Abstract
Assembly of clathrin lattices is mediated by assembly/adaptor proteins which contain domains that bind lipids or membrane bound cargo proteins, and clathrin binding domains (CBDs) that recruit clathrin. Here we characterize the interaction between clathrin and a large fragment of the CBD of the clathrin assembly protein AP180. Mutational, NMR chemical shift, and analytical ultracentrifugation analyses allowed us to precisely define two clathrin binding sites within this fragment, each of which is found to bind weakly to the N-terminal domain of the clathrin heavy chain (TD). The locations of the two clathrin binding sites are consistent with predictions from sequence alignments of previously identified clathrin binding elements and, by extension, indicate that the complete AP180 CBD contains ~12 degenerate repeats, each containing a single clathrin binding site. Sequence and circular dichroism analyses have indicated that the AP180 CBD is predominantly unstructured and our NMR analyses confirm that this is largely the case for the AP180 fragment characterized here. Unexpectedly, unlike the many proteins which undergo binding coupled folding upon interaction with their binding partners, the AP180 fragment is similarly unstructured in its bound and free states. Instead, we find that this fragment exhibits localized β turn-like structures at the two clathrin binding sites both when free and bound to clathrin. These observations are incorporated into a model in which weak binding by multiple, pre-structured clathrin binding elements regularly dispersed throughout a largely unstructured CBD allows efficient recruitment of clathrin to endocytic sites and dynamic assembly of the clathrin lattice.
Keywords: Clathrin, Endocytosis, Intrinsically Disordered Protein, Intrinsically Unstructured Protein, AP180
Introduction
Clathrin mediated vesicular trafficking is a fundamental mechanism used by all compartmentalized cells to move proteins between different compartments in the biosynthetic-secretory and endocytic pathways1. This process begins on the membrane, when clathrin assembly/adaptor proteins such as AP2 and AP180 are cooperatively recruited via interactions with the phosphoinositide PIP22 and with membrane bound cargo molecules3. This nascent complex in turn recruits cytosolic clathrin, and nucleates the formation of a clathrin coated pit4. As the coated pit grows, additional accessory proteins are recruited which promote membrane scission, allowing clathrin coated vesicles (CCVs) to detach from the membrane5. The CCVs are then rapidly uncoated via a chaperone mediated reaction that returns the coat proteins to the cytosol and the liberated transport vesicles then deliver their cargo molecules to an appropriate sub-cellular compartment by membrane fusion6; 7.
Most of the molecular players in this process are well characterized. Indeed, we have crystal structures of clathrin8; 9, the domains of the clathrin assembly proteins that interact with phosphoinositides, cargo molecules, and accessory proteins4; 10; 11; 12; 13; 14; 15; 16; 17; 18; 19, as well as many components of the uncoating apparatus20; 21; 22; 23. However, while peptides corresponding to the clathrin binding sites of the assembly proteins have been co-crystallized with the clathrin terminal domain24; 25, the clathrin binding domains of these proteins have never been crystallized, probably because these domains appear to be intrinsically disordered26.
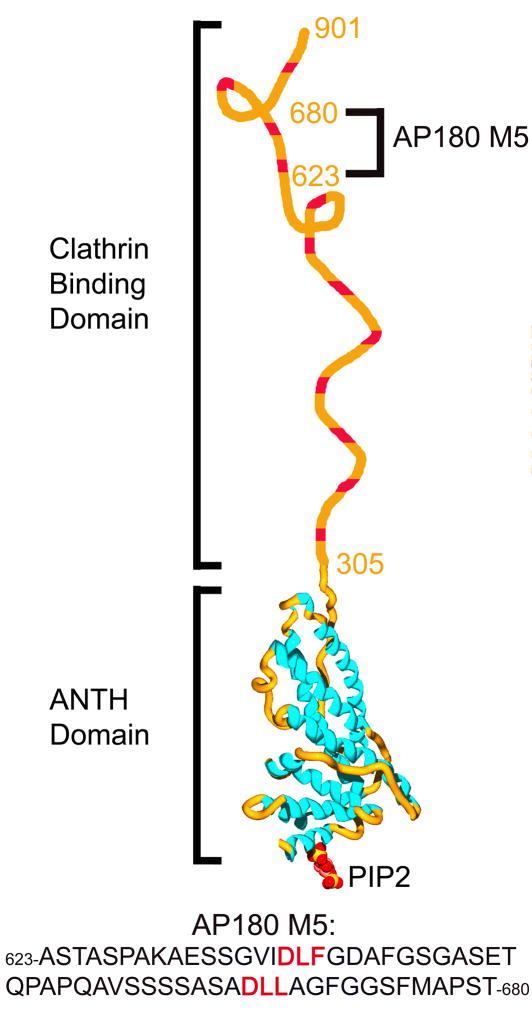
AP180 is a monomeric clathrin assembly protein27 with a ~33kD N-terminal domain (NTD) with an amino acid sequence typical of a globular protein, and an ~58kD C-terminal domain (CTD) with a highly repetitive structure which is unusually acidic and rich in proline, serine, threonine, and alanine residues, and a low propensity to form amphipathic α helices28; 29. AP180 migrates anomalously on SDS-PAGE, due to the highly acidic CTD29. These features of the AP180 CTD are typical for intrinsically disordered proteins (IDPs)30. Further evidence that the CTD of AP180 is an IDP came from circular dichroism (CD) measurements31, and from programs designed to identify IDPs, which led to its inclusion in the DisProt database (DP0025)32. Further work revealed that the 33kD NTD has the structure of an ANTH domain, and is involved in phosphoinositide binding4; 13; 33, while the 58kD CTD was shown to be involved in clathrin binding and assembly34 (Fig. 1). A self-homology analysis of the CTD revealed it to contain 12 degenerate repeats, each approximately 23aa in length34. Each of these repeats contains a central DLL/DLF sequence, which has been hypothesized27 to be a variation of the clathrin-box motif first described in the linker region of the tetrameric adaptor protein AP324; 35. The CTD of AP180 binds to the N-terminal domain (TD) of the clathrin heavy chain34, which contains binding sites for the clathrin-box peptides24.
Figure 1.
The domain structure of AP180. AP180 is a 92kD clathrin assembly protein with a structured N-terminal ANTH domain which interacts with membrane phospholipids, and a disordered clathrin binding domain that interacts with the N-terminal domain of the clathrin heavy chain (TD). The putative clathrin binding motifs are shown in red. The ANTH domain bound to PIP2 was modeled using the coordinates in PDB file 1hfa4. Indicated is the location and sequence of AP180 M5, the recombinant fragment of AP180 containing 2 putative clathrin binding sites that was used in this study.
It has recently become recognized that as many as 25–30% of all eukaryotic proteins are intrinsically disordered36. One of the most useful tools that has been applied to the study of these intrinsically disordered proteins is NMR spectroscopy, since it is suitable for the study of flexible as well as ordered proteins, and provides measures of protein dynamics37. In this study we utilized NMR spectroscopy, together with other solution approaches, to characterize the interaction between a large fragment of the AP180 CTD and the clathrin TD. This ~5kD fragment contains 2 putative clathrin binding sites and encompasses a middle segment (aa 623-680) of the AP180 CTD, and was therefore designated AP180 M538 (Fig. 1). We demonstrate that the DLL and DLF sequences within the putative clathrin binding sites are, indeed, critical for clathrin binding and measure their dissociation constants and binding rates. Furthermore, our studies reveal that while AP180 M5 is largely unstructured, there are local β turn-like structures at the clathrin binding sites, in both the free and clathrin TD bound states. The AP180 clathrin binding domain therefore appears to be neither an example of a fully structured protein, nor a completely disordered protein that assumes structure upon binding its partner. Instead, it is a largely disordered protein interspersed with multiple, short elements pre-structured to bind the clathrin TD. Therefore AP180 seems to be an example of a ‘fuzzy’ IDP, that is to say a protein which is disordered even in the partner-bound state39. At the same time, it is also an example of an IDP in which preformed structural elements play a key role in partner recognition40. These features make AP180 particularly well suited to its function as a clathrin assembly protein. By analogy to the fishing technique in which a long flexible line is baited at regular intervals with multiple hooks, this has been described as a line fishing mechanism for recruitment of clathrin to endocytic sites26; 31; 41. We find that the interaction of each of these ‘hooks’ with clathrin is weak and characterized by extremely fast association and dissociation rates, but the concentration of multiple clathrin binding elements at an endocytic site nevertheless provides for efficient clathrin recruitment, while the weakness of the individual interactions allows the recruited clathrin to have the motional freedom required for the dynamic assembly of the clathrin lattice at the endocytic site.
Results
Backbone Sequential Resonance Assignments for AP180 M5
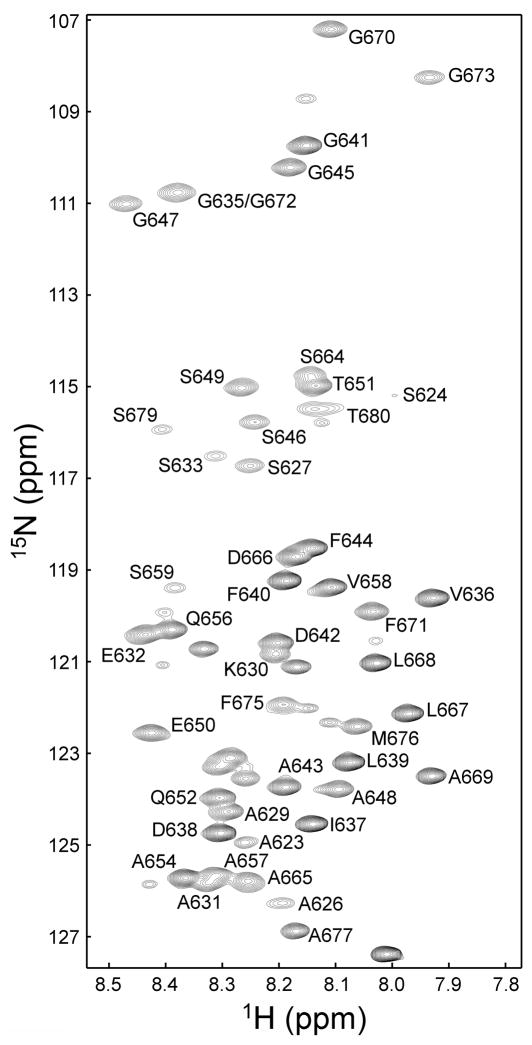
The peaks in the two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of 15N-13C labeled AP180 M5 are well separated, and peaks for 90% of the non-proline residues were successfully assigned using standard triple-resonance-based methods (HNCA, HNCACB, CBCA(CO)NH and HNCO42; 43; 44; Fig. 2). In this spectrum all of the peaks representing the backbone of AP180 M5 are located within the region corresponding to backbone amide 1H chemical shifts from 7.9–8.5 ppm, indicating that AP180 M5 is predominantly unstructured in solution45, consistent with CD measurements31.
Figure 2.
Backbone sequential resonance assignments for AP180 M5. Two-dimensional 1H-15N HSQC spectrum of AP180 M5 showing the backbone assignments. Peaks representing the side chain signals are not shown.
AP180 M5 undergoes chemical shift perturbations in the presence of Clathrin TD
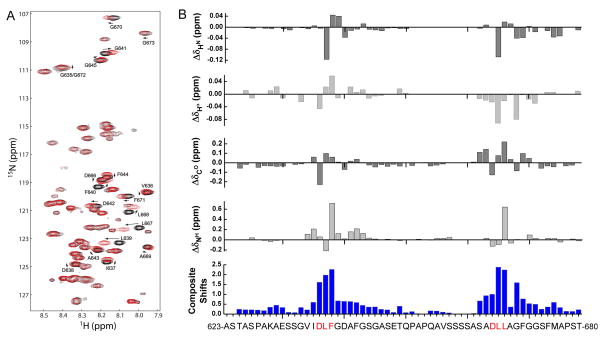
NMR chemical shift perturbation allows mapping of binding sites on a protein, as well as monitoring of structural transitions that occur upon ligand binding. We compared a 2-dimensional 1H-15N HSQC spectrum of 500 μM 15N-13C-AP180 M5 to that of a spectrum of 500 μM 15N-13C-AP180 M5 mixed with 500 μM unlabeled clathrin TD (molar ratio 1:1) (Fig. 3). Shifts in the positions of many of the AP180 M5 peaks could be clearly identified, indicating that the clathrin TD was binding to the AP180 M5 (Fig. 3A). However, the dispersion of the AP180 M5 spectrum did not expand in the 1H dimension upon addition of clathrin TD, indicating that AP180 M5 remains predominantly unstructured, even as it binds to clathrin TD. HSQC, HNCO and HNHA spectra were all collected on the same samples, and the shifts for each indicated atom type were determined (Fig. 3B). The composite absolute shift perturbations were determined by first normalizing the maximum of the absolute shift for all residues of a given atom type to a value of 1.0, and then summing over all the absolute shifts (Fig. 3B)46. The residues showing significant chemical shift changes in the NMR spectra are centered around the two sequence motifs that were previously identified as putative clathrin binding sites34. We carried out a similar analysis of AP180 M5 mutants in which each binding site was independently mutated. Mutation of the putative binding site 2 (more C-terminal) eliminated all significant chemical shift changes during the titration experiment from residues within site 2, but not site 1 (‘AP180 M5 site 1’; Fig. S1A). Similarly, mutation of site 1 eliminated all significant clathrin TD induced chemical shift changes at site 1, but not site 2 (‘AP180 M5 site 2’; Fig. S1B). We conclude that these two regions of AP180 M5 are the clathrin TD binding sites.
Figure 3.
AP180 M5 undergoes chemical shift perturbations in the presence of clathrin TD. A. An HSQC spectrum of 500 μM 15N-13C labeled AP180 M5 is shown in black while a spectrum of 500 μM 15N-13C labeled AP180 M5 with 500 μM unlabeled clathrin TD is shown in red. B. Chemical shift differences between AP180 M5 in the free and TD bound states. The composite absolute shift perturbations shown in blue include the backbone HN, Hα, CO and NH shift perturbations. The regions showing significant chemical shift changes are consistent with the putative clathrin binding sites identified by our previous studies34.
The two clathrin binding sites in AP180 M5 bind clathrin TD weakly, and with fast association and dissociation rates
To quantitatively study the interaction between AP180 M5 and clathrin TD, unlabeled clathrin TD (900 μM) was titrated into 700 μM 15N labeled AP180 M5 (WT or indicated mutants) for a total of nine titration points, giving [clathrin TD]total/[AP180 M5]total ratios from 0 to 2.8. A two-dimensional 1H-15N HSQC NMR spectrum was collected at each titration point. Examples of representative peaks shifts in binding site 1 (Fig. 4A) and binding site 2 (Fig. 4E) are shown. Quantitative analysis of the peak shifts over the course of each titration are plotted (Fig. 4B,C,F,G). While some peaks exhibited significant broadening upon addition of saturating amounts of TD, we did not observe any peak splitting indicative of slow exchange processes, indicating that binding and dissociation kinetics are in the intermediate to fast exchange regime. KDs for each binding site were determined by global fitting as described in materials and methods. The KD of binding site 1 was found to be 160 ± 23 μM in the WT AP180 M5 with two clathrin binding sites (Fig. 4B) and 168 ± 34 μM in the AP180 M5 with only one binding site (Fig. 4C), which are not statistically different. Likewise the KD of binding site 2 was in the same range, and similar in the two-site (224 ± 48 μM) (Fig. 4F) and single-site (246 ± 78 μM) AP180 M5 (Fig. 4G). Since similar KD values are obtained for these interactions, irrespective of whether the AP180 M5 has one or two clathrin binding sites, we conclude that the presence of two sites does not markedly enhance binding to clathrin TD.
Figure 4.
Analysis of the chemical shift changes in 15N-AP180 M5 upon titration with unlabeled clathrin TD. A,E: HSQC spectra of two representative amino acid residues within clathrin binding site 1 (A) and clathrin binding site 2 (E). As the [clathrin TD]total/[AP180 M5]total ratio increased, the peak positions shifted as indicated by progression from black to red to green to magenta colored peaks. The asterisks below the sequences indicate the residues used for the subsequent KD determination. Peaks that broadened extensively were omitted from the KD analysis. B,C. Determination of the KD of clathrin binding site 1 in the WT AP180 M5 (B), and in a single-site AP180 M5 in which binding site 2 was mutated (C). F,G: Determination of the KD of clathrin binding site 2 in WT AP180 M5 (F), and in a single-site AP180 M5 in which binding site 1 was mutated (G). Plotted (B,C,F,G) are the weighted average chemical shift changes of the 1H and 15N resonance of the amino acid residues. The data shown in each panel was globally fit to a hyperbolic equation, and fits are indicated with red traces. D,H: Determination of the dissociation rate constant koff of clathrin binding site 1 (D) and site 2 (H) in the WT AP180 M5. The line width analysis was performed as described in materials and methods, and global fits are indicated with red traces.
The relatively weak binding revealed by these dissociation constants is consistent with the observation that exchange kinetics lie in the intermediate to fast regime and that the association and dissociation rates of the AP180 M5:clathrin TD complex are relatively rapid. To estimate these rates, we performed a line width analysis (Fig. 4D,H), as a function of the fraction of bound site 1 or site 2. This analysis gave dissociation rate constants (koff) of 3614 ± 1070 sec−1 and 2505 ± 454 sec−1 for sites 1 and 2, respectively. The association rate constants (kon values) estimated from the measured KD and koff values (using the equation kon=koff/KD) are 2.3 ± 0.7 × 107 M−1s−1 and 1.1 ± 0.3 × 107 M−1s−1 for sites 1 and 2, respectively. These are very fast rate constants for biomolecular protein:protein association reactions, but are consistent with both the measured KD values and the observation that the NMR exchange kinetics are in the intermediate to fast regime.
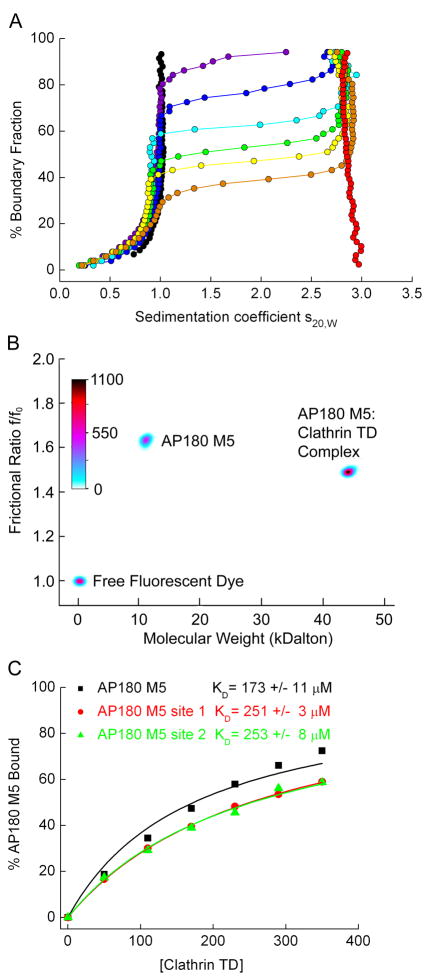
Analytical ultracentrifugation (AUC) was used to independently confirm the KD values of AP180 M5 binding to clathrin TD, and to obtain information on the stoichiometry of clathrin TD binding to the single-site vs. two-site AP180 M5 molecules. In these studies, AP180 M5 was fluorescently labeled with Alexa 488. It was then incubated in limiting amounts with a concentration series of unlabeled clathrin TD, and the reactions were allowed to come to equilibrium. AUC was then used to separate bound from free AP180 M5. AP180 M5 sediments with an S-value of ~1S, clathrin TD at ~ 2.9S, and the AP180 M5:clathrin TD complex at ~3S. As the concentration of clathrin TD in the reaction is increased, an increasing fraction of the fluorescent AP180 M5 is seen to move from the free to the clathrin TD-bound sedimentation position (Fig. 5A). The amount of free versus bound AP180 M5 was quantified by genetic algorithm47 combined with Monte Carlo analysis48 (Fig. 5B), and these values were plotted as a function of the concentration of clathrin TD (Fig. 5C). Similar experiments were carried out on the two single-site AP180 M5 fragments, and KDs were calculated from hyperbolic fits of the binding isotherms (Fig. 5C). The KD of clathrin TD for WT AP180 M5 was found to be 173 ± 11 μM, while for the AP180 M5 site 1 and AP180 M5 site 2 fragments, KD values were found to be 251 ± 3 μM and 253 ± 8 μM, respectively. These values are similar to those measured by the NMR chemical shift analysis (Fig. 4), Though it can be difficult to compare KD determinations from different studies due to variations in buffers and other experimental conditions, the KD values we measure are consistent with the previous conclusions that the interactions between clathrin and the binding motifs found in clathrin binding proteins are weak, with reported KD values ranging 22–400 uM in previous studies that examined clathrin binding to clathrin-box and W-box motifs 25; 34; 49. In these experiments we could detect no species sedimenting significantly faster than 3S, and the sedimentation coefficients of the complexes in the experiment with the two-site AP180 M5 were indistinguishable from those in the experiments with the single-site AP180 M5 molecules, indicating that the two-site AP180 M5 binds just one clathrin TD molecule.
Figure 5.
AUC reveals 1:1 binding of AP180 M5 to clathrin TD and confirms weak binding measured by NMR. 1 μM fluorescently labeled AP180 M5 was incubated with a concentration series of unlabeled clathrin TD. After the reactions were allowed to come to equilibrium, free AP180 M5 was separated from bound by AUC. A. Enhanced van Holde-Weischet integral S-distribution plots for a titration of 1 μM fluorescently labeled AP180 M5, with increasing concentrations of clathrin TD (purple: 50 μM; blue: 110 μM; turquoise: 170 μM; green: 230 μM; yellow: 290 μM; orange: 350 μM). Only AP180 M5 is detectable in the titration experiment, since it is labeled with Alexa 488 and fluorescence emission at 500 nm is followed. Free AP180 M5 (black curve) and free TD (red curve, measured with interference optics) are also shown as controls. AP180 M5 sediments with an S-value of ~1S, clathrin TD sediments with ~2.9S, and the AP180 M5:clathrin TD complex with ~3S. B. Genetic algorithm analysis combined with 50 iterations of a Monte Carlo analysis of a mixture of 1 μM AP180 M5 with 230 μM TD reveals the more extended (high f/fo) structure for free AP180 M5 and the more compact AP180 M5:clathrin TD complex. The molecular weight for both species agrees well with the molecular weight predicted from sequence, and indicates that one clathrin TD is binding per AP180 M5. Also visible is a signal from a minor species which represents the free fluorescent dye. The relative signal of each species is shown as a color gradient in units of counts of fluorescence emission. C. Using the analysis method described in panel B to quantify the amount of bound AP180 M5, plots of binding as a function of clathrin TD concentration were generated for experiments carried out with either WT AP180 M5, the single-site AP180 site 1 mutant, or the AP180 M5 site 2 mutant. KDs were determined by fitting the data to a hyperbolic equation, and are in good agreement with the KDs determined by the NMR analysis.
The Clathrin TD binding sites within AP180 have limited structure in both the free and clathrin TD bound state
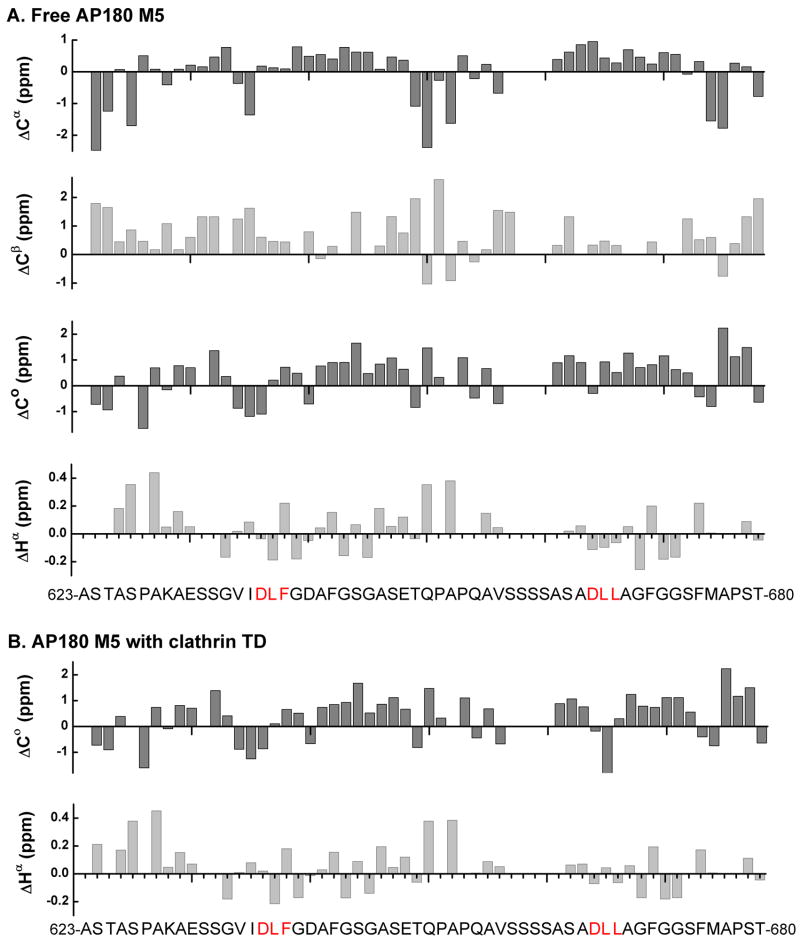
As previously mentioned, the finding that all of the peaks in the HSQC spectrum of AP180 are in the region corresponding to backbone amide 1H chemical shifts from 7.9 to 8.5 ppm, in both the free (Fig. 2) and clathrin TD bound (Fig. 3) states, indicates that AP180 M5 is predominantly unstructured whether free or bound to clathrin TD. To examine this in greater detail, we carried out a secondary chemical shift analysis. In this analysis, for each residue along the AP180 M5 polypeptide chain, we subtracted the chemical shift of that residue if it were in a random coil (using the Wishardt database50) from the observed chemical shift using the NMRView program51. The resultant secondary chemical shifts (corrected for temperature, pH and co-solvent effects) were plotted for each atom monitored, along the AP180 M5 sequence (Fig. 6). We found that the secondary chemical shifts for consecutive residues neither approach the threshold for those in stable helices and strands, nor show a consistent pattern of deviation from random coil. This indicates that AP180 M5 contains no extended regions of α helical or β sheet structure, in either the free (Fig. 6A) or clathrin TD bound (Fig. 6B) states. This was consistent with our earlier finding that application of the chemical shift index (CSI) algorithm of Wishart and Sykes50 yielded no detectable regions of α helix or β strand in either the free or clathrin TD bound state.
Figure 6.
Analysis of secondary chemical shifts indicates that AP180 M5 contains no regions of α helical or β street structure, in either the free or bound state. It has been reported that for residues in stable α helices, the average secondary chemical shifts are 2.5 ppm for 13Cα and −0.38 ppm for 1Hα, with near random coil values for 13Cβ and positive values for 13Co. In stable β sheets, the average secondary chemical shifts are −2.0 ppm for 13Cα, 2.5 ppm for 13Cβ and 0.38 ppm for 1Hα, with negative values for 13Co75. A. Secondary chemical shifts of free 500 μM 15N-13C labeled AP180 M5 for 13Cα, 13Cβ, 13Co and 1Hα were calculated by subtracting random coil shifts corrected for sequence-dependent variations from the experimental chemical shifts. No regions with patterns indicative of either α helix or β sheet were identified. B. Secondary chemical shifts of 500 μM 15N-13C labeled AP180 M5 with 500 μM unlabeled clathrin TD for 13Co and 1Hα were calculated in the same way. No regions with patterns indicative of either α helix or β sheet were identified.
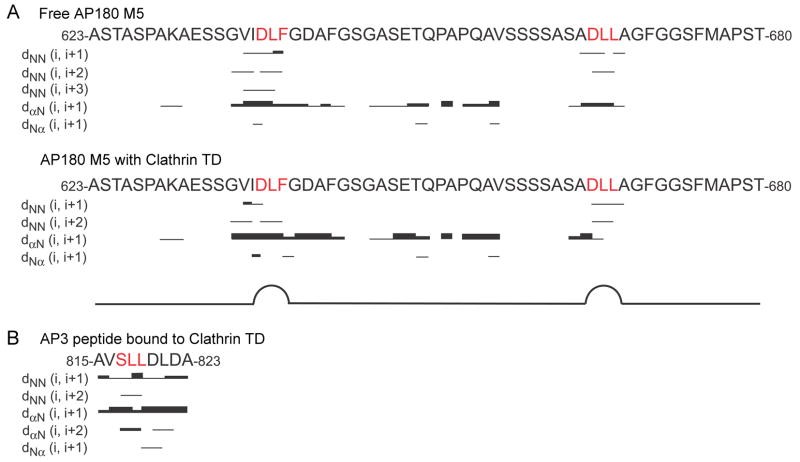
Next, we carried out nuclear overhauser effect spectroscopy (NOESY) experiments, which are more sensitive to subtle structural features than the secondary chemical shift analyses. The 1H-1H nuclear overhauser effects (NOEs) of AP180 M5 in both the free and clathrin TD bound states show no NOEs representing α helices or β sheets (Fig. 7A), indicating that disordered AP180 M5 does not adopt extensive regions of secondary structure upon binding to clathrin TD. However, short-range NOEs observed within both clathrin binding sites indicate that AP180 M5 is less flexible in these regions. Furthermore, dNN(i, i+2) NOEs, which are typically found in β turn conformations, are observed between D638 and F640 in the DLF motif and between D666 and L668 in the DLL motif, indicating that AP180 M5 has limited β turn-like structure at both clathrin binding sites, in both the free and clathrin TD bound states (Fig. 7A). Consistent with this, we measured interatomic distances in the AP3 clathrin box peptide, which assumes a β turn-like structure in a co-crystal with clathrin TD24 and found that these would result in similar dNN(i, i+2) NOEs between S817 and L819 in the SLL motif that interacts with clathrin TD in the co-crystal structure (Fig. 7B).
Figure 7.
NOESY experiments reveal local β turn-like structures in AP180 M5 at the clathrin binding sites. A. Summary of 1H-1H NOEs of 500 μM 15N labeled AP180 M5 (top) and 500 μM 15N labeled AP180 M5 with 500 μM unlabeled clathrin TD (bottom). The different thicknesses of the lines indicate the NOE signal intensities. B. A prediction of the 1H-1H NOEs of the AP3 β3 clathrin box peptide bound to TD.
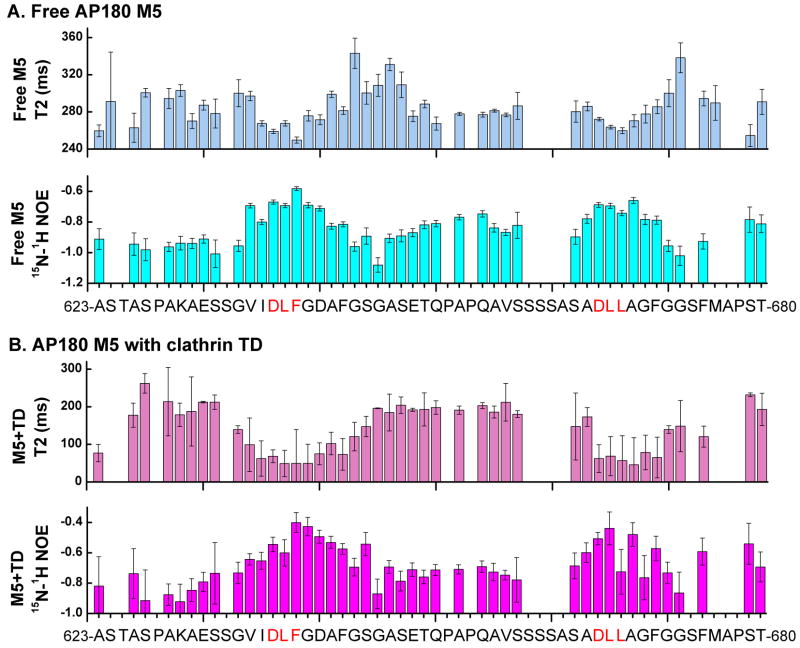
If the clathrin binding sites are forming limited β turn-like structures, we expect these sites should also exhibit more restricted dynamics than the rest of the molecule. Consistent with this, we observed that, relative to the rest of the AP180 M5 residues, residues within the two clathrin binding sites had smaller 15N T2 relaxation times, in both the free (Fig. 8A) and clathrin TD bound states (Fig. 8B) states. Furthermore, we measured larger 1H-15N NOEs within the clathrin binding sites in both the free (Fig. 8A) and clathrin TD bound (Fig. 8B) states, which also indicates reduced polypeptide chain flexibility. The negative values observed for the 1H-15N NOEs in both the free and bound states support our earlier findings that AP180 M5 lacks extensive stretches of either α helices or β sheets. Taken together, our data indicate that while AP180 M5 is predominantly disordered, the two clathrin binding sites contain limited but persistent β turn-like structure in both the free and bound states.
Figure 8.
The AP180 M5 polypeptide chain is less flexible at the clathrin binding sites whether free or bound to clathrin TD. A. The T2 relaxation times and 1H-15N NOEs of 500 μM 15N-AP180 M5 in the free state. B. The T2 relaxation times and 1H-15N NOEs of 500 μM 15N-AP180 M5 with 500 μM unlabeled clathrin TD. Regions showing shorter T2 relaxation times and higher 1H-15N NOEs values represent less flexibility in the polypeptide chain.
Discussion
In this study, we set out to understand how a fragment from the intrinsically disordered clathrin binding domain of AP180 interacts with clathrin TD to gain insight into how AP180 and other clathrin assembly/adaptor proteins mediate assembly of clathrin lattices at endocytic sites. With the successful backbone assignments of AP180 M5 (Fig. 2), we were able to map the binding sites for clathrin TD on AP180 M5 by NMR chemical shift perturbation analysis (Figs. 3 and 4). We found two well defined binding sites, one centered around a DLF motif and one centered around a DLL motif (Fig. 3). This is consistent with predictions based on sequence analysis and clathrin assembly activity studies34. We found that the binding of clathrin TD to each site is relatively weak, with KD values in the ~2×10−4 M range (Figs. 4 and 5).
X-ray crystallography studies have defined multiple distinct ligand binding sites on clathrin TD24; 25; 52. The length of the segment linking the two clathrin binding sites in AP180 M5 would allow these two sites to bind simultaneously to any two of the crystallographically observed binding sites in clathrin TD. Simultaneous binding of both clathrin binding sites in a two-site AP180 M5 molecule to a single clathrin TD would be expected to enhance binding relative to a single-site AP180 M5 molecule. However, the KD values for each clathrin binding site, as measured by NMR chemical shift analysis, were statistically indistinguishable, irrespective of whether each site was present in a mutated AP180 M5 molecule that retained only one functional clathrin binding site, or was embedded in a WT AP180 M5 molecule that contained two clathrin binding sites (Fig. 4). This could therefore indicate that each site in the two-site AP180 M5 molecule binds a different clathrin TD. However, the sedimentation coefficients of the complexes observed in the AUC experiments with the two-site and single-site AP180 M5 molecules are indistinguishable and consistent with binding of only a single TD to each AP180 M5 molecule (Fig. 5). In the AUC experiments we did observe an increase in the affinity of the clathrin TD for a two-site vs. single-site AP180 M5 (Fig. 5), suggesting that both sites may interact simultaneously with a single clathrin TD to enhance the affinity of the two-site AP180 M5. But this enhanced affinity, while within the error range of these experiments, was quantitatively modest (~1.5 fold), and was not seen in the NMR experiments, though analysis of the latter is complicated by fast exchange phenomena and the fact that neither binding partner is in large excess so that it’s free concentration could be treated as an independent parameter. To address the questions raised by these data, we are currently characterizing the effects of the multiple clathrin binding sites in AP180 on binding affinity and interaction stoichiometry by using both NMR and AUC to measure KD values and molecular weights of complexes formed with AP180 fragments containing increasing numbers of clathrin binding sites, and by directly observing AP180 binding to distinct sites on isotopically labeled clathrin TD, which has recently become possible as a consequence of our successful assignment of 92% of the visible non-proline residues in the 290 residue clathrin TD molecule53.
It is often observed that when an IDP interacts with a binding partner, the IDP becomes more ordered, through a process called binding coupled folding54. Our studies of AP180 M5 revealed that AP180 M5 is predominantly unstructured in solution, in both the free and clathrin TD bound states. This conclusion is based on the following observations: 1. All of the peaks representing the backbone of AP180 M5 are located within the region corresponding to backbone amide 1H chemical shifts from 7.9–8.5 ppm in both free and bound states (Figs. 2 and 3). 2. Secondary chemical shift analysis did not reveal patterns associated with α helices or β sheets in either state (Fig. 6). 3. NOESY experiments did not reveal any 1H-1H NOEs that would indicate the presence of α helices or β sheets in AP180 M5 either when free in solution or bound to TD (Fig. 7). 4. 15N-1H NOEs throughout the polypeptide chain are negative in both free and bound state, further indicating a lack of extensive secondary structure in AP180 M5 (Fig. 8). Our finding that AP180 M5 is predominantly unstructured in solution in the free state is consistent with its unusual amino acid composition and previous CD measurements of the entire CBD of AP18029; 31.
Despite the lack of extensive secondary structure in AP180 M5, our studies revealed that AP180 M5 contains β turn-like structures at both clathrin binding sites in the free as well as clathrin TD bound states. This conclusion is based on the results of both NOESY experiments and 15N T2-relaxation measurements. Short-range 1H-1H NOEs observed within both clathrin binding sites indicate that AP180 M5 is less flexible in these regions (Fig. 7). We also observed dNN(i, i+2) NOEs, which are typically found in β turn conformations, between the D and F in the DLF motif and between the D and L in the DLL motif (Fig. 7). Shorter T2 relaxation times, as well as higher 15N-1H NOEs were found within the clathrin binding sites, which also indicate reduced flexibility (Fig. 8). Interestingly, while the similarly narrow chemical shift dispersion of the AP180 M5 HSQC spectra in both the free and bound forms (Fig. 2) indicates that binding is not associated with extensive folding of this fragment, we do observe qualitatively similar, but quantitatively enhanced, NOEs within the clathrin binding sites in the bound versus free AP180 M5 (Fig. 7), suggesting that binding reinforces the β turn-like structures of these sites. The frictional coefficients measured by AUC also support the conclusion that AP180 M5 has partial structure in solution even when it is not bound to clathrin TD. If a polypeptide like AP180 M5, with a MW of 7184 were completely unfolded, then it would be expected to have an f/f0 value of 1.89, whereas if it were folded like a globular protein this ratio would be 1.0–1.255). The experimentally measured f/f0 of 1.66 ± 0.10 (Fig. 5) is intermediate between these two values, though closer to that of a fully unfolded polypeptide. This is consistent with a polypeptide chain that is largely unstructured but contains local structural elements as observed in the NMR studies.
Since the clathrin binding site structures in AP180 are effectively pre-formed in solution they should be able to bind clathrin with relatively fast kinetics. In contrast, binding interactions that involve extensive folding are often associated with slow binding kinetics, likely because they are limited by the rate at which the folded structures form.56; 57. In fact, the weak binding of these sites to the clathrin TD and the observation that chemical exchange kinetics are in the intermediate to fast exchange regime indicates that both association and dissociation rates for these interactions are rapid. This was confirmed by our line width analysis which indicated that the dissociation rate constants are in the range of 2–4×103 sec−1, and association rate constants are in the range of 1–2 × 107 M−1sec−1 (Fig. 4D,H). These are among the fastest association rates that have ever been measured for a protein:protein interaction58.
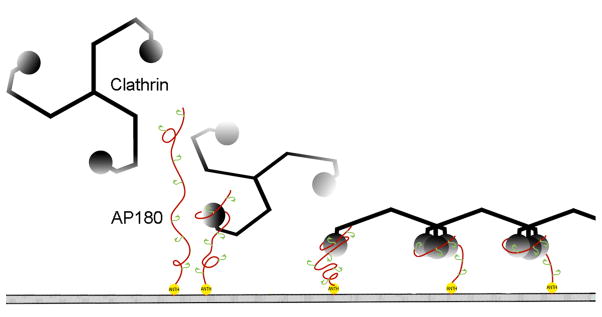
These observations allow us to extend previously proposed models26; 31; 41 for how the structural features of AP180 mediate its biological function in clathrin coat assembly. Within the M5 fragment of AP180, there are two clathrin binding sites. However, there are an additional 10 similar motifs spaced ~23 amino acids apart throughout the entire 58kD clathrin assembly domain of AP180, indicating that the CTD may contain up to 12 clathrin binding sites34. We draw an analogy between these multiple β turn-like clathrin binding sites within the flexible CTD and the multiple hooks that a line fisherman places along his fishing line, and so describe this as a ‘line fishing’ model for the recruitment of clathrin to the membrane during clathrin coated pit formation (Fig. 9). In this model, AP180 docks on the plasma membrane via interactions between its ordered N-terminal ANTH domain and membrane bound PIP2. The extended and flexible CTD of AP180 allows it to scan a large volume of cytosol and to rapidly engage any clathrin molecules encountered via the multiple pre-formed β turn-like ‘hooks’ regularly interspersed throughout this domain. Even though each clathrin binding site binds clathrin weakly and with a rapid dissociation rate, once recruited to the endocytic site the probability of a clathrin triskelion diffusing away is minimized because: 1. Each AP180 contains up to 12 clathrin binding sites so if a clathrin molecule releases one site it may remain engaged via interactions with other sites or is very likely to encounter and bind another such site. 2. This effect is further amplified because multiple AP180s will be concentrated at the endocytic site. 3. In addition to AP180, this endocytic site will contain other proteins that are known to have intrinsically disordered domains which also contain multiple binding sites for clathrin26; 27; 41, further increasing the number of clathrin binding elements concentrated at the endocytic site. 4. Finally, since each clathrin triskelion contains 3 clathrin TDs and each TD may be able to bind more than one clathrin binding element 25; 52, the likelihood of a clathrin triskelion being retained at the endocytic site through multiple, weak interactions is multiplied even further.
Figure 9.
The line fishing model for assembling the endocytic apparatus. After docking to the plasma membrane via interactions between the N-terminal ANTH domain of AP180 (yellow) and membrane bound PIP2, the long and flexible C-terminal domain of AP180 (red) can bind and recruit clathrin (black) from a large volume of cytosol to initiate the formation of a clathrin coated pit. The large number of clathrin binding sites (green) recruit multiple clathrin heavy chains together to form the vertexes of the clathrin lattice (adapted from31 with permission from the Journal of Biological Chemistry).
However, in what may be the most important feature of these interactions, the fact that each AP180 clathrin binding element binds with rapid dissociation kinetics means that the retention of the triskelion at the endocytic site will be highly dynamic, with individual interactions constantly being released and re-formed. This may serve at least two functions: it will limit the formation of non-productive intermediates stuck in off-pathway events (tangled assemblies, nonfunctional aggregates) and it will allow each triskelion to move and reorient itself so that it can establish the interactions with other clathrin triskelia that largely determine the geometry and stability of the clathrin lattice. That the stability and geometry of the clathrin lattice is largely determined by interactions between clathrin molecules, and not dictated by the assembly protein, is indicated by the observation that, even in the absence of assembly/adaptor proteins, clathrin alone can form coat structures that, while more heterogeneous, have the same icosahedral geometry as those formed in the presence of adaptor/assembly proteins59.
This very dynamic assembly mechanism has features similar to the model proposed for the interaction of pSic1 with Cdc4, in which multiple, weakly binding CDP motifs interspersed throughout an intrinsically disordered pSic1 domain constantly exchange with each other for interaction with a single site on Cdc4 60. In this case, this interaction mechanism has been proposed to underlie the requirement for phosphorylation of multiple, weakly binding CPD sites to achieve strong binding to Cdc4, possibly explaining the ultrasensitivity of this interaction to phosphorylation61. In the case of AP180 mediated clathrin lattice assembly, however, the multiple, weak binding elements embedded in a disordered domain are here proposed to be important in a mechanism that can recruit and concentrate clathrin at a specific site without inhibiting the motions that allow the clathrin triskelia to establish the interactions that determine the final stability and geometry of the clathrin coat. These two examples highlight the functional potential of binding domains that combine the features of weak, multivalent binding with intrinsic disorder to create highly dynamic modes of protein:protein interaction.
Materials and Methods
Plasmid Constructs
The construct expressing clathrin TD (aa 1-363) was kindly provided by Dr. Linton Traub62. The construct expressing AP180 M5 (aa 623-680) was reported previously38. The AP180 M5 mutants were constructed from the WT AP180 M5 construct using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). Single-site AP180 M5 site 1 was made by mutating the residues at positions 667-668 into alanines using the sense primer 5′-CTAGTTCATCAGCATCGGCAGATGCAGCAGCTGGATTTGGGGGTTCTTTC-3′ and the antisense primer 5′-GAAAGAACCCCCAAATCCAGCTGCTGCATCTGCCGATGCTGATGAACTA G-3′. Single-site AP180 M5 site 2 was made by mutating the residues at positions 639-642 into alanines using with the sense primer 5′-GAGTCCTCGGGTGTCATAGACGCTGCTGCGGCTGCGTTTGGAAGTGGTGCTTC-3′ and the antisense primer 5′-GAAGCACCACTTCCAAACGCAGCCGCAGCAGCGTCTATGACACCCGAG GACTC-3′.
Expression of Clathrin TD and AP180 M5
Clathrin TD and AP180 M5 (WT and mutants) were expressed as GST fusion proteins in E. coli BL21 (DE3) pLysS host cells (Stratagene, Santa Clara, CA). Fresh transformation on LB plates containing both carbenicillin (25 mg/mL) and chloramphenicol (17 mg/mL) was required for optimal expression. The transformed cells were cultured in 2×YT at 30° C containing 50 mg/mL carbenicillin. Protein expression was induced by the addition of 1 mM Isopropyl-β-D-Galactopyranoside (IPTG) when the OD600 reached 0.6–0.7. Cells were harvested 14–16 hours after induction, and frozen at −80° C. When isotopically labeling AP180 M5, cells were cultured as described above (except using LB instead 2xYT media) until the OD600 reached 0.7–0.8. Then the cells were pelleted by centrifugation at 5,000 × g, 4° C for 6 minutes. Cell pellets from 4L of culture were gently transferred into 1L of M9 minimal medium supplemented with 50 mg/mL carbenicillin in which the NH4Cl and glucose were replaced with 15N-NH4Cl (1g/L), and if required 13C-glucose (3g/L; isotopes obtained from Cambridge Isotope, Andover, MA). The cells were cultured for 1 hour in the minimal medium at 30° C prior to induction by 1 mM IPTG. Cells were harvested 28–30 hours after induction, and frozen at −80 °C.
Purification of Clathrin TD and AP180 M5
Cells from 1L cultures were resuspended in 40 mL of lysis buffer (phosphate-buffered saline (PBS) containing 100 mM Ethylenediaminetetraacetic Acid (EDTA), 3 mM Dithiothreitol (DTT), 1 mM Phenylmethyl-Sulfonyl Fluoride (PMSF), 1 mM Benzamidine, 10 μM Leupeptin and 1 μM Pepstatin) and sonicated. Lysates were mixed with 40 mL of lysis buffer and 4 mL 20% Triton X-100 then centrifuged at 125,000 × g for 30 minutes to remove the debris. The supernatant was loaded onto an 8 mL bed of Glutathione-Sepharose 4B resin equilibrated with lysis buffer. The resin was sequentially washed with 100 mL of lysis buffer, 50 mL of PBS containing 3mM DTT and 50 ml of cleavage buffer (50 mM Tris pH 8.3, 150 mM NaCl, 3 mM DTT). The resin was equilibrated with cleavage buffer containing 0.2 mg/mL thrombin then kept at 4° C overnight. The cleaved protein was eluted by 10 mL of cleavage buffer and the reaction was stopped by the addition of 1 mM PMSF. The eluted protein was dialyzed against buffer containing 20 mM Tris pH 8.0, 3 mM DTT and fractionated on a 6.5 mL Q-Sepharose ion-exchange column with a 0 to 600 mM NaCl gradient. The purified proteins were either dialyzed into storage buffer (10 mM Tris pH 8.0, 3 mM DTT, 50% glycerol) and kept at −20° C, or dialyzed directly into reaction buffer and concentrated using Centricon 10 at 5,000 × g (for clathrin TD) or Centricon 5 at 7,500 × g (for AP180 M5) (Millipore, Billerica, MA).
Analytical Ultracentrifugation
Analytical ultracentrifugation experiments were performed in a Beckman Optima XL-A equipped with an Aviv (Aviv Biomedical, Lakewood, NJ) fluorescence detection system (AU-FDS) in the UTHSCSA Center for Macromolecular Interactions. AP180 M5 was labeled with Alexa 488 (Alexa Fluor 488 succinimidyl ester; Molecular Probes, Carlsbad, CA) by incubating 250 μM AP180 M5 with 2 mM Alexa 488 in 100 mM NaHCO3 pH 8 for 4 hours at 4° C degrees in the dark. Both the amino terminus and the ε-amino group of K630 of AP180 M5 were labeled. Unincorporated dye was removed by dialysis. A limited fixed concentration of Alexa488-AP180 M5 (1 μM) was incubated with the indicated concentration series of unlabeled clathrin TD at 25° C in AUC buffer (50 mM NaCl, 25 mM Na2HPO4 pH 7.5) for 2 hours to allow the reactions to come to equilibrium. The degassed samples were loaded into cells containing either titanium (Nanolytics, Potsdam, Germany) or Sedveloc60K (Spin Analytical, S. Berwick, ME) 2-channel centerpieces with quartz windows. In the first experiment the cells were aligned in an An 60 Ti rotor and spun at 5,000 rpm to determine PMT and PGA gain settings (89% and 2, respectively). These settings were then used for all subsequent experiments. The samples were thoroughly remixed in the cells and allowed to temperature equilibrate to 25° C in the instrument, which was maintained throughout the experiment. All samples were run at 60,000 rpm and scans collected continuously using AOS v1.7 with 488 nm excitation and 505 nm emission wavelengths. Sedimentation velocity data were analyzed with UltraScan63. Calculations were performed on the Lonestar cluster at the Texas Advanced Computing Center at the UT Austin, and on the Jacinto cluster at the Bioinformatics Core Facility at UTHSCSA. The data were analyzed by two-dimensional spectrum analysis with simultaneous removal of time-invariant noise64 and then by van Holde-Weischet analysis65 and genetic algorithm refinement47, combined with Monte Carlo analysis48. We do not believe the hydrodynamic properties of AP180 M5 were affected by the attachment of the fluorophore, since dynamic light scattering (DLS) measurements of the hydrodynamic radii of unlabeled versus labeled AP180 indicate that the f/f0 values are within the confidence limits of the value measured by AUC (1.61 unlabelled, 1.59 labeled).
NMR Spectroscopy
NMR spectra were acquired on Bruker 500 MHz, 600 MHz and 700 MHz spectrometers equipped with either conventional (500 MHz) or cryogenically cooled (600 MHz and 700 MHz) 5 mm 1H probes equipped with 13C and 15N decoupler and pulsed field gradient coils at the UTHSCSA Center for Biomolecular NMR Spectroscopy. All NMR experiments were performed at 300 K in buffer containing 50 mM NaCl, 25 mM Na2HPO4 pH 7.0 and 0.02 % NaN3 with addition of 5% D2O immediately prior to data collection. The spectra were processed with NMRPipe66 and analyzed with NMRView51.
Sequential Backbone Assignments of AP180 M5
The sequential backbone assignments of AP180 M5 were obtained by collecting and analyzing triple-resonance data sets of 15N, 13C labeled AP180 M5, including HNCA, HNCACB, CBCA(CO)NH and HNCO42; 43; 44.
Titration of Clathrin TD into 15N labeled AP180 M5
Unlabeled clathrin TD at 900 μM was titrated into 300 μL of 15N labeled AP180 M5 at 700 μM. Two-dimensional HSQC spectra were collected for each titration point. The weighted average chemical shift changes of the backbone amide 1HN and 15N of AP180 M5 were determined by the equation67; 68:
The dissociation constant of the AP180 M5:clathrin TD interaction was determined by fitting the weighted average chemical shift changes of AP180 M5 proteins upon clathrin TD binding using least square nonlinear curve fitting (ORIGIN v7.0) with the following equation for one-to-one protein-ligand binding:
in which (δb−δf) is the total chemical shift change of AP180 M5 between the bound and free states69; 70. The equilibrium constants (KD) of the two binding sites were determined independently by global fitting the chemical shift changes of the residues within each site.
The dissociation rate constant (koff) of the AP180 M5:clathrin TD interaction was determined by fitting the line widths of AP180 M5 resonances that underwent both chemical shift perturbation and line broadening during the titration with clathrin TD. The observed line widths Δνobs, at half height of the resonance (reported in Hz), were measured (Sparky v3.114) and fitted using least square nonlinear curve fitting (ORIGIN v7.0) with the following equation for a nucleus in moderately fast exchange between the free and bound states:
where Δνf and Δνb are the line widths in the free and bound states, ff and fb are the fractions of AP180 M5 free and bound, (δb−δf) is the total chemical shift change of AP180 M5 between the bound and free states (reported in Hz) 69. Since KDs of the two binding sites were already determined, ff and fb for each titration point and (δb−δf) could be calculated. Δνf was measured from the titration data. The dissociation rate constant, koff, for each of the two binding sites was determined independently, by global fitting the line widths of the residues within each site. The association rate constant kon was then calculated from the KD and koff values, using the equation kon = koff/KD.
Structural Restraints of AP180 M5
Intrermolecular NOE distance restraints of 500 μM 15N labeled AP180 M5, in both the free (500 μM 15N-AP180 M5) and clathrin TD bound (500 μM 15N-AP180 M5 with 500 μM unlabeled clathrin TD) states, were identified through analysis of three-dimensional 15N-edited NOESY experiment performed using mixing time (τm) of 200 ms71.
Backbone 15N Relaxation Parameters
Backbone amide 15N T2 and 1H-15N NOE relaxation parameters of 15N labeled AP180 M5, in both the free (500 μM 15N-AP180 M5) and clathrin TD bound (500 μM 15N-AP180 M5 with 500 μM unlabeled clathrin TD) states, were measured using the 1H-detected pulse schemes72. The data sets consisted of multiple interleaved time points with variable relaxation delays of 4.0, 40.0, 80.0, 160.0, 220.0, 280.0 and 360.0 ms for T2. The 1H-15N NOEs were measured by recording the signal intensities in two experiments, one with a series of proton presaturation pulses and one with the presaturation period replaced by a delay of equal length (2.9 s). The T2 data set was acquired using a 15N 90° pulse width of 40.0 μs with a spacing of 500 μs between 180° pulses in the CPMG pulse train. The data were analyzed by measuring the peak intensities as a function of the variable relaxation delay, which were then fit to a decaying exponential, I(t)=Ioexp(−t/Ti) for each site i, using the conjugate gradient minimization technique73. 1H-15N NOE values were calculated from the ratio of peak intensities from the spectra obtained with and without 1H presaturation modified by a correction factor that takes into account incomplete magnetization recovery during the recovery period74.
NMR Assignments
The AP180 M5 chemical shift assignments have been deposited in the BioMagResBank (BMRB) under accession code 17048. The BRMB has been instructed to release the assignments immediately upon publication.
Supplementary Material
Figure S1. Characterization of AP180 M5 Mutants. Single-site AP180 M5 fragments were prepared by mutating major residues within each binding site as described in materials and methods. An HSQC spectrum of 500 μM of the indicated 15N-AP180 M5 is shown in black while a spectrum of 500 μM of the indicated 15N-AP180 M5 with 500 μM unlabeled clathrin TD is shown in red. A. AP180 M5 Site 1 has the second binding site mutated. The only chemical shift changes observed upon titration with clathrin TD correspond to site 1. B. AP180 M5 Site 2 has the first binding site mutated. The only chemical shift changes observed upon titration with clathrin TD correspond to site 2.
Acknowledgments
This work was supported by NIH-NINDS grant NS029051 to EML. We also gratefully acknowledge the support of the UTHSCSA Center for Biomolecular NMR Spectroscopy, and the UTHSCSA Center for Macromolecular Interactions, both of which are supported by the Cancer Therapy and Research Center through the NIH-NCI P30 award CA054174, as well as by Texas State funds provided through the Office of the Vice President for Research of the UTHSCSA. We would also like to thank Dr. Neal Robinson for helpful discussions of the work.
Abbreviations
- Alexa 488
alexa fluor 488 succinimidyl ester
- AP180 M5
5kD middle segment (aa 623-680) of the AP180 C-terminal domain
- AUC
analytical ultracentrifugation
- CBD
clathrin binding domain
- CCV
clathrin coated vesicle
- CD
circular dichroism
- Clathrin TD
N-terminal domain of the clathrin heavy chain (aa1-363)
- CSI
chemical shift index
- CTD
C-terminal domain
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- HSQC
heteronuclear single quantum coherence
- IDP
intrinsically disordered protein
- IPTG
isopropyl-β-D-galactopyranoside
- NMR
magnetic resonance spectroscopy
- NOE
nuclear overhauser effect
- NOESY
nuclear overhauser effect spectroscopy
- NTD
N-terminal domain
- PBS
phosphate-buffered saline
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PMSF
phenylmethyl-sulfonyl fluoride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 2.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–7. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 3.Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–96. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- 4.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–5. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 5.Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- 6.Sousa R, Lafer EM. Keep the traffic moving: mechanism of the Hsp70 motor. Traffic. 2006;7:1596–603. doi: 10.1111/j.1600-0854.2006.00497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ungewickell EJ, Hinrichsen L. Endocytosis: clathrin-mediated membrane budding. Curr Opin Cell Biol. 2007;19:417–25. doi: 10.1016/j.ceb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 8.ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–73. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ybe JA, Brodsky FM, Hofmann K, Lin K, Liu SH, Chen L, Earnest TN, Fletterick RJ, Hwang PK. Clathrin self-assembly is mediated by a tandemly repeated superhelix. Nature. 1999;399:371–5. doi: 10.1038/20708. [DOI] [PubMed] [Google Scholar]
- 10.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–32. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owen DJ, Vallis Y, Noble ME, Hunter JB, Dafforn TR, Evans PR, McMahon HT. A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell. 1999;97:805–15. doi: 10.1016/s0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- 12.Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the beta 2-adaptin appendage domain. Embo J. 2000;19:4216–27. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao Y, Chen J, Maynard JA, Zhang B, Quiocho FA. A novel all helix fold of the AP180 amino-terminal domain for phosphoinositide binding and clathrin assembly in synaptic vesicle endocytosis. Cell. 2001;104:433–40. doi: 10.1016/s0092-8674(01)00230-6. [DOI] [PubMed] [Google Scholar]
- 14.Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–35. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- 15.Kent H, McMahon H, Evans P, Benmerah A, Owen D. Gamma-adaptin appendage domain. Structure and binding site for Eps15 and gamma-synergin. Structure (Camb) 2002;10:1139. doi: 10.1016/s0969-2126(02)00801-8. [DOI] [PubMed] [Google Scholar]
- 16.Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev Cell. 2006;10:329–42. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Schmid EM, Ford MG, Burtey A, Praefcke GJ, Peak-Chew SY, Mills IG, Benmerah A, McMahon HT. Role of the AP2 beta-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006;4:e262. doi: 10.1371/journal.pbio.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly BT, McCoy AJ, Spate K, Miller SE, Evans PR, Honing S, Owen DJ. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Honing S, Evans PR, Owen DJ. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell. 2010;141:1220–9. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang J, Taylor AB, Prasad K, Ishikawa-Brush Y, Hart PJ, Lafer EM, Sousa R. Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface. Biochemistry. 2003;42:5748–53. doi: 10.1021/bi034270g. [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–24. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang J, Lafer EM, Sousa R. Crystallization of a functionally intact Hsc70 chaperone. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2006;62:39–43. doi: 10.1107/S1744309105040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenez LE, Jin S, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31:232–43. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci U S A. 2000;97:1096–100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miele AE, Watson PJ, Evans PR, Traub LM, Owen DJ. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nat Struct Mol Biol. 2004;11:242–8. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- 26.Dafforn TR, Smith CJ. Natively unfolded domains in endocytosis: hooks, lines and linkers. EMBO Rep. 2004;5:1046–52. doi: 10.1038/sj.embor.7400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafer EM. Clathrin-protein interactions. Traffic. 2002;3:513–20. doi: 10.1034/j.1600-0854.2002.30801.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhou S, Sousa R, Tannery NH, Lafer EM. Characterization of a novel synapse-specific protein. II. cDNA cloning and sequence analysis of the F1–20 protein. J Neurosci. 1992;12:2144–55. doi: 10.1523/JNEUROSCI.12-06-02144.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou S, Tannery NH, Yang J, Puszkin S, Lafer EM. The synapse-specific phosphoprotein F1-20 is identical to the clathrin assembly protein AP-3. J Biol Chem. 1993;268:12655–62. [PubMed] [Google Scholar]
- 30.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–64. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell EJ. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem. 2002;277:8209–16. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- 32.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the database of disordered proteins. Nucleic Acids Res. 2007;35:D786–93. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao W, Tan Z, Prasad K, Reddy KK, Chen J, Prestwich GD, Falck JR, Shears SB, Lafer EM. Regulation of AP-3 function by inositides. Identification of phosphatidylinositol 3,4,5-trisphosphate as a potent ligand. J Biol Chem. 1997;272:6393–8. doi: 10.1074/jbc.272.10.6393. [DOI] [PubMed] [Google Scholar]
- 34.Morgan JR, Prasad K, Hao W, Augustine GJ, Lafer EM. A conserved clathrin assembly motif essential for synaptic vesicle endocytosis. J Neurosci. 2000;20:8667–76. doi: 10.1523/JNEUROSCI.20-23-08667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dell’Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–4. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- 36.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 37.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–64. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao W, Luo Z, Zheng L, Prasad K, Lafer EM. AP180 and AP-2 interact directly in a complex that cooperatively assembles clathrin. J Biol Chem. 1999;274:22785–94. doi: 10.1074/jbc.274.32.22785. [DOI] [PubMed] [Google Scholar]
- 39.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–26. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 41.Evans PR, Owen DJ. Endocytosis and vesicle trafficking. Curr Opin Struct Biol. 2002;12:814–21. doi: 10.1016/s0959-440x(02)00395-0. [DOI] [PubMed] [Google Scholar]
- 42.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 43.Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A suite of triple resonance NMR experiments for the backbone assignment of 15N, 13C, 2H labeled proteins with high sensitivity. J Am Chem Soc. 1994;116:11655–66. [Google Scholar]
- 44.Yu J, Simplaceanu V, Tjandra NL, Cottam PF, Lukin JA, Ho C. 1H, 13C, and 15N NMR backbone assignments and chemical-shift-derived secondary structure of glutamine-binding protein of Escherichia coli. J Biomol NMR. 1997;9:167–80. doi: 10.1023/a:1018606304131. [DOI] [PubMed] [Google Scholar]
- 45.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222:311–33. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 46.Hinck AP, Markus MA, Huang S, Grzesiek S, Kustonovich I, Draper DE. The RNA binding domain of ribosomal protein L11: three-dimensional structure of the RNA-bound form of the protein and its interaction with 23 S rRNA. Journal of Molecular Biology. 1997;274:101–13. doi: 10.1006/jmbi.1997.1379. [DOI] [PubMed] [Google Scholar]
- 47.Brookes E, Demeler B. Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments. GECCO Proceedings ACM. 2007 978-1-59593-697-4/07/0007. [Google Scholar]
- 48.Demeler B, Brookes E. Monte Carlo analysis of sedimentation experiments. Colloid Polym Sci. 2008;286:129–137. [Google Scholar]
- 49.Di Pietro SM, Cascio D, Feliciano D, Bowie JU, Payne GS. Regulation of clathrin adaptor function in endocytosis: novel role for the SAM domain. Embo J. 2010;29:1033–44. doi: 10.1038/emboj.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–80. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 51.Johnson NMR View: A computer program for the visualization and analysis of NMR data. Journal of Biomolecular NMR. 1994;4:603–14. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 52.Kang DS, Kern RC, Puthenveedu MA, von Zastrow M, Williams JC, Benovic JL. Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. J Biol Chem. 2009;284:29860–72. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lafer EM, Zhuo Y, Ilangovan U, Hinck AP. 2009 Neuroscience Meeting Planner. Chicago, IL: Society for Neuroscience; 2009. The assembly of the endocytic apparatus during synaptic vesicle recycling. 2009 Program No. 420.16/D6 Online. [Google Scholar]
- 54.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 55.Cantor CR, Schimmel PR. Biophysical Chemistry Part III: The behavior of biological macromolecules. W.H. Freeman and Company; New York: 1980. [Google Scholar]
- 56.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–84. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 57.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol. 2004;11:358–64. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 58.Schlosshauer M, Baker D. Realistic protein-protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 2004;13:1660–9. doi: 10.1110/ps.03517304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye W, Lafer EM. Bacterially expressed F1-20/AP-3 assembles clathrin into cages with a narrow size distribution: implications for the regulation of quantal size during neurotransmission. J Neurosci Res. 1995;41:15–26. doi: 10.1002/jnr.490410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci U S A. 2008;105:17772–7. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–21. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 62.Drake MT, Traub LM. Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J Biol Chem. 2001;276:28700–9. doi: 10.1074/jbc.M104226200. [DOI] [PubMed] [Google Scholar]
- 63.Demeler B. Modern Analytical Ultracentrifugation: Techniques and Methods. Royal Society of Chemistry; Cambridge, UK: 2005. [Google Scholar]
- 64.Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39:405–14. doi: 10.1007/s00249-009-0413-5. [DOI] [PubMed] [Google Scholar]
- 65.Demeler B. Sedimentation velocity analysis of highly heterogeneous systems. Analytical biochemistry. 2004;335:279–88. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 66.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 67.Garrett DS, Seok YJ, Peterkofsky A, Clore GM, Gronenborn AM. Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry. 1997;36:4393–8. doi: 10.1021/bi970221q. [DOI] [PubMed] [Google Scholar]
- 68.Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu JS, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol. 1996;3:340–5. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 69.Kingston RL, Hamel DJ, Gay LS, Dahlquist FW, Matthews BW. Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc Natl Acad Sci U S A. 2004;101:8301–6. doi: 10.1073/pnas.0402690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McKenna S, Hu J, Moraes T, Xiao W, Ellison MJ, Spyracopoulos L. Energetics and specificity of interactions within Ub. Uev Ubc13 human ubiquitin conjugation complexes. Biochemistry. 2003;42:7922–30. doi: 10.1021/bi034480t. [DOI] [PubMed] [Google Scholar]
- 71.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage lambda N-peptide/boxB RNA complex. J Am Chem Soc. 1997;119:6711–6721. [Google Scholar]
- 72.Kay LE, Nicholson LK, Delaglio F, Bax A, Torchia DA. Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J Magn Reson. 1992;97:359–73. [Google Scholar]
- 73.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. C: The Art of Scientific Computing. 2. Cambridge University Press; 1992. Numerical Recipes. [Google Scholar]
- 74.Grzesiek S, Bax A. The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–4. [Google Scholar]
- 75.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Characterization of AP180 M5 Mutants. Single-site AP180 M5 fragments were prepared by mutating major residues within each binding site as described in materials and methods. An HSQC spectrum of 500 μM of the indicated 15N-AP180 M5 is shown in black while a spectrum of 500 μM of the indicated 15N-AP180 M5 with 500 μM unlabeled clathrin TD is shown in red. A. AP180 M5 Site 1 has the second binding site mutated. The only chemical shift changes observed upon titration with clathrin TD correspond to site 1. B. AP180 M5 Site 2 has the first binding site mutated. The only chemical shift changes observed upon titration with clathrin TD correspond to site 2.