, Pollard D. Rapid Cenozoic glaciation of Antarctica induced by declining atmospheric CO2. Nature. 2003;421:245–9. doi: 10.1038/nature01290. [DOI] [PubMed] [Google Scholar]
- Detrich HW, III, Fitzgerald TJ, Dinsmore JH, Marchese-Ragona SP. Brain and egg tubulins from Antarctic fishes are functionally and structurally distinct. J Biol Chem. 1992;267:18766–75. [PubMed] [Google Scholar]
- Detrich HW, III, Johnson KA, Marchese-Ragona SP. Polymerization of Antarctic fish tubulins at low temperatures: energetic aspects. Biochemistry. 1989;28:10085–93. doi: 10.1021/bi00452a031. [DOI] [PubMed] [Google Scholar]
- Detrich HW, III, Parker SK, Williams RC, Jr, Nogales E, Downing KH. Cold adaptation of microtubule assembly and dynamics: structural interpretation of primary sequence changes present in the α- and β-tubulins of Antarctic fishes. J Biol Chem. 2000;275:37038–47. doi: 10.1074/jbc.M005699200. [DOI] [PubMed] [Google Scholar]
- Detrich HW, III, Stuart A, Schoenborn M, Parker SK, Methe BA, Amemiya CT. Genome enablement of the Notothenioidei: genome size estimates from eleven species and BAC libraries from two representative taxa. J Exp Zool B Mol Dev Evol. 2010;314:369–81. doi: 10.1002/jez.b.21341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettäi A, Lecointre G. In search of notothenioid (Teleostei) relatives. Antarctic Sci. 2004;16:71–85. [Google Scholar]
- DeWitt HH. Coastal and deep-water benthic fishes of the Antarctic. In: Bushnell VC, editor. Antarctic Map Folio Series, Folio 15. New York: American Geographical Society; 1971. pp. 1–10. [Google Scholar]
- Eastman JT. San Diego: Academic Press; 1993. Antarctic fish biology: evolution in a unique environment. [Google Scholar]
- Eastman JT. Phyletic divergence and specialization for pelagic life in the Antarctic notothenioid fish Pleuragramma antarcticum. Comp Biochem Physiol. 1997;118A:1095–101. [Google Scholar]
- Eastman JT. Aspects of the biology of the icefish Dacodraco hunteri (Notothenioidei, Channichthyidae) in the Ross Sea, Antarctica. Polar Biol. 1999;21:194–6. [Google Scholar]
- Eastman JT. Antarctic notothenioid fishes as subjects for research in evolutionary biology. Antarctic Sci. 2000;12:276–87. [Google Scholar]
- Eastman JT. The nature of the diversity of Antarctic fishes. Polar Biol. 2005;28:93–107. [Google Scholar]
- Eastman JT, Clarke A. A comparison of adaptive radiations of Antarctic fish with those of non-Antarctic fish. In: di Prisco G, Pisano E, Clarke A, editors. Fishes of Antartica: a biological overview. Milano: Springer-Verlag; 1998. pp. 3–26. [Google Scholar]
- Eastman JT, McCune AR. Fishes on the Antarctic continental shelf: evolution of a marine species flock? J Fish Biol. 2000;57A:84–102. [Google Scholar]
- Gould SJ. Ontogeny and phylogeny–revisited and reunited. Bioessays. 1992;14:275–279. doi: 10.1002/bies.950140413. [DOI] [PubMed] [Google Scholar]
- Hardie DC, Hebert PD. The nucleotypic effects of cellular DNA content in cartilaginous and ray-finned fishes. Genome. 2003;46:683–706. doi: 10.1139/g03-040. [DOI] [PubMed] [Google Scholar]
- Hardie DC, Hebert PD. Genome-size evolution in fishes. Can J Fish Aq Sci. 2004;61:1636–46. [Google Scholar]
- Hinegardner R, Rosen DE. Cellular DNA content and the evolution of teleostean fishes. Am Naturalist. 1972;106:621–44. [Google Scholar]
- Hofmann GE, Buckley BA, Airaksinen S, Keen JE, Somero GN. Heat-shock protein expression is absent in the antarctic fish Trematomus bernacchii (family Nototheniidae) J Exp Biol. 2000;203:2331–9. doi: 10.1242/jeb.203.15.2331. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kennett JP. Cenozoic evolution of Antarctic glaciation, the circum-Antarctic ocean, and their impact on global paleoceanography. J Geophys Res. 1977;82:3843–60. [Google Scholar]
- Kiss AJ, Mirarefi AY, Ramakrishnan S, Zukoski CF, DeVries AL, Cheng CH. Cold-stable eye lens crystallins of the Antarctic nototheniid toothfish Dissostichus mawsoni Norman. J Exp Biol. 2004;207:4633–49. doi: 10.1242/jeb.01312. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecointre G, Bonillo C, Ozouf-Costaz C, Hureau J-C. Molecular evidence for the origins of Antarctic fishes: paraphyly of the Bovichtidae and no indication for the monophyly of the Notothenioidei (Teleostei) Polar Biol. 1997;18:193–208. [Google Scholar]
- Livermore R, Nankivell A, Eagles G, Morris P. Paleogene opening of the Drake Passage. Earth Planet Sci Lett. 2005;236:459–70. [Google Scholar]
- Logue JA, De Vries AL, Fodor E, Cossins AR. Lipid compositional correlates of temperature-adaptive interspecific differences in membrane physical structure. J Exp Biol. 2000;203:2105–15. doi: 10.1242/jeb.203.14.2105. [DOI] [PubMed] [Google Scholar]
- Miyake T, Amemiya CT. BAC libraries and comparative genomics of aquatic chordate species. Comp Biochem Physiol C Toxicol Pharmacol. 2004;138:233–44. doi: 10.1016/j.cca.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Clements K. Disaptation and recovery in the evolution of Antarctic fishes. Trends Ecol Evol. 2000;15:267–71. doi: 10.1016/s0169-5347(00)01896-6. [DOI] [PubMed] [Google Scholar]
- Morescalchi A, Hureau JC, Olmo E, Ozouf-Costaz C, Pisano E, Stanyon R. A multiple sex-chromosome system in Antarctic ice-fishes. Polar Biol. 1992;11:655–61. [Google Scholar]
- Morescalchi A, Morescalchi MA, Odierna G, Stingo V, Capriglione T. Karyotype and genome size of zoarcids and notothenioids (Teleostei, Perciformes) from the Ross Sea: cytotaxonomic implications. Polar Biol. 1996;16:559–64. [Google Scholar]
- Near TJ, Cheng CH. Phylogenetics of notothenioid fishes (Teleostei: Acanthomorpha): inferences from mitochondrial and nuclear gene sequences. Mol Phylogenet Evol. 2008;47:832–40. doi: 10.1016/j.ympev.2007.11.027. [DOI] [PubMed] [Google Scholar]
- Near TJ, Kendrick BJ, Detrich HW, III, Jones CD. Confirmation of neutral buoyancy in Aethotaxis mitopteryx DeWitt (Notothenioidei: Nototheniidae) Polar Biol. 2007;30:443–7. [Google Scholar]
- Near TJ, Parker SK, Detrich HW., III A genomic fossil reveals key steps in hemoglobin loss by the Antarctic icefishes. Mol Biol Evol. 2006;23:2008–16. doi: 10.1093/molbev/msl071. [DOI] [PubMed] [Google Scholar]
- Near TJ, Pesavento JJ, Cheng CH. Phylogenetic investigations of Antarctic notothenioid fishes (Perciformes: Notothenioidei) using complete gene sequences of the mitochondrial encoded 16S rRNA. Mol Phylogenet Evol. 2004;32:881–91. doi: 10.1016/j.ympev.2004.01.002. [DOI] [PubMed] [Google Scholar]
- O’Brien KM, Sidell BD. The interplay among cardiac ultrastructure, metabolism and the expression of oxygen-binding proteins in Antarctic fishes. J Exp Biol. 2000;203:1287–97. doi: 10.1242/jeb.203.8.1287. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, Woon PY, Zhao B, Frengen E, Tateno M, Catanese JJ, de Jong PJ. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Paluh JL, Killilea AN, Detrich HW, III, Downing KH. Meiosis-specific failure of cell cycle progression in fission yeast by mutation of a conserved β-tubulin residue. Mol Biol Cell. 2004;15:1160–71. doi: 10.1091/mbc.E03-06-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place SP, Hofmann GE. Constitutive expression of a stress-inducible heat shock protein gene, hsp70, in phylogenetically distant Antarctic fish. Polar Biol. 2005;28:261–7. [Google Scholar]
- Place SP, Zippay ML, Hofmann GE. Constitutive roles for inducible genes: evidence for the alteration in expression of the inducible hsp70 gene in Antarctic notothenioid fishes. Am J Physiol Regul Integr Comp Physiol. 2004;287:R429–36. doi: 10.1152/ajpregu.00223.2004. [DOI] [PubMed] [Google Scholar]
- Romisch K, Collie N, Soto N, Logue J, Lindsay M, Scheper W, Cheng C-HC. Protein translocation across the endoplasmic reticulum membrane in cold-adapted organisms. J Cell Sci. 2003;116:2875–83. doi: 10.1242/jcs.00597. [DOI] [PubMed] [Google Scholar]
- Ruud JT. Vertebrates without erythrocytes and blood pigment. Nature. 1954;173:848–50. doi: 10.1038/173848a0. [DOI] [PubMed] [Google Scholar]
- Scher HD, Martin EE. Timing and climatic consequences of the opening of Drake Passage. Science. 2006;312:428–30. doi: 10.1126/science.1120044. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Botka CW, Zhao S, Shetty J, Zhang T, Liu JS, Deschavanne PJ, Edwards SV. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc Natl Acad Sci USA. 2007;104:2767–72. doi: 10.1073/pnas.0606204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–7. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidell BD, O’Brien KM. When bad things happen to good fish: the loss of hemoglobin and myoglobin expression in Antarctic icefishes. J Exp Biol. 2006;209:1791–802. doi: 10.1242/jeb.02091. [DOI] [PubMed] [Google Scholar]
- Sidell BD, Vayda ME, Small DJ, Moylan TJ, Londraville RL, Yuan ML, Rodnick KJ, Eppley ZA, Costello L. Variable expression of myoglobin among the hemoglobinless Antarctic icefishes. Proc Natl Acad Sci USA. 1997;94:3420–4. doi: 10.1073/pnas.94.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 2004 1996–2010, available from: http://www.repeatmasker.org. [Google Scholar]
- Todgham AE, Hoaglund EA, Hofmann GE. Is cold the new hot? Elevated ubiquitin-conjugated protein levels in tissues of Antarctic fish as evidence for cold-denaturation of proteins in vivo. J Comp Physiol [B] 2007;177:857–66. doi: 10.1007/s00360-007-0183-2. [DOI] [PubMed] [Google Scholar]
- Vialli M. Volume et contenu en ADN par noyau. Exp Cell Res Suppl. 1957;4:284–93. [PubMed] [Google Scholar]
- Vinograd AE. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry. 1998;31:100–9. doi: 10.1002/(sici)1097-0320(19980201)31:2<100::aid-cyto5>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Williams RC, Jr, Correia JJ, DeVries AL. Formation of microtubules at low temperature by tubulin from Antarctic fish. Biochemistry. 1985;24:2790–8. doi: 10.1021/bi00332a029. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–65. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–93. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ratnayake-Lecamwasam M, Parker SK, Cocca E, Camardella L, di Prisco G, Detrich HW., III The major adult α-globin gene of Antarctic teleosts and its remnants in the hemoglobinless icefishes: calibration of the mutational clock for nuclear genes. J Biol Chem. 1998;273:14745–52. doi: 10.1074/jbc.273.24.14745. [DOI] [PubMed] [Google Scholar]