Abstract
Nevirapine (NVP) is a non-nucleoside reverse transcriptase inhibitor used against the human immunodeficiency virus type-1 (HIV-1), mostly to prevent mother-to-child HIV-1 transmission in developing countries. Despite its clinical efficacy, NVP administration is associated with a variety of toxic responses that include hepatotoxicity and skin rash. Although the reasons for the adverse effects of NVP administration are still unclear, increasing evidence supports the involvement of metabolic activation to reactive electrophiles. In particular, Phase II activation of the NVP metabolite 12-hydroxy-NVP is thought to mediate NVP binding to bionucleophiles, which may be at the onset of toxicity. In the present study, we investigated the nature and specific locations of the covalent adducts produced in human serum albumin and human hemoglobin by reaction in vitro with the synthetic model electrophile 12-mesyloxy-NVP, used as a surrogate for the Phase II metabolite 12-sulfoxy-NVP. Multiple sites of modification were identified by two different mass spectrometry-based methodologies, liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) and matrix-assisted laser desorption ionization tandem mass spectrometry (MALDI-TOF-TOF-MS). These two distinct methodologies, which in some instances afforded complementary information, allowed the identification of multiple adducts involving cysteine, lysine, tryptophan, histidine, serine, and the N-terminal valine of hemoglobin. Tryptophan, which is not a common site of covalent protein modification, was the NVP-modified amino acid residue detected in the two proteins and consistently identified by both LC-ESI-MS/MS and MALDI-TOF-TOF-MS. The propensity of tryptophan to react with the NVP-derived electrophile is further emphasized by the fact that human serum albumin possesses a single tryptophan residue, which suggests a remarkable selectivity that may be useful for biomonitoring purposes. Likewise, the NVP adduct with the terminal valine of hemoglobin, detected by LC-ESI-MS/MS after N-alkyl Edman degradation, appears as an easily assessed marker of NVP binding to proteins. Our results demonstrate the merits and complementarity of the two MS-based methodologies for the characterization of protein binding by NVP and suggest a series of plausible biomarkers of NVP toxicity, that should be useful in the monitoring of toxicity effects in patients administered NVP.
Keywords: biomarker, HIV-1, mass spectrometry, nevirapine, non-nucleoside, reverse transcriptase inhibitor, protein adducts, toxicity
Introduction
Nevirapine (11-cyclopropyl-5,11-dihydro-4-methyl-6H-dipyrido[3,2-b:2′,3′-e][1,4]diazepin-6-one, NVP1, Scheme 1) was the first non-nucleoside reverse transcriptase inhibitor approved for use in combination therapy of HIV-1 infection (1). NVP-based regimens have since gained a significant role in HIV-treatment guidelines (2), due to the high efficacy levels of the drug, favorable lipid profile, and suitability for use during pregnancy (3). NVP is currently one of the most prescribed antiretrovirals in the developing world, both to prevent mother-to-child HIV transmission and in combination therapy (4-7). Moreover, regimens that include non-nucleoside reverse transcriptase inhibitors allow holding protease inhibitors for later use, with the advantage of delaying exposure to some protease inhibitor-associated adverse effects, such as metabolic disease (8). Whereas a disadvantage of chronic NVP regimens is a twice-daily dosage schedule, which reduces patient compliance causing treatment failure and drug resistance, prospects of a new extended-release formulation aimed for a once-daily treatment regimen have recently been reported (9). Should this more convenient dosing schedule become available, worldwide NVP use will likely increase, especially considering the low cost of the drug (4).
Scheme 1.
Structures of NVP, NVP metabolites, and other NVP derivatives mentioned in the text.
Despite its clinical efficacy, and although individual susceptibilities to adverse effects differ among patients, NVP produces a variety of toxic responses (10-12). The most severe is an occasionally fatal hepatotoxicity, while the most common side effect is skin rash, which may be life threatening and lead to discontinuation of the drug. Although NVP is neither mutagenic nor clastogenic in standard in vitro assays, it induces hepato-neoplasias in rodents (13). Moreover, whereas no direct correlation between NVP administration and the development of cancer in humans has been reported, an association between the use of non-nucleoside reverse transcriptase inhibitors and an increased incidence of Non-AIDS-Defining Cancers in HIV-1-positive patients has been suggested (14). These toxic events raise concerns about the chronic administration of the drug, particularly in perinatal and pediatric settings.
While the reasons for the adverse effects of NVP are still unclear, the involvement of metabolic activation to reactive electrophiles is supported by recent studies, such as the detection of covalent binding of [14C]nevirapine to rat and human liver microsomal proteins in vitro, and to rat liver tissue and plasma proteins in vivo (15). In all species investigated, including humans, NVP metabolism consistently involves cytochrome P450-mediated oxidation to 2-, 3-, and 8-hydroxy-NVP, 4-hydroxymethyl-NVP (12-hydroxy-NVP), and 4-carboxy-NVP (Scheme 1); these metabolites typically undergo subsequent glucuronidation and excretion (16-20). In humans, the formation of 2-hydroxy-NVP is attributed to the CYP3A subfamily, 3-hydroxy-NVP to CYP2B6, 8-hydroxy-NVP to CYP3A4, CYP2B6, and CYP2D6, and 12-hydroxy-NVP to CYP3A4 and possibly CYP2D6 and CYP2C9 (18). Phase II pathways other than glucuronidation (e.g., acetylation, sulfonation, or the generation of quinone/semi-quinone species) (21) are not excluded; such pathways can generate electrophilic metabolites with the potential to react with bionucleophiles and initiate toxic events. Consistent with this interpretation is the evidence for NVP sulfonation in vivo presented by Chen et al. (22), who detected 12-sulfoxy-NVP (Scheme 1) by LC-MS in urine and bile samples from female Brown Norway rats administered NVP. Moreover, the same study provided significant support for the involvement of 12-hydroxy-NVP in an idiosyncratic NVP-induced skin rash observed in this animal model and resembling the rash that occurs in humans treated with NVP (23-25). A quinone methide metabolite (Scheme 1) was proposed as the reactive intermediate in this process (22), and also as the precursor of an NVP-glutathione conjugate through NVP-C12 that was detected upon incubation of NVP with human liver microsomes supplemented of glutathione (26). More recently, two NVP mercapturates, through NVP-C3 and NVP-C12, were identified in the bile and urine of NVP-treated rats, as well as in the urine of HIV-positive patients treated with a standard antiretroviral therapeutic regimen that included NVP (27). Taken together, these data provide evidence that the Phase I metabolite, 12-hydroxy-NVP, has the ability to react with bionucleophiles yielding covalent adducts. Given that sulfotransferases are present in the skin (28, 29), one plausible mechanism accounting for the idiosyncratic skin rash associated with NVP administration may involve the sulfation of 12-hydroxy-NVP, possibly followed by elimination of hydrogen sulfate to yield the quinone methide, subsequent binding to skin proteins and the onset of an immune response (30).
Protein binding by reactive electrophiles has received much attention in recent years (31, 32). Although the interpretation of covalent binding data is often confounded by the lack of obvious causal relationships between the occurrence of protein adduction and toxicity, the available data have consistently indicated that protein modification by a specific electrophile tends to be reproducible and highly selective, which may in turn lead to selective organ toxicity (33-36). Moreover, stable protein adducts are convenient biomarkers of exposure because, unlike DNA adducts, they are not prone to repair and accumulate over the lifespan of the protein (37). Abundant blood proteins, such as human serum albumin (HSA) and human hemoglobin (Hb) are used extensively in this context, and mass spectrometry (MS)-based methodologies have become essential tools for qualitative, quantitative, and mechanistic studies of the interaction between reactive metabolites and proteins (38, 39).
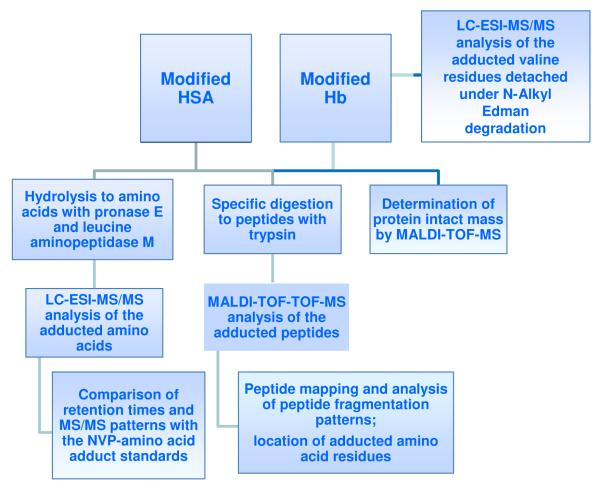
Using 12-mesyloxy-NVP (Scheme 1) as a synthetic surrogate for 12-sulfoxy-NVP, we have demonstrated direct reaction in vitro with both DNA and amino acids and characterized a series of covalent NVP-DNA and NVP-amino acid adducts (40, 41). Moreover, by isolating glutathione and N-acetylcysteine conjugates through NVP C12 that were identical to those reported to be formed in vivo (26, 27), we established the validity of our synthetic model (41). The aim of the current study was to explore this model further by characterizing the chemical structures and binding sites of the NVP-amino acid adducts formed upon in vitro reaction of HSA and human Hb with 12-mesyloxy-NVP. The strategies adopted toward this goal are summarized in Scheme 2. The approach encompassed i) enzymatic hydrolysis of the modified proteins to amino acids, followed by liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS), ii) specific detachment of N-terminal valine adducts in Hb by an N-alkyl Edman procedure, followed by LC-ESI-MS/MS, iii) determination of the intact protein mass by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS), and iv) specific digestion of the modified proteins with trypsin, followed by MALDI-TOF-TOF-MS analysis of the obtained peptides. By using a combination of MS-based methodologies, we sought to identify the amino acids and specific sites in both proteins more prone to react with NVP-derived electrophiles and, ultimately, to establish which adducts and/or tryptic peptides may potentially be used as biomarkers to monitor NVP adduction in blood proteins from NVP-treated patients.
Scheme 2.
Experimental approach used to identify the adducted amino acid residues in HSA and Hb following incubation of the proteins with 12-mesyloxy-NVP.
Materials and Methods
Caution
NVP and its derivatives are potentially carcinogenic. They should be handled with protective clothing in a well-ventilated fume hood.
Chemicals
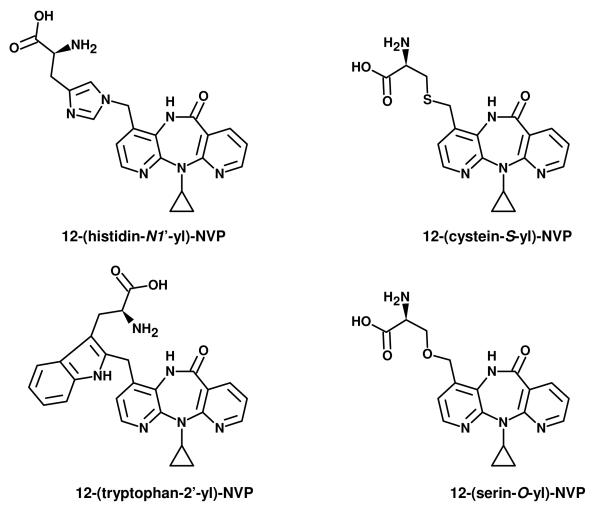
NVP was purchased from Cipla (Mumbai, India). HSA, human Hb, and all other commercially available reagents and enzymes were acquired from Sigma-Aldrich Química, S.A. (Madrid, Spain), unless specified otherwise, and used as received. Whenever necessary, solvents were purified by standard methods (42). 12-Hydroxy-NVP and 12-mesyloxy-NVP were prepared as described in Antunes et al. (40). The NVP-amino acid adduct standards, 12-(histidin-N1′-yl)-NVP, 12-(cystein-S-yl)-NVP, 12-(tryptophan-2′-yl)-NVP (Scheme 3) and 12-[5-isopropyl-4-oxo-3-phenyl-2-thioxoimidazolidin-1-yl]-NVP (Scheme 4) were prepared by reaction of 12-mesyloxy-NVP with histidine, N-acetyl cysteine, tryptophan, and ethyl valinate, respectively, as described in Antunes et al. (41).
Scheme 3.
Structures of the NVP-amino acid adducts detected in the NVP-modified proteins. 12-(Histidin-N1′-yl)-NVP, 12-(cystein-S-yl)-NVP, and 12-(tryptophan-2′-yl)-NVP were identified by LC-ESI-MS/MS in the enzymatic hydrolysate obtained by treatment of NVP-modified HSA with pronase E and leucine aminopeptidase M. Following a similar treatment, 12-(cystein-S-yl)-NVP and 12-(tryptophan-2′-yl)-NVP were also identified in the hydrolysate from NVP-modified Hb. The Hb hydrolysate contained an additional adduct, presumed to be 12-(serin-O-yl)-NVP.
Scheme 4.
Structure of the derivatized valine adduct, 12-[5-isopropyl-4-oxo-3-phenyl-2-thioxoimidazolidin-1-yl]-NVP, identified by LC-ESI-MS/MS in the ethyl acetate extract obtained after N-alkyl Edman degradation of NVP-modified Hb.
Protein Modification Reactions
1.1. Modification of HSA with two different amounts of 12-mesyloxy-NVP
1.1.1. A solution of 12-mesyloxy-NVP (5 mg, 14 μmol) in THF (500 μL) was added to a solution of HSA (10 mg) in PBS (10 mL). After an overnight incubation at 37°C, the mixture was dialyzed for 24 h against deionized water (2 L). The resulting solution was dispensed into 1 mL aliquots that were evaporated separately under a nitrogen stream.
1.1.2. After an overnight incubation at 37°C, as described above, a second portion of 12-mesyloxy-NVP (5 mg, 14 μmol) in THF (500 μL) was added, and the incubation was continued at 37°C for 72 h. Non-bonded materials were again removed by dialysis against deionized water (2 L) for 24 h and the resulting solution was aliquoted and dried as indicated above.
1.2. Modification of Hb with 12-mesyloxy-NVP
A solution of 12-mesyloxy-NVP (6 mg, 17 μmol) in acetonitrile (260 μL) was added to a solution of human Hb (10 mg) in 100 mM phosphate buffer, pH 7.4 (10 mL). The resulting solution was incubated overnight at 37°C and the non-bonded materials were subsequently removed by extraction with ethyl acetate (2 × 1 mL).
2. Adduct isolation from the NVP-modified proteins
2.1. Detachment of N-terminal valine adducts from NVP-modified Hb by N-Alkyl Edman Degradation
An aliquot (2.5 mL) of the modified Hb solution was lyophilized and then subjected to an N-alkyl Edman procedure (43, 44). Briefly, the sample was dissolved in DMF (1 mL), followed by addition of 1 M NaOH (40 μL) and phenyl isothiocyanate (7 μL, 58.6 μmol). The sample was subsequently stirred for 2 h at 37°C and then for 1.5 h at 45°C. Upon cooling to room temperature, water (2 mL) was added and the adducts were extracted with ethyl acetate (2 × 1 mL). The organic phase was dried under reduced pressure and the contents were analyzed by LC-ESI-MS.
2.2. Hydrolysis of NVP-modified HSA to amino acids
Pronase E (19 μL, 530 μg/mL) and leucine aminopeptidase M (8 μL, 130 μg/mL) were added to each aliquot containing approximately 1 mg of NVP-modified HSA in PBS (350 μL). The solution was incubated at 37°C overnight. The enzymatic hydrolysate thus obtained was analyzed directly by LC-ESI-MS with no further treatment.
2.3. Hydrolysis of NVP-modified Hb to amino acids
One aliquot (4 mL) of the modified Hb was hydrolyzed enzymatically to amino acids by treatment with pronase E (76 μL, 530 μg/mL) and leucine aminopeptidase M (32 μL, 130 μg/mL). The solution was incubated overnight at 37°C. One aliquot (500 μL) of the enzymatic hydrolysate was analyzed directly by LC-ESI-MS with no further treatment. The remaining portion of the enzymatic hydrolysate (3.5 mL) was purified using a C-18 Sep-Pak cartridge (Waters Associates, Milford, MA). The cartridge was conditioned with methanol (4 mL), followed by water (4 mL). The sample was then loaded and the cartridge was rinsed with water (2 mL) and methanol (2 mL). The methanolic eluate was concentrated to 300 μL with a nitrogen stream and analyzed by LC-ESI-MS.
2.4. In gel trypsin digestion of NVP-modified HSA and Hb
The NVP-modified proteins were separated by 1D-polyacrylamide gel electrophoresis (1D-PAGE). Specifically, five μg of modified HSA and Hb in 1D-PAGE sample buffer [62.5 mM Tris-HCl, pH 6.8, containing 20% (v/v) glycerol, 2% (w/v) β-mercaptoethanol, and a trace of bromophenol blue] were separated on 12% (w/v) polyacrylamide gels and stained with Colloidal Coomassie Blue (45). In gel trypsin digestion of the modified proteins was performed according to a protocol described in Gomes et al. (46). Briefly, the HSA and Hb gel bands were excised for in-gel digestion and destained with 50% (v/v) acetonitrile. After dehydration of the excised gel pieces by treatment with 100% acetonitrile and vacuum-drying, the proteins were reduced with 10 mM dithiothreitol, subsequently alkylated with 55 mM iodoacetamide, and then digested overnight at 37°C with sequencing-grade modified trypsin (6.7 ng/μL in 50 mM ammonium bicarbonate). The supernatant was then recovered and analyzed by MALDI-TOF-TOF-MS after desalting and concentration of the tryptic peptides (vide infra).
Instrumentation and Analytical Procedures
1. Liquid Chromatography-Electrospray Ionization-Tandem Mass Spectrometry (LC-ESI-MS/MS)
LC-ESI-MS/MS analyses were performed with a ProStar 410 autosampler, two 210-LC chromatography pumps, a ProStar 335 diode array detector, and a 500-MS ion trap mass spectrometer, with an ESI ion source (Varian, Inc., Palo Alto, CA). Data acquisition and processing were performed using Varian MS Control 6.9 software. The samples were injected onto the column via a Rheodyne injector with a 20 μL loop. Separations were conducted at 30°C, using a Luna C18 (2) column (150 mm × 2 mm, 3 μm; Phenomenex, Torrance, CA). The mobile phase was delivered at a flow rate of 200 μL/min, using a 5-min isocratic elution with 5% acetonitrile in 0.1% aqueous formic acid, followed by a 30-min linear gradient from 5-70% acetonitrile, a 2-min linear gradient to 100% acetonitrile, and an 8-min isocratic elution with acetonitrile. The mass spectrometer was operated in the positive ESI mode; the optimized operating parameters were: ion spray voltage, +5.2 kV; capillary voltage, 20 V; and RF loading, 90%. Nitrogen was used as the nebulizing and drying gas, at pressures of 50 and 30 psi, respectively; the drying gas temperature was 350°C. MS/MS spectra were obtained with an isolation window of 1.5 Da, excitation energy values between 0.9 and 1.2 V, and an excitation time of 10 ms.
2. Matrix-Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS)
Desalting and concentration of the NVP-modified proteins and tryptic peptides were performed with home-made chromatographic microcolumns using GELoader tips packed with POROS R1 (for NVP-modified proteins) and POROS R2 (for tryptic peptides) (Applied Biosystems, Foster City, CA). The proteins and peptides were eluted directly from the microcolumns onto the MALDI plate using 50% (v/v) aqueous acetonitrile containing 5% (v/v) formic acid and either 10 mg/mL sinapinic acid (for proteins) or 5 mg/mL α-cyano-4-hydroxy-trans-cinnamic acid (for peptides) (46). The sample-matrix mixtures were then allowed to air dry.
MS experiments were performed using a MALDI-TOF/TOF 4800 plus mass spectrometer (Applied Biosystems) in the positive linear mode for intact protein mass determinations and the positive reflectron mode for peptide mass measurements. For peptide mass measurements, the mass spectrometer was externally calibrated using des-Arg-Bradykinin (904.468 Da), angiotensin 1 (1296.685 Da), Glu-Fibrinopeptide B (1570.677 Da), adrenocorticotropic hormone (1-17) (2093.087 Da), and adrenocorticotropic hormone (18-39) (2465.199 Da) (4700 Calibration Mix, Applied Biosystems). For intact protein mass measurements, the mass spectrometer was calibrated using bovine serum albumin (66431 and 33216 Da for the 1+ and 2+ charge ions, respectively), yeast enolase (46672 and 23336 Da for the 1+ and 2+ charge ions, respectively), and trypsinogen (23982 Da for the 1+ charge ion) (Promix 3 calibration mixture, LaserBio Labs, Sophia-Antipolis, France). MS spectra were collected in a result-independent acquisition mode, typically using 1000 laser shots per spectrum and a fixed laser intensity of 3500V.
The peptides of interest (i.e., having a mass consistent with modification by NVP) were selected for MS/MS experiments. The MS/MS analyses were performed using Collision Induced Dissociation (CID), with 1 kV collision energy and an air pressure of 106 torr. Two thousand laser shots were collected for each MS/MS spectrum using a fixed laser intensity of 4500 V. Raw data were generated by the 4000 Series Explorer Software v3.0 RC1 (Applied Biosystems); contaminant m/z tryptic peptide peaks resulting from the autodigestion of trypsin (842.51, 1045.56, 2211.11, and 2225.12 Da) were excluded when generating the peptide mass list used for comparison with the theoretical NVP-modified HSA and Hb tryptic peptide masses. The identification of NVP-modified peptide and amino acid residues was further validated using Peaks Studio 4.5 software (Bioinformatic Solutions Inc., Waterloo, ON, Canada) for auto de novo sequencing of the MS/MS spectra, combined with manual inspection of the assigned sequence.
Results
We have previously demonstrated the validity of the synthetic model ester, 12-mesyloxy-NVP, as a surrogate for the NVP metabolite, 12-sulfoxy-NVP and/or its putative derivative, NVP quinone methide, and prepared a series of well-characterized NVP-DNA and NVP-amino acid adduct standards (40, 41). In the present study, we have further explored the same model electrophile to investigate its reaction in vitro with HSA and Hb and determine both the chemical identity of the NVP-modified amino acid residues and their specific locations in each protein by using a combination of MS-based methodologies.
HSA (1 mg/mL in PBS) was incubated with a THF solution of 12-mesyloxy-NVP (10 μg/μL) at a ratio of 1:0.5 (HSA:electrophile, w/w). To investigate if a larger amount of 12-mesyloxy-NVP would lead to more extensive protein modification and/or greater diversity of amino acid targets, a second experiment was performed using a total 1:1 ratio (w/w), with 12-mesyloxy-NVP being added in two sequential equimolar fractions to minimize competitive hydrolysis. The modification of Hb required an adaptation of the experimental conditions to prevent protein denaturation. We were able to minimize this effect by adding an acetonitrile solution of 12-mesyloxy-NVP (23 μg/μL) to an Hb solution in 100 mM phosphate, pH 7.4 (1 mg/mL) at a 5:3 ratio (Hb:electrophile, w/w). Upon removal of the non-bonded materials, either by dialysis or ethyl acetate extraction, the covalently bound modifications through the NVP C12 were identified in each protein by two different methodologies: enzymatic hydrolysis of the modified protein to amino acids, followed by LC-ESI-MS/MS analysis, and trypsin digestion of the modified protein to peptides, followed by MALDI-TOF-TOF-MS analysis. Moreover, an N-alkyl Edman degradation of the NVP-modified Hb was also performed, to search for adducted N-terminal valine residues by LC-ESI-MS/MS (Scheme 2).
LC-ESI-MS/MS identification of the NVP-modified amino acids
The NVP-modified proteins were hydrolyzed to free amino acids by an adaptation of reported methodologies, using a combination of pronase E and leucine aminopeptidase M, to ensure the endo- and exopeptidase activities required for complete hydrolysis (47, 48). The adducts released from the proteins were then characterized by comparison of LC-ESI-MS/MS patterns with those of the corresponding NVP-amino acid adduct standards previously prepared from reaction in vitro of 12-mesyloxy-NVP with individual amino acids containing nucleophilic side chains (e.g. histidine, cysteine, and tryptophan) and fully characterized by NMR and MS (41).
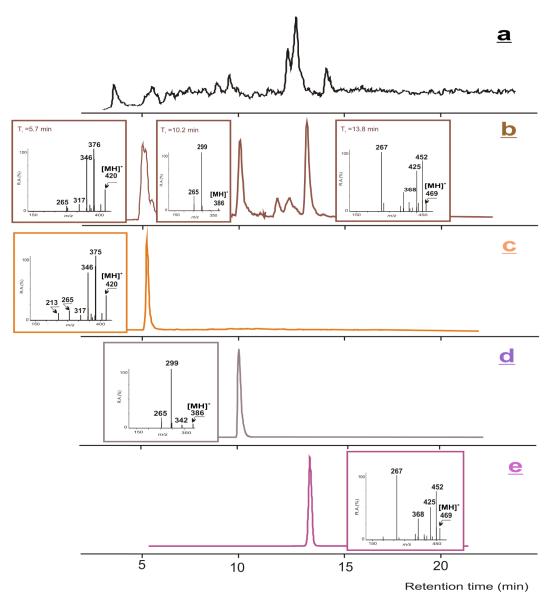
The enzymatic hydrolysates obtained from HSA modification with two different amounts of 12-mesyloxy-NVP led to very similar results regarding the identity of the NVP-modified amino acid residues. Thus, based upon comparison with the available synthetic standards (Scheme 3), three distinct adducts, 12-(histidin-N1′-yl)-NVP (retention time = 5.7 min) , 12-(cystein-S-yl)-NVP (retention time = 10.2 min), and 12-(tryptophan-2′-yl)-NVP (retention time = 13.8 min), having protonated molecules at m/z 420, 386, and 469, respectively, were identified in both mixtures by LC-ESI-MS/MS (Figure 1).
Figure 1.
Analysis of the enzymatic hydrolysate of NVP-modified HSA, obtained upon reaction with 12-mesyloxy-NVP and subsequent hydrolysis with pronase E and leucine aminopeptidase M. (a) HPLC-ESI/MS total ion chromatogram; (b) HPLC-ESI-MS/MS ion chromatograms and product ion mass spectra of the m/z 420, 386, and 469 ions; (c)-(e) HPLC-ESI-MS/MS ion chromatograms and product ion mass spectra of the NVP adduct standards with (c) histidine, (d) cysteine, and (e), tryptophan.
LC-ESI-MS/MS analysis of the NVP-modified Hb hydrolysate (Figure 2) allowed the identification of a tryptophan and a cysteine adduct, both structurally identical to those found in the NVP-modified HSA. One additional putative adduct (retention time = 10.6 min), eluting very close to the NVP-cysteine standard, was detected in the Hb hydrolysate. The extracted ion chromatogram (m/z 370) suggests a serine adduct, presumably 12-(serin-O-yl)-NVP (Scheme 3). This interpretation is also consistent with the similarity of elution characteristics with those of the cysteine adduct, as expected from two structural analogues, differing only by one heteroatom (O versus S) on the amino acid side chain. Nonetheless, due to the lack of a synthetic NVP-serine standard, the structural assignment is tentative at this stage. In addition, it should be noted that we obtained the same results when analyzing either the crude Hb hydrolysate or a concentrated methanolic eluate obtained after Sep-Pak cleanup. Thus, concentrating the sample did not lead to the detection of any other adducted amino acids in Hb.
Figure 2.
Analysis of the enzymatic hydrolysate of NVP-modified Hb, obtained upon reaction with 12-mesyloxy-NVP and subsequent hydrolysis with pronase E and leucine aminopeptidase M. (a) HPLC-ESI/MS total ion chromatogram; (b) extracted HPLC-ESI/MS ion chromatogram and mass spectrum of the m/z 370 ion, presumed to be a serine adduct; (c) HPLC-ESI-MS/MS ion chromatograms and product ion mass spectra of the m/z 386 and 469 ions; (d)-(e) HPLC-ESI-MS/MS ion chromatograms and product ion mass spectra of the NVP adduct standards with (d) cysteine, and (e) tryptophan.
N-Alkyl Edman degradation of NVP-modified Hb and LC-ESI-MS/MS analysis of the detached NVP-valine residues
The α-nitrogen atoms of the N-terminal valine residues in human Hb A and B are primary nucleophilic sites, typically capable of reacting with several classes of electrophiles. The binding products thus formed can be analyzed either as valine adducts, upon total hydrolysis of the protein, or as hydantoins (e.g., phenylthiohydantoins), upon an “N-alkyl Edman” procedure that leads to the specific detachment of the adducted valine residues from the protein (38, 43). This approach was originally developed for GC-MS analysis, which explains the common use of a fluorinated phenyl isothiocyanate, but its application to polar, thermolabile and high-molecular-weight adducts was limited. However, recent modifications, using LC-ESI-MS/MS for measuring valine adducts after derivatization with phenyl isothiocyanate have successfully broadened the scope of the method (44).
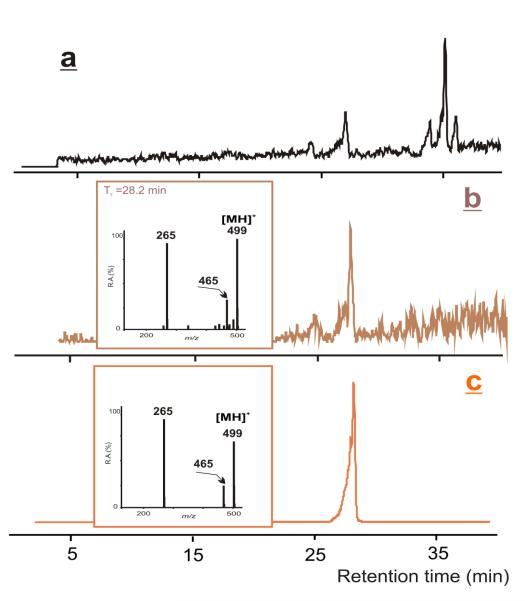
Due to the anticipated low volatility of NVP-derived phenylthiohydantoins, LC-ESI-MS/MS is a suitable methodology for the prospective analysis of these species, following ex vivo derivatization of NVP-modified Hb. Thus, to determine whether or not the N-terminal valines reacted with 12-mesyloxy-NVP, we subjected the NVP-modified Hb to N-alkyl Edman degradation using phenyl isothiocyanate as the derivatization agent (43). LC-ESI-MS/MS analysis of the ethyl acetate extract (Figure 3) confirmed the presence of the NVP-valine-derived thiohydantoin (Scheme 4), with a retention time of 28.2 min and m/z 499 for the protonated molecule, which was identified by comparison with a fully characterized synthetic standard (41).
Figure 3.
Analysis of the ethyl acetate extract obtained after N-alkyl Edman degradation of the NVP-modified Hb. (a) HPLC-ESI/MS total ion chromatogram; (b) HPLC-ESI-MS/MS ion chromatogram and product ion mass spectrum of the m/z 499 ion; (c) HPLC-ESI-MS/MS ion chromatogram and product ion mass spectrum of the derivatized valine adduct standard.
Determination of the intact mass of the NVP-modified HSA and Hb by MALDI-TOF-MS
In order to estimate the extent of modification obtained upon reaction of 12-mesyloxy-NVP with HSA and Hb, we used MALDI-TOF-MS to compare the intact mass of the control and the NVP-modified proteins. The mass spectrum of unmodified HSA had two major m/z peaks at 66509 and 33327 Da, corresponding to the single (1+) and double (2+) charged molecular ions, respectively. A 1092 Da mass increment, calculated as the difference between the molecular masses of the single charged molecular ions, was detected upon HSA modification with two sequential additions of 12-mesyloxy-NVP; this implies that an average of four amino acid residues were modified since the mass increment imposed by one NVP modification (C15H12N4O) is 264 Da. Incubation with one single addition of 12-mesyloxy-NVP resulted in a mass increment of 711 Da (calculated as indicated above), corresponding to an average of three amino acid modifications (Figure 4a).
Figure 4.
Intact mass analysis of (a) unmodified and NVP-modified HSA, and (b) unmodified and NVP-modifed Hb. A clear mass increase is observed upon modification. This increment provides an estimation of the extent of modification.
A more complex result was obtained for Hb. The mass spectrum of unmodified Hb had two major m/z peaks at 15060 and 15811 Da, corresponding to the molecular masses of the Hb A and B polypeptide chains, respectively. By contrast, several new peaks appeared in the MS spectrum of NVP-modified Hb, corresponding to various modified forms of the Hb A and B chains. For instance, the m/z peaks at 15331, 15594, and 15856 Da correspond to incorporation of one, two, and three NVP residues, respectively, on the A chain of Hb (Figure 4b). Likewise, the m/z peak series at 16072, 16345, and 16608 Da corresponds to the addition of one, two, and three NVP residues, respectively, to the B chain of Hb (Figure 4b).
MALDI-TOF-TOF-MS analysis of the adducted peptides
MALDI-TOF-TOF-MS of tryptic digests was used to identify which amino acid residues were bound to NVP upon incubation of HSA and Hb with 12-mesyloxy-NVP. The adopted strategy consisted of: i) comparison of the MS spectra of unmodified and NVP-modified HSA and Hb digests. The presence of new m/z peaks in the latter was presumed to correspond to potential NVP-amino acid adducts; ii) the m/z values observed exclusively in the MS spectra of the tryptic digests of the modified proteins were compared with the theoretical tryptic peptide mass list for each protein, taking into account the mass increase characteristic of NVP modification. This would impart a 264.28 Da increment to amino acid residues bearing nucleophilic side chains (i.e., cysteine, serine, histidine, tryptophan, lysine, or threonine); iii) this information was used to construct an inclusion list of possible NVP-modified peptides to be fragmented by an additional MS/MS experiment using the MALDI-TOF-TOF instrument. The obtained MS/MS spectra were then carefully analyzed with the assistance of Peaks software. The amino acid sequence information thus obtained allowed the unequivocal identification of NVP-modified peptides and the assignment of the specific NVP-modified amino acid.
A comparative analysis of the tryptic peptide mass spectra from NVP-modified and unmodified proteins revealed noticeable differences, with several peptides appearing exclusively in the modified HSA and Hb (Figure 5 and Table I). To identify NVP-modified peptides and assign the target amino acid residues, a theoretical trypsin digestion of both proteins under investigation was performed considering up to three missed cleavages (49); the mass increment imposed by one NVP modification (264.28 Da) was then added to the theoretically generated peptide masses. Using this approach, several peptides, appearing only in the peptide mass spectra of the modified proteins and showing the NVP-specific mass increment, were observed. For instance, the species at m/z of 937.55 in the spectrum of NVP-modified HSA (Figure 5a) appeared to correspond to the 237-242 peptide (AWAVAR), with the observed m/z peak (673.38 + 264.17 Da) bearing a mass increment consistent with NVP modification. Given that tryptophan (W) is the only amino acid in this peptide sequence reasonably anticipated to react with electrophiles, this observation strongly suggests that the W238 residue in HSA is a specific target for NVP modification.
Figure 5.
Detection and location of NVP-modified peptides. The panels show representative sections of the MALDI-TOF spectra of tryptic digests of (a) unmodified and NVP-modified HSA, and (b) unmodified and NVP-modified Hb. New m/z peaks, absent from the controls, are clearly detected in the mass spectra of the NVP-modified proteins (highlighted in the figure). These new m/z values correspond to the mass of a peptide from HSA or Hb plus the mass increment characteristic of NVP modification.
Table I.
Assignment of the NVP-modified amino acid residues from HSA and Hb that were identified by MALDI-TOF-TOF-MS analyses of the tryptic peptides.
| Protein | Observed massa(Da) |
Theoretical mass of the unmodified peptide(Da) |
Mass increase (Da) |
Peptide sequenceb | NVP- modified residue |
|---|---|---|---|---|---|
| HSA | 937.55 | 673.38 | 264.17 | AWAVAR (237-242) |
W238 |
| 1731.99 | 1467.84 | 264.15 | RHPDYSVVLLLR (361-372) |
H362 | |
| 1782.98 | 1518.78 | 264.20 | LDELRDEGKASSAK (206-219) |
K214 | |
| 2047.12 | 1783.08 | 264.04 | ERQIKKQTALVELVK (544-558) |
K548 or K 549 |
|
|
| |||||
| Hb | 1538.91 | 1274.73 | 264.18 | LLVVYPWTQR (32-41; Hb B) |
W38 |
| 1793.93 | 1529.74 | 264.19 | VGAHAGEYGAEALER (18-32; Hb A) |
H21 | |
| 2793.43 | 2529.22 | 264.21 | GTFATLSELHCDKLHVDPENFR (84-105; Hb B) |
S90 | |
Compared to the theoretical mass of the corresponding unmodified peptide, each peptide had an NVP-specific mass increment (ca. 264.28 Da).
The modified amino acid residues are highlighted in each peptide sequence.
To obtain unequivocal confirmation that the new peptides observed in the MS spectra of the modified proteins were in fact modified by NVP, tandem MS experiments were performed to provide sequence information. When using the CID fragmentation technique, bond cleavage occurs mainly through the lowest energy pathways, i.e., the peptide bond. This leads to b-ions, in which the charge is retained by the amino-terminal fragment, and y-ions, in which the charge is retained by the carboxy-terminal fragment. Thus, if an amino acid residue is modified by NVP, the specific complementary y and b ions encompassing the modification site will have the mass value of that particular amino acid increased by 264.28 Da. Considering the peptide with m/z 937.55 mentioned above [A(NVP)WAVAR], we observed (Figure 6) a b1 ion corresponding to an alanine (A) residue (m/z 72.04); however, the b2 ion (m/z 522.23) did not correspond to the addition of a tryptophan residue (186.08 Da) but rather to the addition of NVP-W (450.19 Da in total). The subsequent b3-b6 ions also showed the NVP-specific mass increment (Table II). The same feature was observed for the y ions. Thus, the y1 ion (m/z 175.12) corresponded to an arginine (R) residue, and the y2-y4 ions also displayed the m/z values expected from a non-modified peptide; however, the y5 ion (m/z 866.44) had the NVP-characteristic mass increment, as did the y6 ion (m/z 937.48). These results provide unambiguous confirmation that W238 was modified by NVP. The same approach was followed for all the potential NVP-modified HSA- and Hb-derived peptides; the results are summarized in Table I and the corresponding spectra are displayed as Supporting Information. Only modified amino acid residues with confirmed sequence information were considered.
Figure 6.
MS/MS spectrum of a representative NVP-modified peptide from HSA. The panels show the y and b fragment ions of the peptide with m/z 937.55. The detected fragment ions arise from the amino acid sequence AWAVAR, with an NVP-modification on W.
Table II.
MALDI-TOF-TOF-MS analysis of the tryptic peptide with m/z 937.55 obtained from NVP-modified HSA.
| Peptide sequence | ||||||
|---|---|---|---|---|---|---|
| A | W | A | V | A | R | |
| Iona | 1 | 2 | 3 | 4 | 5 | 6 |
| b | 72.04 | 522.23 | 593.27 | 692.23 | 763.37 b | 919.47 |
| y | 175.12 | 246.16 | 345.22 | 416.26 | 866.44 | 937.48 |
All possible b and y ion masses are shown. The fragment ions encompassing the modification site are highlighted in bold.
Not observed.
Although MALDI-TOF-MS analyses of the intact proteins indicated a dose-dependent extent of HSA modification (vide supra), the amino acid targets identified were the same, regardless of the amount of 12-mesyloxy-NVP (one or two additions) used to modify the protein. Thus, four modified amino acid residues were detected in HSA: one tryptophan (W238), one histidine (H362), and two lysines (K214 and K548 or K549). While the modifications of W238, H362, and K214 were identified unequivocally, the adducted lysine in the E544-K558 pentadecapeptide (m/z 2047.12) could not be assigned unambiguously. In this instance, the sequential information was insufficient because the y and b ions stemming from peptide bond cleavage between the adjacent lysines (K548 and K549) were not observed in the MS/MS spectrum. A similar MALDI-TOF-TOF-MS analysis of the tryptic digest from NVP-modified Hb indicated one modified tryptophan residue (W38) and one modified serine residue (S90), both in Hb B, in addition to one histidine residue (H21) in Hb A (Table I).
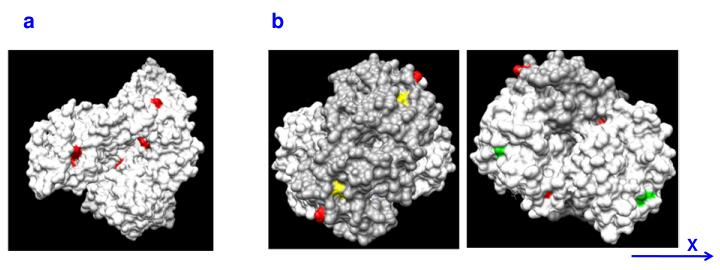
In summary, the MALDI-TOF-MS results indicate that HSA and Hb are both prone to NVP-induced modification and that tryptophan, histidine, lysine, and serine residues are major targets for reaction. Inspection of 3D structures of the two proteins (Figure 7) shows that all the amino acids found to be modified are solvent-exposed, which may explain their susceptibility. Although one additional solvent-exposed tryptophan residue is present in both Hb chains (W15A and W16B, Figure 7b), the corresponding tryptic peptides (AAWGK and SAVTALWGK, in Hb A and Hb B, respectively) were not detected, possibly due to poor ionization.
Figure 7.
Surface landscape of (a) HSA and (b) Hb, showing the solvent-exposed amino acid residues found to be NVP-modified (red). For Hb, the non-modified solvent-exposed tryptophan residues are displayed in yellow (chain A) and green (chain B). For greater clarity, the surface of one Hb subunit is shown in dark grey. Two views of the same molecule, rotated by 180° along the indicated axis, are depicted. Atomic coordinates retrieved from the Protein Data Bank archive (http://www.pdb.org) were used to represent the tridimensional structures of HSA (ID 1E78) and Hb (ID 2H35).
Discussion
The relationships between protein adduct formation by reactive metabolites of both endogenous and xenobiotic species and the onset of toxicity have been the focus of an increasing number of studies in the last decades, largely on account of the progress made in highly sensitive analytical methodologies, particularly mass spectrometry (31-39). Currently, protein adducts are extensively investigated as biomarkers of exposure to carcinogens and allergens, in search for potential dose-toxicity correlations. The abundant blood proteins HSA and Hb are often the models of choice for protein binding studies. Hb has a relatively long and well-controlled lifespan (ca. 120 days in humans); in addition, reactive intermediates have to cross cell membranes to reach the protein, a reason underlying the frequent assessment of Hb adducts as surrogates for DNA adducts of genotoxic carcinogens (37, 38). Moreover, despite having a shorter half-life than Hb (ca. 20 days), HSA is a well-characterized protein and approximately 40% of extravascular HSA is located in the skin (50), which supports its use as a tool to investigate protein haptenation mechanisms, particularly by skin allergens.
In the current study we demonstrate that both HSA and Hb undergo covalent modification upon incubation with 12-mesyloxy-NVP. The two MS-based methodologies selected for the present work afforded similar results regarding the structural identity of multiple amino acid targets for NVP modification; in addition, complementary information was obtained by combining the data from each approach. Thus, although the adopted LC-ESI-MS/MS strategy did not provide sequence information, both techniques indicated that the NVP-modified amino acids in HSA were of the same type, regardless of the amount of 12-mesyloxy-NVP used in the incubations. Likewise, the results from both techniques were in agreement regarding the amino acid residues modified in Hb (vide infra).
Comparison of the MALDI-TOF-MS results obtained for the intact NVP-modified HSA and for the corresponding tryptic peptides revealed a noteworthy consistency. In fact, the assay conducted on the intact protein using the linear mode suggested the modification of, on average, four amino acid residues when the protein was treated with two sequential additions of 12-mesyloxy-NVP; this was confirmed with the MALDI-TOF-TOF-MS experiment, which identified four distinct adducts. Interestingly, the same four amino acid residues were found to be modified after a single addition of 12-mesyloxy-NVP, although the extent of modification corresponded to an average of three amino acid residues per protein molecule. Thus, the reaction with this model ester was not only reproducible but also highly selective.
Tryptophan and histidine modifications in HSA were clearly detected both by LC-ESI-MS/MS and MALDI-TOF-TOF-MS. Previous studies of selective covalent modification of HSA by different chemicals have reported adduction at several key residues, including one cysteine (C34) and several lysines and histidines (33). Thus, histidine modification by 12-mesyloxy-NVP was expected. By contrast, the identification of a modified tryptophan residue (W238) was somewhat surprising, since W238 is the only tryptophan present in HSA (51) and, with the exception of binding to this residue in HSA by activated N-aryl hydroxamic acids (52), reports of tryptophan adducts in proteins are virtually non-existent. Assuming that 12-mesyloxy-NVP is a good model for the in vivo situation, this suggests a remarkable affinity of the indole ring of tryptophan toward NVP-derived electrophiles, which may provide a good biomarker of NVP exposure.
Cysteine modification in HSA was readily identified by LC-ESI-MS/MS but not confirmed by MALDI-TOF-TOF-MS. HSA has 35 cysteine residues, of which 34 are paired in 17 intramolecular disulfide linkages (33). Thus, only one cysteine (C34) maintains a free sulfhydryl group, which exists primarily in the highly nucleophilic thiolate form due to the vicinity of three ionizable residues, D38, H39, and Y84, within the tertiary structure of the protein (53). As indicated above, this residue is a frequent target for electrophiles (33), which makes our detection of cysteine modification by LC-ESI-MS/MS entirely credible. The reasons for our failure to detect the modified cysteine by MALDI-TOF-TOF-MS are not clear. Prior to trypsinization, the protein was subjected to reduction of the disulfide bonds with dithiothreitol and alkylation to prevent reoxidation of the sulfhydryl groups. It is possible that the microenvironment in the vicinity of the adducted C34 catalyzed an interchange of the NVP fragment with dithiothreitol, thereby destroying the adduct. Alternatively, ionization of the tryptic peptide containing the modified cysteine may have been inefficient under the MALDI experimental conditions that were used. By contrast, the LC-ESI-MS/MS analysis did not detect the lysine modifications identified by MALDI-TOF-TOF-MS. This may have been due to deficient hydrolysis at the adducted lysine residues and/or lack of sensitivity of the analytical method when compared to MALDI-TOF-TOF-MS. Taken together, the results from a combination of MS methodologies provided complementary information, which allowed the identification of five amino acid targets for NVP modification in HSA: W238, H362, K214, K548 or 549, and C34.
As indicated above, analysis of the intact protein mass by MALDI-TOF-MS gave a complex pattern for Hb, with several clusters of m/z peaks suggesting that some molecules of each strand were modified with at least up to three NVP residues. However, the results obtained by MALDI-TOF-TOF-MS (Table I), which identified one modified residue in strand A and two modified residues in strand B, imply that some of the specific adduction sites were not detected.
The ability of tryptophan to be modified by 12-mesyloxy-NVP was further confirmed in the analysis of NVP-modified Hb. Thus, evidence for the presence of 12-(tryptophan-2′-yl)-NVP was achieved unequivocally by LC-ESI-MS/MS through comparison with an authentic standard, and corroborated by MALDI-TOF-TOF-MS, which provided the identification of W38 in strand B as a specific site of modification. The MALDI-TOF-TOF-MS results also gave clear indication that a serine residue (S90, in the B chain) was modified in Hb. This confirmed the tentative ascription of serine modification made by LC-ESI-MS/MS; although the lack of a synthetic standard precluded a definite assignment of the adduct structure, it can be reasonably anticipated to involve binding of the serine oxygen through the NVP-C12 (Scheme 3).
As observed for HSA, the two MS methodologies provided complementary information regarding Hb modification by NVP. Again, cysteine modification was confirmed unequivocally by LC-ESI-MS/MS but not indicated by MALDI-TOF-TOF-MS. In contrast, MALDI-TOF-TOF-MS gave clear indication of histidine modification (H21 in strand A) but no histidine adducts were detected by LC-ESI-MS/MS. Since histidine modification was readily detected by this technique in the HSA hydrolysate, the failure to detect this adduct in Hb was presumably due to deficient hydrolysis, rather than lack of sensitivity of the LC-ESI-MS/MS method.
The N-terminal valine residues of Hb were an expected modification site, since they are primary sites of reaction with several classes of electrophiles (38). However, the adopted general procedures, involving Hb hydrolysis to amino-acids followed by LC-ESI-MS/MS or MALDI-TOF-TOF-MS of the tryptic peptides, gave no indication of valine modification in Hb. Several factors may have been at play, from inefficient hydrolysis and poor sensitivity of the LC-ESI-MS/MS method to poor ionization under the MALDI-TOF-TOF-MS conditions, as discussed above. However, use of an N-alkyl Edman procedure to detach specifically (and thus enrich) the NVP-modified N-terminal valine residues as a phenyl thiohydantoin derivative, followed by LC-ESI-MS/MS analysis, showed unequivocally that modification of the N-terminal valine occurred upon treatment of Hb with 12-O-mesyl-NVP. This suggests that, similarly to what has been extensively demonstrated for numerous other electrophiles (38), NVP adducts through the N-terminal valine of Hb may be very convenient biomarkers of NVP activation.
In summary, the present work investigated the ability of the synthetic model ester 12-mesyloxy-NVP to bind to the blood proteins, HSA and Hb, in vitro. The sites of modification in HSA and Hb were determined by two different MS-based methodologies that involved the enzymatic hydrolysis of the modified proteins to amino-acids, followed by LC-ESI-MS/MS, and trypsin digestion of the modified proteins to peptides, followed by MALDI-TOF-TOF-MS. Multiple sites of Hb and HSA modification through the NVP-C12 were identified, consistently involving adduction at amino acids bearing side chains reasonably anticipated to have nucleophilic character, such as cysteine, lysine, tryptophan, histidine, and serine. With the exception of tryptophan, all other amino acids are common sites of covalent protein modification by electrophilic species (32-39). Some of the modified residues were identified by the two methodologies but in other instances complementary information was obtained by combining the results from both methods. One significant feature is the fact that tryptophan was the adducted amino acid systematically identified in the two proteins by both LC-ESI-MS/MS and MALDI-TOF-TOF-MS. Also noteworthy is the detection of N-terminal valine adducts in Hb by LC-ESI-MS/MS following an N-alkyl Edman degradation procedure. The usefulness of the two MS-based methodologies developed in this work is currently being explored in the analysis of serum proteins from rodents administered NVP and 12-hydroxy-NVP to investigate the feasibility of this approach in the assessment of in vivo modified samples. Ultimately, the goal is to use these methods in the biomonitoring of NVP-treated patients by taking advantage from both the high sensitivity of MALDI-TOF-TOF-MS to detect protein adducts present in low concentrations and determine their specific location in the protein sequence, and from the potential quantitative applications presented by LC-ESI-MS/MS.
Supplementary Material
Acknowledgments
This work was supported in part by a research grant from Fundação para a Ciência e a Tecnologia (FCT), Portugal and FEDER (POCI/56582/QUI/2004; PPCDT/QUI/56582/2004), by a postdoctoral fellowship from FCT to R.A.G. (SFRH/BPD/41037/2007), and by Interagency Agreement No. 224-93-0001 between the National Center for Toxicological Research/Food and Drug Administration and the National Institute for Environmental Health Sciences/National Toxicology Program. The LC-MS equipment used in this work is located at the IST-UTL node of the Portuguese National Mass Spectrometry Network (RNEM) and was acquired within the framework of the National Re-equipment Program sponsored by FCT. The opinions expressed in this paper do not necessarily represent those of the U.S. Food and Drug Administration.
Footnotes
- 1D-PAGE
- one-dimensional polyacrylamide gel electrophoresis
- CID
- collision-induced dissociation
- ESI
- electrospray ionization
- Hb
- hemoglobin
- HIV-1
- human immunodeficiency virus type 1
- HSA
- human serum albumin
- MALDI-TOF-MS
- matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- MALDI-TOF-TOF-MS
- matrix-assisted laser desorption ionization tandem mass spectrometry
- MS/MS
- tandem mass spectrometry
- NVP
- nevirapine
- PBS
- phosphate-buffered saline
- Ph
- phenyl
References
- (1).FDA FDA approves nevirapine to treat HIV. News release T96-44. 1996 June 24; 1996. [Google Scholar]
- (2).Hammer SM, Eron JJ, Jr., Reiss P, Schooley RT, Thompson MA, Walmsley S, Cahn P, Fischl MA, Gatell JM, Hirsch MS, Jacobsen DM, Montaner JSG, Richman DD, Yeni PG, Volberding PA. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society-USA panel. JAMA. 2008;300:555–570. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- (3).Perinatal HIV Guidelines Working Group [Accessed April 20, 2010];Public Health Service task force recommendations for use of antiretroviral drugs in pregnant HIV-infected women for maternal health and interventions to reduce perinatal HIV transmission in the United States. 2009 April 29;:1–90. Available at http://aidsinfo.nih.gov/ContentFiles/PerinatalGL.pdf.
- (4).Marseille E, Kahn JG, Mmiro F, Guay L, Musoke P, Fowler MG, Jackson JB. Cost effectiveness of single-dose nevirapine regimen for mothers and babies to decrease vertical HIV-1 transmission in sub-Saharan Africa. Lancet. 1999;354:803–809. doi: 10.1016/S0140-6736(99)80009-9. [DOI] [PubMed] [Google Scholar]
- (5).Jackson JB, Musoke P, Fleming T, Guay LA, Bagenda D, Allen M, Nakabiito C, Sherman J, Bakaki P, Owor M, Ducar C, Deseyve M, Mwatha A, Emel L, Duefield C, Mirochnick M, Fowler MG, Mofenson L, Miotti P, Gigliotti M, Bray D, Mmiro F. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18-month follow-up of the HIVNET 012 randomised trial. Lancet. 2003;362:859–868. doi: 10.1016/S0140-6736(03)14341-3. [DOI] [PubMed] [Google Scholar]
- (6).Lallemant M, Jourdain G, Le Coeur S, Mary JY, Ngo-Giang-Huong N, Koetsawang S, Kanshana S, McInstosh K, Thaineua V, for the Perinatal HIV Prevention Trial (Thailand) Investigators Single-dose perinatal nevirapine plus standard zidovudine to prevent mother-to-child transmission of HIV-1 in Thailand. N. Engl. J. Med. 2004;351:217–228. doi: 10.1056/NEJMoa033500. [DOI] [PubMed] [Google Scholar]
- (7).Lockman S, Shapiro RL, Smeaton LM, Wester C, Thior I, Stevens L, Chand F, Makhema J, Moffat C, Asmelash A, Ndase P, Arimi P, van Widenfelt E, Mazhani L, Novitsky V, Lagakos S, Essex M. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N. Engl. J. Med. 2007;356:135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- (8).Lundgren JD, Battegay M, Behrens G, De Wit S, Guaraldi G, Katlama C, Martinez E, Nair D, Powderly WG, Reiss P, Sutinen J, Vigano A, the EACS Executive Committee European AIDS Clinical Society (EACS) guidelines on the prevention and management of metabolic diseases in HIV. HIV Med. 2008;9:72–81. doi: 10.1111/j.1468-1293.2007.00534.x. [DOI] [PubMed] [Google Scholar]
- (9).Quinson A, Arasteh K, Plettenberg A, Bogner J, Boue F, Livrozet JM, Witt M, Van Steenberge E, Sauce C, Yong CL, Macha S, Wu J, Waldhauser L, Mensa FJ, Berger F, Stern J, Robinson P, Battegay M. Steady state evaluation of two extended release (XR) nevirapine (NVP) tablets 400 mg QD compared with immediate release (IR) NVP tablets 200 mg BID in HIV-1 Infected Patients. 49th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC 2009); San Francisco, CA. September 12-15, 2009; [Accessed April 20, 2010]. 2009. paper #2879. Available at http://www.hivandhepatitis.com/2009icr/icaac/pdfs/quinson.pdf. [Google Scholar]
- (10).Pollard RB, Robinson P, Dransfield K. Safety profile of nevirapine, a nonnucleoside reverse transcriptase inhibitor for the treatment of human immunodeficiency virus infection. Clin. Ther. 1998;20:1071–1092. doi: 10.1016/s0149-2918(98)80105-7. [DOI] [PubMed] [Google Scholar]
- (11).Mirochnick M, Clarke DF, Dorenbaum A. -Nevirapine: pharmacokinetic considerations in children and pregnant women. Clin. Pharmacokinet. 2000;39:281–293. doi: 10.2165/00003088-200039040-00004. [DOI] [PubMed] [Google Scholar]
- (12).Waters L, John L, Nelson M. Non-nucleoside reverse transcriptase inhibitors: a review. Int. J. Clin. Pract. 2007;61:105–118. doi: 10.1111/j.1742-1241.2006.01146.x. [DOI] [PubMed] [Google Scholar]
- (13).Anonymous . Physicians’ Desk Reference. 63rd ed Physicians’ Desk Reference Inc.; Montvale, NJ: 2009. VIRAMUNE® (nevirapine) pp. 873–881. [Google Scholar]
- (14).Powles T, Robinson D, Stebbing J, Shamash J, Nelson M, Gazzard B, Mandelia S, Møller H, Bower M. Highly active antiretroviral therapy and the incidence of non–AIDS-defining cancers in people with HIV infection. J. Clin. Oncol. 2009;27:884–890. doi: 10.1200/JCO.2008.19.6626. [DOI] [PubMed] [Google Scholar]
- (15).Takakusa H, Masumoto H, Yukinaga H, Makino C, Nakayama S, Okazaki O, Sudo K. Covalent binding and tissue distribution/retention assessment of drugs associated with idiosyncratic drug toxicity. Drug Metab. Dispos. 2008;36:1770–1779. doi: 10.1124/dmd.108.021725. [DOI] [PubMed] [Google Scholar]
- (16).Riska P, Lamson M, MacGregor T, Sabo J, Hattox S, Pav J, Keirns J. Disposition and biotransformation of the antiretroviral drug nevirapine in humans. Drug. Metab. Dispos. 1999;27:895–901. [PubMed] [Google Scholar]
- (17).Riska PS, Joseph DP, Dinallo RM, Davidson WC, Keirns JJ, Hattox SE. Biotransformation of nevirapine, a non-nucleoside HIV-1 reverse transcriptase inhibitor, in mice, rats, rabbits, dogs, monkeys, and chimpanzees. Drug. Metab. Dispos. 1999;27:1434–1447. [PubMed] [Google Scholar]
- (18).Erickson DA, Mather G, Trager WF, Levy RH, Keirns JJ. Characterization of the in vitro biotransformation of the HIV-1 reverse transcriptase inhibitor nevirapine by human hepatic cytochromes P-450. Drug. Metab. Dispos. 1999;27:1488–1495. [PubMed] [Google Scholar]
- (19).Liu Z, Fan-Havard P, Xie Z, Ren C, Chan KK. A liquid chromatography/atmospheric pressure ionization tandem mass spectrometry quantitation method for nevirapine and its two oxidative metabolites, 2-hydroxynevirapine and nevirapine 4-carboxylic acid, and pharmacokinetics in baboons. Rapid Commun. Mass Spectrom. 2007;21:2734–2742. doi: 10.1002/rcm.3136. [DOI] [PubMed] [Google Scholar]
- (20).Ren C, Fan-Havard P, Schlabritz-Loutsevitch N, Ling Y, Chan KK, Liu Z. A sensitive and specific liquid chromatography/tandem mass spectrometry method for quantification of nevirapine and its five metabolites and their pharmacokinetics in baboons. Biomed. Chromatogr. 2010 doi: 10.1002/bmc.1353. DOI: 10.1002/bmc.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Silverman RB. The Organic Chemistry of Drug Design and Drug Action. 2nd. ed Elsevier Academic Press; London, U.K.: 2004. pp. 1–617. [Google Scholar]
- (22).Chen J, Mannargudi BM, Xu L, Uetrecht J. Demonstration of the metabolic pathway responsible for nevirapine-induced skin rash. Chem. Res. Toxicol. 2008;21:1862–1870. doi: 10.1021/tx800177k. [DOI] [PubMed] [Google Scholar]
- (23).Shenton JM, Teranishi M, Abu-Asab MS, Yager JA, Uetrecht JP. Characterization of a potential animal model of an idiosyncratic drug reaction: nevirapine-induced skin rash in the rat. Chem. Res. Toxicol. 2003;16:1078–1089. doi: 10.1021/tx034064+. [DOI] [PubMed] [Google Scholar]
- (24).Shenton JM, Popovic M, Chen J, Masson MJ, Uetrecht JP. Evidence of an immune-mediated mechanism for an idiosyncratic nevirapine-induced reaction in the female Brown Norway rat. Chem. Res. Toxicol. 2005;18:1799–1813. doi: 10.1021/tx0501132. [DOI] [PubMed] [Google Scholar]
- (25).Popovic M, Caswell JL, Mannargudi B, Shenton JM, Uetrecht JP. Study of the sequence of events involved in nevirapine-induced skin rash in Brown Norway rats. Chem. Res. Toxicol. 2006;19:1205–1214. doi: 10.1021/tx0601152. [DOI] [PubMed] [Google Scholar]
- (26).Wen B, Chen Y, Fitch WL. Metabolic activation of nevirapine in human liver microsomes: dehydrogenation and inactivation of cytochrome P450 3A4. Drug Metab. Dispos. 2009;37:1557–1562. doi: 10.1124/dmd.108.024851. [DOI] [PubMed] [Google Scholar]
- (27).Srivastava A, Lian L-Y, Maggs JL, Chaponda M, Pirmohamed M, Williams DP, Park BK. Quantifying the metabolic activation of nevirapine in patients by integrated applications of NMR and mass spectrometries. Drug Metab. Dispos. 2010;38:122–132. doi: 10.1124/dmd.109.028688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Meisheri KD, Johnson GA, Puddington L. Enzymatic and non-enzymatic sulfation mechanisms in the biological actions of minoxidil. Biochem. Pharmacol. 1993;45:271–279. doi: 10.1016/0006-2952(93)90061-z. [DOI] [PubMed] [Google Scholar]
- (29).Merk HF. Drug skin metabolites and allergic drug reactions. Curr. Opin. Allergy Clin. Immunol. 2009;9:311–315. doi: 10.1097/ACI.0b013e32832dd13c. [DOI] [PubMed] [Google Scholar]
- (30).Uetrecht J. Idiosyncratic drug reactions: current understanding. Annu. Rev. Pharmacol. Toxicol. 2007;47:513–539. doi: 10.1146/annurev.pharmtox.47.120505.105150. [DOI] [PubMed] [Google Scholar]
- (31).Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem. Res. Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Aleksic M, Thain E, Roger D, Saib O, Davies M, Li J, Aptula A, Zazzeroni R. Reactivity profiling: covalent modification of single nucleophile peptides for skin sensitization risk assessment. Toxicol. Sci. 2009;108:401–411. doi: 10.1093/toxsci/kfp030. [DOI] [PubMed] [Google Scholar]
- (33).Divkovic M, Pease CK, Gerberick GF, Basketter DA. Hapten-protein binding: from theory to practical application in the in vitro prediction of skin sensitization. Contact Dermatitis. 2005;53:189–200. doi: 10.1111/j.0105-1873.2005.00683.x. [DOI] [PubMed] [Google Scholar]
- (34).Park BK, Kitteringham NR, Maggs JL, Pirmohamed M, Williams DP. The role of metabolic activation in drug-induced hepatotoxicity. Annu. Rev. Pharmacol. Toxicol. 2005;45:177–202. doi: 10.1146/annurev.pharmtox.45.120403.100058. [DOI] [PubMed] [Google Scholar]
- (35).Zhou S, Chan E, Duan W, Huang M, Chen Y-Z. Drug bioactivation, covalent binding to target proteins and toxicity relevance. Drug Metab. Rev. 2005;37:41–213. doi: 10.1081/dmr-200028812. [DOI] [PubMed] [Google Scholar]
- (36).Baillie TA. Future of toxicology – metabolic activation and drug design: challenges and opportunities in chemical toxicology. Chem. Res. Toxicol. 2006;19:889–893. doi: 10.1021/tx060062o. [DOI] [PubMed] [Google Scholar]
- (37).Angerer J, Ewers U, Wilhelm M. Human biomonitoring: state of the art. Int. J. Hyg. Environ. Health. 2007;210:201–228. doi: 10.1016/j.ijheh.2007.01.024. [DOI] [PubMed] [Google Scholar]
- (38).Törnqvist M, Fred C, Haglund J, Helleberg H, Paulsson B, Rydberg P. Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications -J. Chromatogr. B. 2002;778:279–308. doi: 10.1016/s1570-0232(02)00172-1. [DOI] [PubMed] [Google Scholar]
- (39).Rubino FM, Pitton M, Di Fabio D, Colombi A. -Toward an “omic” physiopathology of reactive chemicals: thirty years of mass spectrometric study of protein adducts with endogenous and xenobiotic compounds. Mass Spectrom. Rev. 2009;28:725–784. doi: 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- (40).Antunes AMM, Duarte MP, Santos PP, da Costa G. Gamboa, Heinze TM, Beland FA, Marques MM. Synthesis and characterization of DNA adducts from the HIV reverse transcriptase inhibitor nevirapine. Chem. Res. Toxicol. 2008;21:1443–1456. doi: 10.1021/tx8000972. [DOI] [PubMed] [Google Scholar]
- (41).Antunes AMM, Godinho ALA, Martins IL, Justino GC, Beland FA, Marques MM. Amino acid adduct formation by the nevirapine metabolite, 12-hydroxynevirapine – a possible factor in nevirapine toxicity. Chem. Res. Toxicol. 2010;23:889–899. doi: 10.1021/tx900443z. [DOI] [PubMed] [Google Scholar]
- (42).Perrin DD, Armarego WLF. Purification of Laboratory Chemicals. 3rd ed Pergamon Press; Oxford, U. K.: 1988. pp. 1–391. [Google Scholar]
- (43).Törnqvist M. Epoxide adducts to N-terminal valine of hemoglobin. Meth. Enzymol. 1994;231:650–657. doi: 10.1016/0076-6879(94)31045-9. [DOI] [PubMed] [Google Scholar]
- (44).Chevolleau S, Jacques C, Canlet C, Tulliez J, Debrauwer L. Analysis of hemoglobin adducts of acrylamide and glycidamide by liquid chromatography–electrospray ionization tandem mass spectrometry, as exposure biomarkers in French population. J. Chromatogr. A. 2007;1167:125–134. doi: 10.1016/j.chroma.2007.07.044. [DOI] [PubMed] [Google Scholar]
- (45).Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- (46).Gomes RA, Oliveira LMA, Silva M, Ascenso C, Quintas A, Costa G, Coelho AV, Silva M. Sousa, Ferreira AEN, Freire A. Ponces, Cordeiro C. Protein glycation in vivo: functional and structural effects on yeast enolase. Biochem J. 2008;416:317–26. doi: 10.1042/BJ20080632. [DOI] [PubMed] [Google Scholar]
- (47).Tsao M, Otter DE. Quantification of glutamine in proteins and peptides using enzymatic hydrolysis and reverse-phase high-performance liquid chromatography. Anal. Biochem. 1999;269:143–148. doi: 10.1006/abio.1998.3091. [DOI] [PubMed] [Google Scholar]
- (48).Baxter JH, Lai C-S, Phillips RR, Dowlati L, Chio JJ, Luebbers ST, Dimler SR, Johns PW. Direct determination of methionine sulfoxide in milk proteins by enzyme hydrolysis/high-performance liquid chromatography. J. Chromatogr. A. 2007;1157:10–16. doi: 10.1016/j.chroma.2007.04.035. [DOI] [PubMed] [Google Scholar]
- (49).PeptideMass, Expasy. http://www.expasy.ch/tools/peptide-mass.html.
- (50).Rothshild MA, Oratz M, Schreiber SS. Albumin synthesis. N. Engl. J. Med. 1972;286:748–757. doi: 10.1056/NEJM197204062861404. [DOI] [PubMed] [Google Scholar]
- (51).Minghetti PP, Ruffner DE, Kuang W-J, Dennison OE, Hawkins JW, Beattie WG, Dugaiczyk A. Molecular structure of the human albumin gene is revealed by nucleotide sequence within q11-22 of chromosome 4. J. Biol. Chem. 1986;261:6747–6757. [PubMed] [Google Scholar]
- (52).Tannenbaum SR, Skipper PL, Wishnok JS, Stillwell WG, Day BW, Taghizadeh K. Characterization of various classes of protein adducts. Environ. Health Perspect. 1993;99:51–55. doi: 10.1289/ehp.939951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Stewart AJ, Blindauer CA, Berezenko S, Sleep D, Tooth D, Sadler PJ. Role of Tyr84 in controlling the reactivity of Cys34 of human albumin. FEBS J. 2005;272:353–362. doi: 10.1111/j.1742-4658.2004.04474.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.