Abstract
Based on the crystal structure of lactose permease (LacY) open to the cytoplasm, a hybrid molecular simulation approach with self-guided Langevin dynamics (SGLD) is used to describe conformational changes that lead to a periplasmic-open state. This hybrid approach consists of implicit (IM) and explicit (EX) membrane simulations and requires SGLD to enhance protein motions during the IM simulations. The pore radius of the lumen increases by 3.5 Å on the periplasmic side and decreases by 2.5 Å on the cytoplasmic side (relative to the crystal structure), which suggest a lumen that is fully open to the periplasm to allow for extracellular sugar transport and closed to the cytoplasm. Based on our simulations, the mechanism that triggers this conformational change to the periplasmic-open state is the protonation Glu269 and binding of the disaccharide. Then, helix packing is destabilized by breaking of several side chains involved in hydrogen bonding (Asn245, Ser41, Glu374, Lys42 and Gln242). For the periplasmic-open conformations obtained from our simulations, helix-helix distances agree well with experimental measurements using double electron-electron resonance, fluorescence resonance energy transfer, and varying sized cross-linkers. The periplasmic-open conformations are also in compliance with various substrate accessibility/reactivity measurements that indicate an opening of the protein lumen on the periplasmic side on sugar binding. The comparison with these measurements suggests a possible incomplete closure of the cytoplasmic half in our simulations. However, the closure is sufficient to prevent the disaccharide from transporting to the cytoplasm, which is in accordance with the well-established alternating access model. Ser53, Gln60, and Phe354 are determined to be important in sugar transport during the periplasmic-open stage of the sugar transport cycle and the sugar is found to undergo an orientational change in order to escape the protein lumen.
Keywords: Major Facilitator Superfamily, Secondary Active Transporters, Protein Folding, Enhanced Conformational Sampling, Membrane Protein
INTRODUCTION
In biological cells, one of the most important functions of membranes is to control the molecular traffic in and out of the cell. This transport is primarily controlled by transmembrane proteins and these can either be classified as primary or secondary transporters. Primary transporters directly utilize energy, such as, adenosine triphosphate or light, to induce conformational changes in proteins and transport small molecules “uphill”.1 Secondary active transporters couple the chemical gradient or movement of ions, such as H+ or Na+, with the movement of the solute2 and are expressed in all species from single-celled organisms to mammals.3 The Major Facilitator Superfamily (MFS) is an important class of secondary active membrane transporters. Members of MFS can be found in almost all types of biota, from unicellular organisms to humans,4 and are very diverse in terms of substrate transport. The molecules transported by MFS range from simple disaccharides to complex drug molecules.4
One of the most extensively studied MFS proteins, lactose permease (LacY) of Escherichia coli, uses the free energy released from the from downhill translocation of protons across the plasma membrane to carry out stoichiometric symport of galactosides against a concentration gradient.5; 6 In this active transport, the direction of proton electrochemical gradient (ΔµH+) determines the direction of galactoside transport. In the absence of ΔµH+, LacY uses the free energy released from the downhill translocation of a galactoside to generate ΔµH+.5; 6; 7
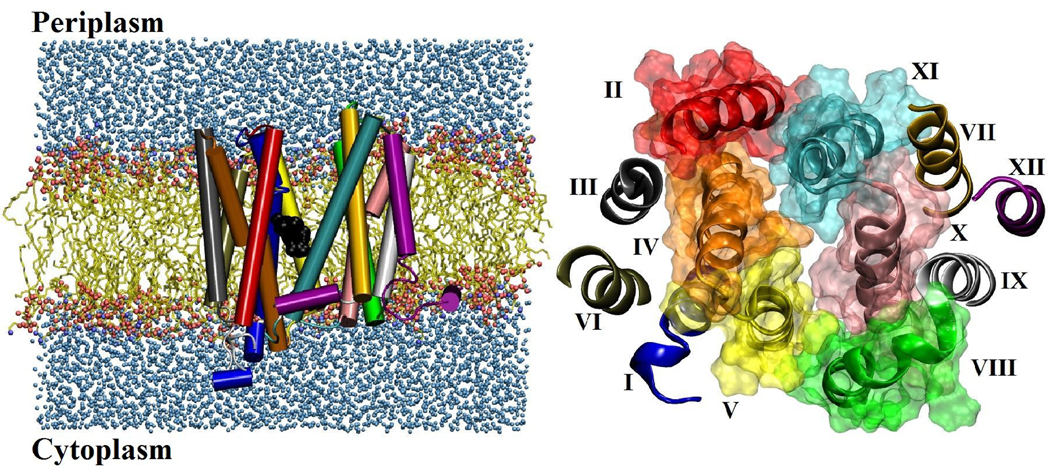
LacY consists of twelve transmembrane α-helices that form two pseudo-symmetric domains and a hydrophilic cavity that exists between these two domains.5 It shares this structural motif with other MFS proteins, i.e., glycerol 3-phosphate transporter (GlpT)8 and EmrD9, suggesting that it can be used as a structural and functional model for this superfamily of proteins. The x-ray crystal structure of LacY was first measured with a C154G mutant that was arrested in the cytoplasmic-open state5 and later the structure of the wild type protein10 was determined (also open to the cytoplasm). There is a negligible Cα root mean squared deviation (RMSD) of 1.2 Å between these two structures and both have a tightly packed periplasmic half preventing movement of the substrate. The pseudo two domain symmetry (Figure S1) suggests that these domains could rotate and form the outward-facing structure of LacY, but the exact details are unknown.11; 12 Although attempts have been made to crystallize the periplasmic-open state, these have not been successful and suggest that at the conditions for crystallization the cytoplasmic-open state is the most stable or that with the lowest free energy.10 The crystal structures of two other MFS proteins (GlpT8 and EmrD9) are each in states with a tightly packed and closed periplasmic half.
Since atomic-level descriptions of the periplasmic-open state of LacY have been unsuccessful, various other experimental techniques have been used to indirectly measure how LacY structurally changes to allow for sugar transport. Double electron-electron resonance (DEER)13 and single molecule fluorescence resonance energy transfer (FRET)12 experiments that use labeled residue pairs to measure interhelical distances indicated a decrease in cytoplasmic interhelical distances and increase in periplasmic interhelical distances for the wild-type LacY (LacY-wt) compared to the inward-facing state. Site directed alkylation11; 14 experiments that measure changes in reactivity for single-Cys replacement mutants for nearly all the 417 residues in LacY indicated an increase in the reactivity on the periplasmic side upon sugar binding. Thiol cross-linking15 experiments also showed that long and flexible cross-linking agents exhibit sugar transport, while no transport activity was observed for short cross-linking agents. All these experiments indicate a mechanism that involves significant opening and closing of the hydrophilic cavity on either sides of the membrane.
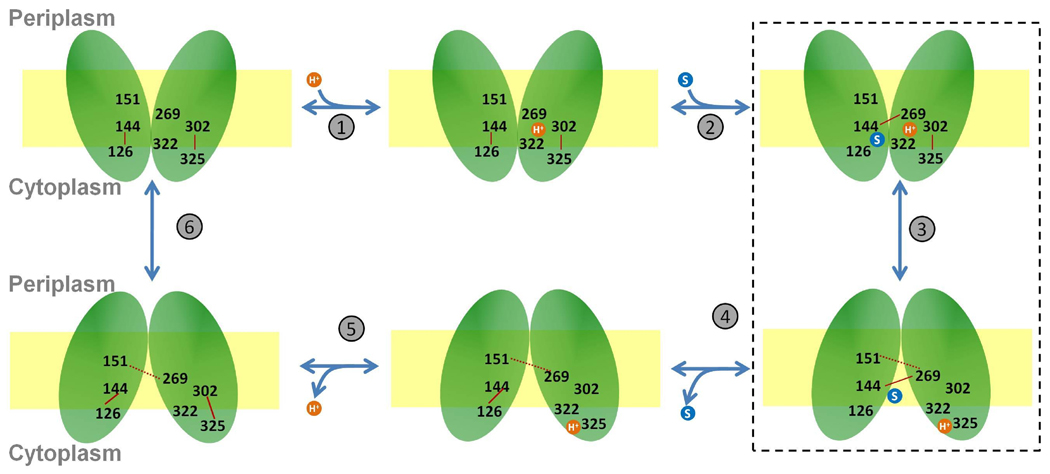
The mechanism for the sugar/proton transport by LacY based on the experimental findings is shown as a simple kinetic scheme similar to Guan and Kaback6 (Figure 1). The proton coupled influx of a sugar consists of the following six steps: (1) LacY in the outward-facing state is protonated at Glu269 or the proton is shared between Glu269 and His322 (2) sugar enters LacY from periplasmic side (3) proton is transferred to Glu325 through His322 and the protein undergoes conformational change to the inward-facing state (4) sugar is then released on the cytoplasmic side (5) proton is released on the cytoplasmic side (6) LacY transitions back to the outward-facing state. According to this mechanism, LacY undergoes a global conformational change from a periplasmic-open to a cytoplasmic-open state on sugar binding with the same binding site available for sugar in both these states.6 Although this mechanism explains transitions between the inward- and outward-facing states of LacY, it lacks structural details of the proposed outward-facing state as well as the transitions between the inward- and outward-facing states (step 3 in Fig. 1). A different model for the sugar transport by LacY states that the specific interplay between the above mentioned residues, Glu269, His322 and Glu325, may not be a requirement for the sugar transport, but several residues including Asp240, Glu269, Arg302, Lys319, His322 and Glu325 facilitate sugar transport without essentially being directly involved in proton/sugar binding.16
Figure 1.
A mechanism for sugar/proton symport based on the experimental findings and similar to Guan and Kaback.6 Important residues are labeled. Interactions between protein residues are represented in red and protein-sugar interactions are represented in black. Salt bridges and hydrogen bonds are represented as solid and dotted lines respectively. The dotted box encompassing step 3 represents the stage of the overall transport mechanism studied in this work.
Several simulation studies were performed on LacY and other MFS proteins to throw light on the transport mechanism. These focused on helix dynamics and stability,17 substrate binding,18; 19; 20; 21 and substrate transport pathway.19; 21; 22 Yin et al.19 were the first to report any structural changes in LacY using a simulation approach. They carried out molecular dynamics (MD) simulations of LacY embedded in a fully hydrated lipid bilayer to study the conformational changes in LacY on translocation of a proton from Glu325 to Glu269. In the Glu269 protonated state, slight closing was observed in the cytoplasmic half during the 10-ns simulation. Similar cytoplasmic closure was observed during the 50-ns simulations by Holyoake et al.22 with Glu325 protonated. This suggests that closing of the cytoplasmic half is not dependent on the protonation state of LacY. Steered molecular dynamics (SMD) simulations by Jensen et al.23 in which sugar was pulled through the protein lumen resulted in small periplasmic conformational changes. Although transport of the disaccharide from the binding pocket to the periplasm was observed, the periplasmic pore did not open in a manner consistent with experimental studies.12; 13; 15 This indicates that the observed structural changes could be an artifact of the SMD approach, which may not allow enough time for the protein to respond to the substrate being pulled through the lumen. Recent MD simulations on an MFS protein, GlpT, also resulted in partial closure of the cytoplasmic half while no significant periplasmic changes were observed.24
The focus of this work is to understand the atomic-level details of the outward-facing state of LacY and the mechanism for structural changes between the inward- and outward-facing states (Fig. 1, step 3). A two-step hybrid simulation approach is used (see methods). In the first step, self guided Langevin dynamics (SGLD)25 simulations are performed with LacY in an implicit bilayer to enhance the conformational changes that MD cannot attain. SGLD uses local averaged momentum, which is averaged over the neighboring conformational space, to guide the conformational transition.25 The local averaging time and the guiding factor determine the low frequency motions to be enhanced and the strength of guiding, respectively.25 Previously, SGLD has been shown to significantly improve the ability to search protein conformations without the loss of accuracy in peptide folding25 and various conformational changes in aqueous proteins.26; 27 This method has been tested extensively in small molecule and peptide folding simulations with the density of states, energies (kinetic and potential), and energy fluctuations all unaltered compared to long unguided simulations. A reasonable choice in SGLD parameters maintains this stability while significantly enhancing conformational sampling.25 In the second step, MD simulations of structures screened from the first step are carried out in a fully explicit and hydrated lipid bilayer. The binding of the disaccharide is studied with a focus on changes associated with periplasmic-open conformations of LacY. Residues and interhelical interactions that are important during protein conformational changes to a periplasmic-open state are also determined. We conclude by suggesting mutations of specific residues to aid in crystallize the outward-facing state and describe a mechanism for this conformational change.
RESULTS
This section is divided in three subsections. The first subsection explains the rationale behind the use of this hybrid simulation approach (IM-EX) for obtaining structural changes in LacY. The details of this approach are given in the Methods section, but the nomenclature, system size and simulation times are in Table 1. Structural changes are described by means of Root Mean Square Deviation (RMSD) from 1PV7 crystal structure and pore radius analysis. Structural information about the proposed outward-facing LacY is also described in this section, such as helix-helix distances, domain angle changes, and structure stabilizing residue-residue interactions. In the second subsection, the data obtained from the simulations is compared with the results from FRET,12 DEER,13 cross-linking,15 and substrate accessibility experiments.14 The final subsection describes the binding of a sugar molecule in the inward- and outward-facing states.
Table 1.
System sizes and simulation times for the lactose permease simulations. MD was used in the EX and IM-MD simulations and SGLD for the other IM simulations. Unless otherwise noted, the simulations are with Glu269 protonated.
| Run name |
Simulation Time (ns) |
Initial structure | Total atoms |
Water molecules |
Lipid molecules |
|---|---|---|---|---|---|
| IM-MD | 20ns | Snapshot from Klauda & Brooks, 200718 |
35,370 | 9,563 | - |
| IM-r1a | 10ns | ||||
| IM-r2 | 10ns | ||||
| IM-r3 | 10ns | ||||
| EX-r1 | 25ns | 3.4ns snapshot | 91,471 | 17,055 | 269 |
| EX-r2 | 20ns | from IM-r1 | |||
| EX-r3 | 20ns | 5.5ns snapshot | 94,876 | 18,440 | 263 |
| EX-r4 | 40ns | from IM-r1 | |||
For this conformation three simulations were run with Glu269, His322, and Glu325 protonated.
Structural changes from the IM-EX simulation approach
Implicit bilayer simulations
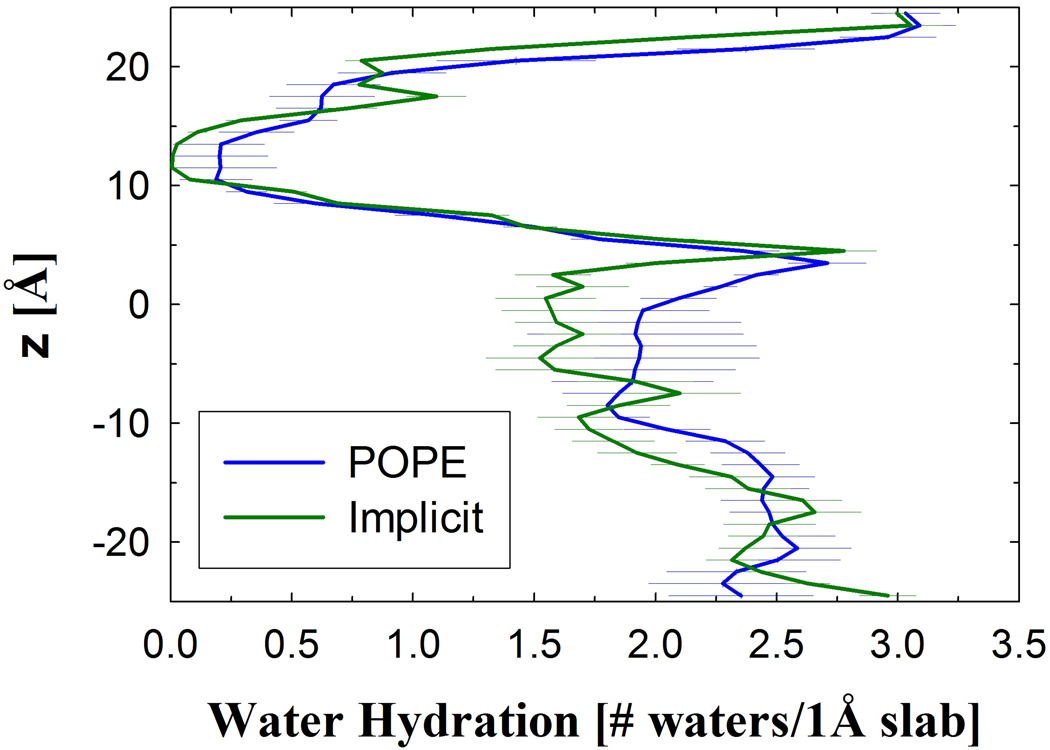
Initially, a MD implicit membrane simulation and a previous explicit simulation18 were compared to verify that the implicit membrane did not influence the protein structure and that of the water in the lumen. (Unless otherwise noted IM simulations are with Glu269 protonated.) Since the implicit membrane only acts on the protein, waters that are in the protein lumen are not directly influenced by this implicit membrane. If there is no significant conformational change in the protein compared to previous explicit simulations,18 then the hydration of the protein lumen should be similar for the EX and IM simulations. As described below, the IM-MD simulations resulted in no significant change in protein structure and can be compared with previous EX simulations. The water profile is nearly identical between the EX and IM-MD simulations (Figure 2), which suggests a negligible influence on waters in the protein lumen.
Figure 2.
The hydration of LacY’s lumen for the average of six simulations in an explicit POPE membrane (POPE)18 and the implicit membrane simulation with MD (Implicit). The number of water molecules within bins of 1 Å is reported with the periplasmic half as positive z.
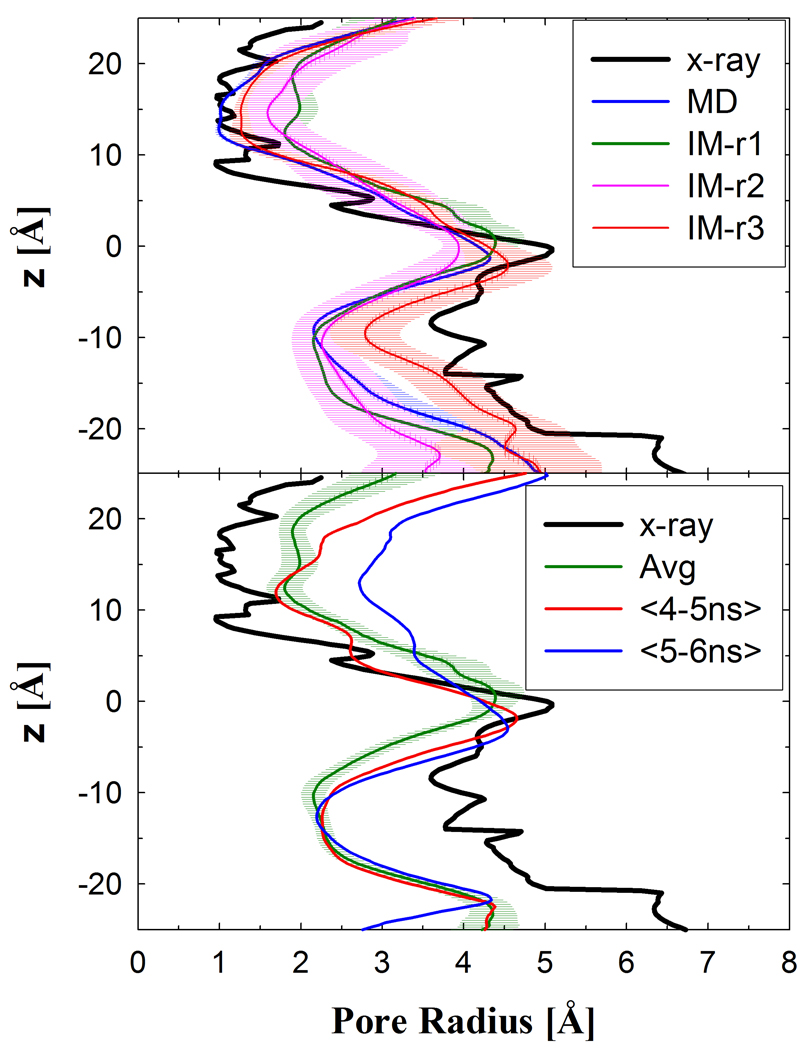
Conformational changes on the periplasmic half of LacY in the IM-MD simulation were negligible compared to previous EX simulations and the x-ray crystal structure. The distances between helices at the membrane interface (Figure S2) only showed slight changes from the x-ray structure and consistent with fluctuations seen in previous EX simulations.18 The pore radius at the periplasmic constriction (Figure 3) did not change from the x-ray crystal structure, and for the cytoplasmic half, it decreased by ~2 Å which is consistent with previous simulations.18; 19 Multiple simulations will be required to determine if the IM-MD simulations result in the conformational changes similar to EX simulations, which is not the focus of present study. The details of the helix-helix distance and pore radius calculations are given in the Methods.
Figure 3.
The pore radius profile of LacY’s lumen is reported for the implicit membrane simulations, where the periplasmic half is for z>0. The top panel is a comparison of several IM simulations with the x-ray structure pore radius. The pore radius profiles reported in the top panel are calculated based on the averages over the entire simulation described in Table 1. The bottom panel is for IM-r1 for the average over the simulation of 10 ns and averages of the 5 and 6 one nanosecond blocks.
Since MD was not successful in obtaining conformational changes in the periplasmic half of LacY, SGLD was used to enhance conformational sampling. Since we are starting from a state open to the cytoplasm, the mechanism6 in Figure 1 suggest that proton translocation from Glu325 to Glu269 facilitates this structural change. Therefore, there are two controls in our simulations that should not result in periplasmic conformational changes if protonation of Glu269 triggers such a change, i.e., IM-r1 simulations with Glu325 and His322 protonated. In addition, simulations were carried out with apo-LacY (without sugar) to understand the effect of sugar binding on conformational changes11; 13 and a likely third control.
All simulations resulted in a significant closure of the cytoplasmic half but only the simulation with Glu269 protonated resulted in opening on the periplasmic side (Figure S3). The cytoplasmic closure was observed to be independent of the protonation states of Glu269, His322 and Glu325 which is in accordance with the findings by Holyoake et al.22 All SGLD simulations with Glu269 protonated resulted in conformational changes in the periplasmic half of LacY (Figures 3, S2, and S4) indicating that protonation of Glu269 and binding of sugar triggers opening of LacY on the periplasmic side.
The remaining presentation of the results will be based on simulations with Glu269 protonated and a disaccharide bound because these were the only to show periplasmic conformational changes. IM-r1 resulted in the largest conformational change with an increase in pore radius ~3 Å (Figure 3, bottom). This transition occurred between 4–6 ns and resulted in a large increase in helix-helix distances between the C- and N-terminus domains (Figure S4). For IM-r2 and -r3, the opening of the periplasmic half is reduced compared to IM-r1. Consistent for all these runs is that H-V/VI and H-IX/X remain packed with only minor changes in helix-helix distances.
The cytoplasmic half of the protein remained partially closed with a pore radius of ~2.25 Å for each run except IM-r3. Similar to the periplasmic half, this involved a decrease in helix-helix distances between the C- and N-terminus domains of LacY (Figure S2 and S4). The largest change from the x-ray crystal structure involved H-V. Overall, H-IV and VIII had the largest RMSD for the runs with SGLD and the disaccharide, i.e., 2.1±0.2 and 1.9±0.4 Å, respectively. Moreover, the C-terminus domain of LacY had a larger RMSD (5.4±0.5 Å) compared to the N-terminus domain (3.3±0.3 Å).
Based on the results above, significant conformational changes in LacY exist for the cyto- and periplasmic halves of the protein with the SGLD implicit membrane simulations with Glu269 protonated and a disaccharide bound. Since the implicit membrane was used to enhance simulation time scales, two protein conformations (Figure S5) were selected and inserted into an explicit POPE membrane to obtain the final conformations of LacY. The protein is simulated in an explicit membrane to prevent any possible artifacts due to the implicit membrane by obtaining structures in a more realistic environment (see Methods). These protein conformations were obtained from the 3.4 and 5.5 ns snapshots of the IM-r1 simulation and represent an intermediate and periplasmic-open state based on the lumen radii, respectively. These were chosen carefully based on the comparison with the distances measured from the DEER experiments13 to see if initial states in the explicit membrane are important to obtain final protein conformations open to the periplasm.
Explicit membrane simulations
The EX-r1 and EX-r2 simulations were carried out starting with the intermediate state and EX-r3 and EX-r4 were carried out starting with the periplasmic-open state. Protein backbone RMSDs from 1PV7 were calculated over the course of the trajectories. For EX-r4, RMSD of the N- and C-terminal domains were 4.1±0.2 and 5.1±0.2 Å, respectively, which is consistent with the IM simulations in which the C-terminal domain shows more structural deviation from the crystal structure than the N-terminal domain. Locally, H-IV with Glu126 and H-VIII with the protonated Glu269 showed the maximum structural drift with average RMSDs of 1.5±0.1 Å and 1.9±0.2 Å, respectively.
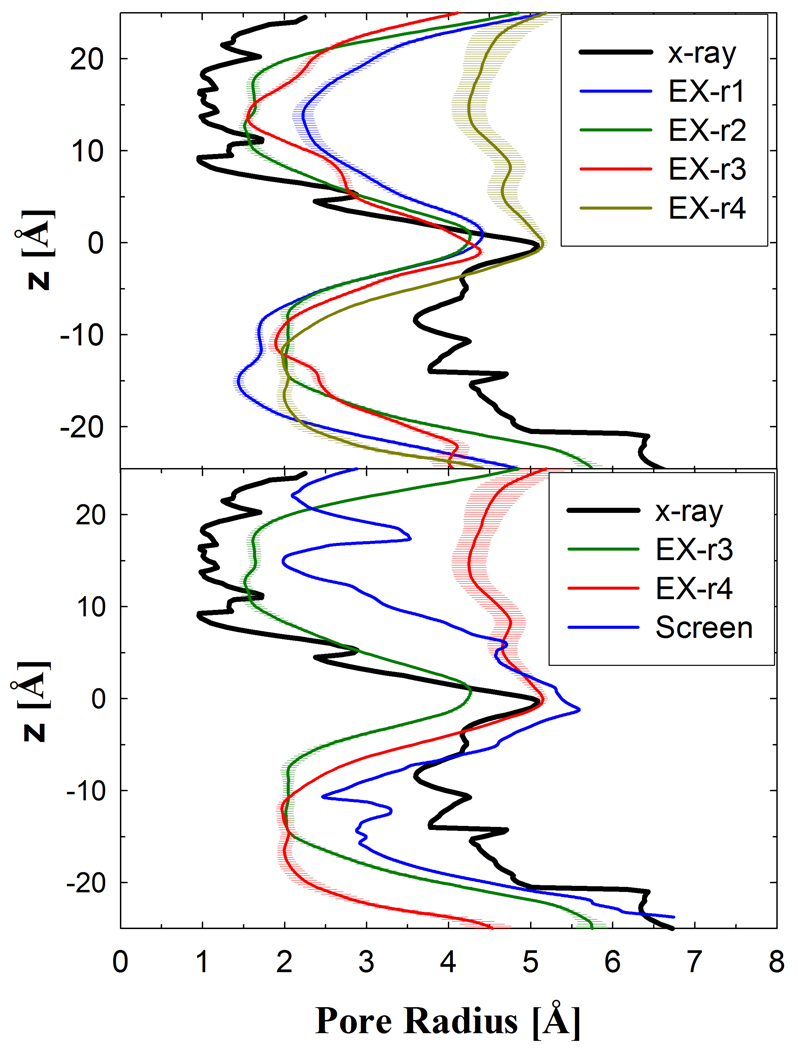
All the simulation runs showed a decrease in the pore radius on the cytoplasmic side by ~2 Å from the crystal structure (Figure 4, top), which is similar to the implicit simulations. The pore radius increase on the periplasmic side was 1–1.5 and 2.5–3 Å for EX-r1 and -r4, respectively. The average pore radius profiles for EX-r3 and EX-r4 were compared with the profile of the periplasmic-open state screened from IM-r1 (Figure 4, bottom). EX-r4 showed an increase of 1–1.2 Å in the pore radius while EX-r3 showed a decrease of ~1 Å from the periplasmic-open snapshot structure. Similarly, EX-r1 showed opening beyond the intermediate snapshot while EX-r2 showed closing (data not shown). In the inward-facing state of LacY (1PV7), the pore radius in the tightly closed periplasmic half is ~1 Å. For EX-r1 and EX-r4, the pore radius at the narrowest section in the cytoplasmic half is ~1.5 and ~2 Å, respectively. For these runs, the cytoplasmic side is not as tightly packed as the periplasmic side in the inward-facing crystal structure, but is sufficient to prevent the disaccharide, which has a minimum span of ~7 Å, to transport to the cytoplasm. Only water can transport from the cytoplasm to the protein lumen in this state and this occurs rarely for this nearly closed cytoplasmic structure.
Figure 4.
The pore radius profile of LacY lumen is reported for the explicit membrane simulations. The top panel is the comparison of the average pore radius profiles for the four explicit simulation suns with the x-ray structure pore radius. Bottom panel is the comparison of the pore radius profiles of EX-r3 and EX-r4 with the pore radius profiles of the periplasmic-open screened structure and the x-ray crystal structure.
On the cytoplasmic side, H-V showed the largest change in separation for all EX runs (Figure 5) and is consistent with the IM simulations. For EX-r1 and EX-r4, several helices showed significant increases for the periplasmic half of the protein (H-II/III/IV/VI/VII/XI). For EX-r2 and EX-r3, no significant change in the helix-helix separation was observed on the periplasmic side. For these runs, H-IV was the only helix to show some increase in separation with respect to the helices in C-terminal domain.
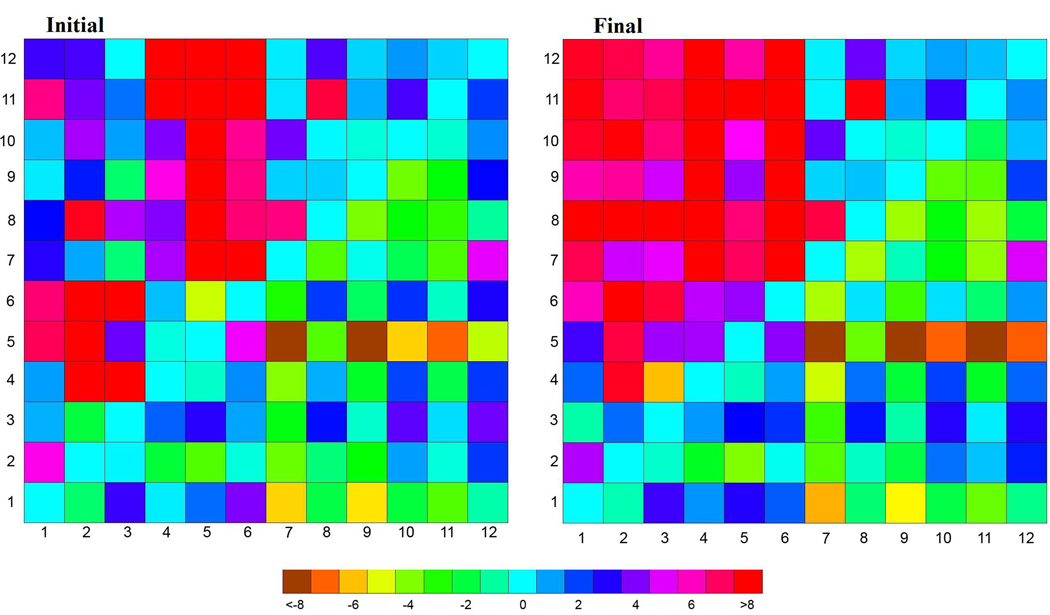
Figure 5.
Average helix center distance deviation from the x-ray crystal structure (1PV7) for (left) first and (right) last nanosecond from the trajectory of EX-r4. All helix-helix pairs (# in rows and columns) are shown with the distances below the diagonal corresponding to the cytoplasmic half and periplasmic above. The legend or colorbar is shown on the bottom and is in Å.
To summarize, for EX-r1 and EX-r4, the protein underwent major conformational changes beyond IM-r1 resulting in a significant periplasmic opening (Figure 6, left). The protein lumen closed on the cytoplasmic side (Figure 6, right) and blocked the access of sugar on the cytoplasmic side. This is in accordance with the well-established alternating access model,11 which states that the sugar binding site is accessible only from the periplasm in the outward-facing state of LacY.
Figure 6.
The outward-facing structure of LacY from EX-r4. (left) Snapshot of LacY embedded in a fully hydrated lipid bilayer. (right) The cytoplasmic halves of the transmembrane helices (z < 0 from the center of mass (COM) of the protein) viewed from the cytoplasmic end in the direction perpendicular to the membrane. The residues on the helices forming the lumen are represented as molecular surfaces based on the van der waals radii of the individual atoms. The color-coding of the helices is shown in right.
Description of the transitions to the Outward-facing State of LacY
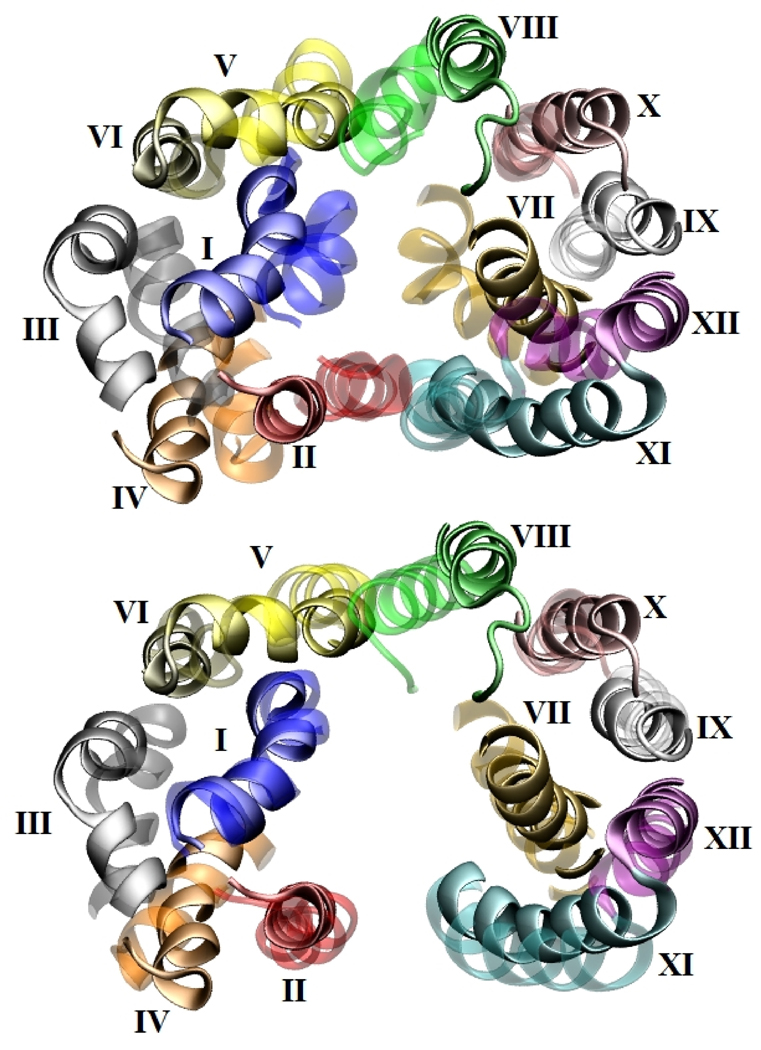
A full description of the of transition between the open states of this protein (Fig. 1, step 3) is left for the discussion section but important structural and stabilizing interactions are briefly presented in this section. The pore radius profiles clearly indicate that two out of four explicit simulation runs (EX-r1 and -r4) resulted in opening of the protein structure on the periplasmic side (movie in supplementary material), while the other runs lacked any significant periplasmic opening. Figure 7 (top) shows the separation transmembrane helices of the periplasmic half (z>0) and opening of the protein lumen for EX-r4. Although the conformational changes of LacY involve a mechanism more complex than rigid-body rotation of the two domains,6 the angle between the principal axis vector of the entire domain obtained from simulations and that from the crystal structure is calculated to quantify the pseudo-rotation of the domain. The principal axes of the N- and C-terminal domain are calculated based on the backbone atoms of the first six and the last six helices, respectively. For EX-r1, principal axis vectors of the N- and C-terminal domains made 11.3±1.6° and 11.4±1.6° angles with the corresponding crystal structure vectors. For EX-r4, the N-terminal domain rotated by ~11°, whereas the C-terminal domain rotated by ~20° from the crystal structure. The domains rotate in directions opposite to each other so that the protein lumen closes on the cytoplasmic side and opens on the periplasmic side. PDB of the outward-facing model of LacY obtained from EX-r4 is included in the supplementary materials.
Figure 7.
The periplasmic halves of the transmembrane helices (z > 0 from the COM of the protein) viewed from the periplasmic end in the direction perpendicular to membrane in which the solid helices indicate the periplasmic-open structure from EX-r4. Transparent helices represent (top) the crystal structure (1PV7) and (bottom) the structure from EX-r1.
Packing of some helices on the periplasmic side was stabilized by hydrogen bond interactions, which are listed in the Table 2. These interdomain hydrogen bonds are broken in EX-r1 and EX-r4 resulting in the opening of LacY on the periplasmic side. The backbone oxygen of Pro31 was involved in hydrogen bonding in EX-r2 and EX-r3 indicating the existence of close tertiary packing between H-I/H-VII near the periplasmic end. The other residue involved in hydrogen binding with Pro31, Asn245, has been shown to play an important role in gating the periplasmic pathway of LacY.28
Table 2.
Hydrogen bond (HB) pairs that stabilize the cytoplasmic-open state. These hydrogen bonds existed for the runs that were periplasmic-closed (EX-r2/r3) and destabilized in the runs with a periplasmic-open state (EX-r1/r4). (SC-side chain, BB-backbone)
| Residue Pairs | Donor-Acceptor Atoms |
Helices |
|---|---|---|
| N245-P31 | NSC –OBB | H-VII/HI |
| S41-E374 | OSC–OBB | H-II/XI |
| K42-Q242 | NBB– OSC | H-II/VII |
| NSC–OBB |
Comparison with the experimental data
Since no periplasmic-open structure exists for this or other MFS proteins a detailed comparison with our simulations is needed to justify the above described periplasmic opening and cytoplasmic closing. Although atomic-level detail is not available, lower-resolution experimental techniques will be compared with our simulations, i.e., FRET,12 DEER,13 cross-linking,15 and site-directed alkylation experiments.11; 14
DEER and FRET Experiments
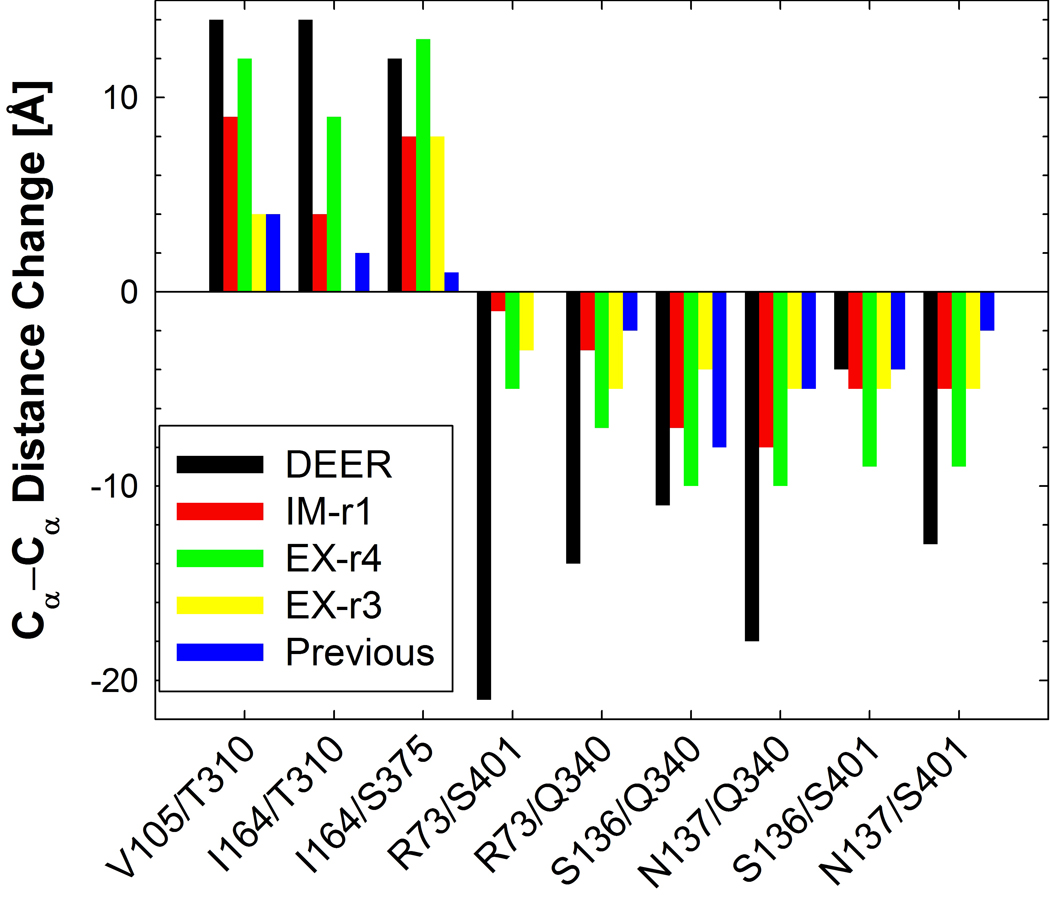
Previous DEER experiments were carried out on Cys mutants of LacY labeled with (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (MTSL). The distances between the spin labels measured in the experiments were compared with the corresponding Cα-Cα distances calculated from the simulations. The correspondence between the two sets of distances is not one to one. However, the comparison throws light on cytoplasmic and periplasmic structural changes and distance changes between cytoplasmic- and periplasmic-open states may cancel out the effect of this bulky spin label if the orientation remains constant. The Cα-Cα distance changes from the cytoplasmic-open (x-ray structure) to the periplasmic-open state (DEER-based) are compared with the LacY simulations in Figure 8 and Table S1. The values reported are based on Gaussian fits to distance distributions of the pairs (Figure S6).
Figure 8.
The Cα-Cα difference in distance between residue pairs relative to the x-ray crystal structure. Negative values indicate that the helix-helix distance has decreased. The distance is measured as the mean of the Gaussian peak fit to the distance distributions (see Figure S6 as an example).
On the periplasmic side, the distances increased significantly from IM-r1 for EX-r1 (Table S1) and EX-r4 (Figure 8). For EX-r4, increase in the distances from IM-r1 was 3–5 Å. The pairs Val105/Thr310 (H-IV/H-IX) and Ile164/Ser375 (H-V/H-XI) showed the maximum separation from the x-ray crystal structure by 12–13 Å. These two distances match considerably well with the DEER experiments. The Ile164/Thr310 distance (H-V/H-X) increased over the IM-r1 run by ~3 Å and ~7 Å from the cytoplasmic-open state but was slightly lower than experiment. For EX-r2 and EX-r3, the distances remained the same or decreased from IM-r1 for all periplasmic pairs.
On the cytoplasmic side, the distance between the pairs decreased for EX-r1 and EX-r4 compared to IM-r1. For EX-r4, this decrease was 2–5 Å for all the pairs. The distances Ser136/Gln340 and Asn137/Gln340 (both H-IV/H-X) showed the maximum decrease of 10 Å from the crystal structure. The distance change for Arg73/Ser401 (H-III/H-XII) was 5 Å which was considerably smaller than the experimental value of 21 Å, but all other distances from EX-r4 compared favorably with the experimental data. For EX-r3, most distances increased from IM-r1, but a few show no significant change or a slight decrease (Figure 8).
These results also qualitatively comply with the findings from the FRET experiments in which the cytoplasmic ends of H-III and H-XII move closer while the periplasmic ends of H-V and H-XI move apart upon sugar binding.12
Cross-linking Experiments
Three paired double-Cys mutants (I40C/N245C, T45C/N245C and I32C/N245C) located at the interface of the N- and C-terminal domains near the periplasmic end were used in previous cross-linking experiments.15 Homobifunctional thiol cross-linking reagents of different lengths and flexibilities were used to test the influence of cross-linking on the transport activity of a disaccharide, TDG. For the I40C/N245C double-Cys mutant, transport activity of sugar was almost entirely blocked with cross-linking reagents of length less than ~15 Å. With the flexible reagents MTS-14-O4-MTS (~17 Å) and MTS-17-O5-MTS (~22 Å), full transport activity of the disaccharide was observed. These cross-linkers suggest that the periplasmic opening of the protein should be between 15 – 17 Å for the outward-facing state.
To determine the size of periplasmic opening, Cβ-Cβ distances between the three residue pairs (I40/N245, T45/N245 and I32/N245) were calculated from our simulations. The experimentally-based periplasmic open distances are the spacer arm distances of the cross-linking reagent and not the distance between Cβ atoms. The distance between the Cβ atoms will be higher than the spacer arm distance and the equilibrium bond distance between Cβ-S of cysteine (1.8 Å) is used as an offset. Therefore, Cβ-Cβ distances between the residue pairs should be between 18.6 and 20.6 Å. The results (Table 3) show that for EX-r1 and EX-r4, the separations for the residue pairs I32/N245, T45/N245 and I32/N245 agree favorably with the size of the opening indicated by the cross-linking experiments. These cross-linking experiments also indicate that the inter-thiol distance between I32C/N245C is close to ~6 Å in absence of the ligand, which is similar to Cβ-Cβ distance for the inward-facing crystal structure (1PV7).5 For EX-r2 and EX-r3 (periplasmic-closed runs), the mean values for Cβ-Cβ distance between I32/N245 are 5.8 Å and 6.5 Å, respectively. Also, the mean Cβ-Cβ distances between the other two pairs are close to the crystal structure.
Table 3.
Mean Cβ-Cβ distances between residues (Å) used in the double-Cys mutant experimental studies.15 The suggested Cβ-Cβ distance in the outward-facing state from cross-linking experiments are 18.6–20.6 Å.
| Residue | Calculated mean distances | 1PV7 | |||
|---|---|---|---|---|---|
| Pairs | EX-r1 | EX-r2 | EX-r3 | EX-r4 | |
| I40-N245 | 20.5 | 10.4 | 12.1 | 21.2 | 8.9 |
| T45-N245 | 17.0 | 8.1 | 9.7 | 17.7 | 8.1 |
| I32-N245 | 17.2 | 5.8 | 6.5 | 18.3 | 6.9 |
Accessibility and Reactivity Experiments
Site directed alkylation of single-Cys mutants of LacY at various positions with small membrane-permeant alkylating reagents, N-ethylmaleimide (NEM)11; 29; 30; 31; 32; 33; 34 and tetra-methylrhodamine-5-maleimide (TMRM),14 was previously measured to investigate the conformational changes in LacY upon sugar binding by quantifying the accessibility and/or reactivity at those residues. The reactivity of a Cys residue can be limited by close contacts between transmembrane helices in its surroundings and a change in the reactivity on sugar binding indicates an alteration in the environment at that position in the sequence.11; 14 Sugar binding was shown to result in an increase in the reactivity of Cys-mutants that predominantly lie in the periplasmic half while a decrease in the reactivity was observed at positions predominantly lying in the cytoplasmic half. For the protein conformations obtained from our simulations, the contact surface areas were calculated for the residues at these positions as a measure of the close contacts (see Methods). These areas were compared with the contact surface areas of the respective residues in the crystal structure to assess the change in environment of these residues (Table S2). As the experiments were performed on the Cys-mutants, the bulky side chain atoms beyond the Cβ were neglected when calculating the surface areas.
On the periplasmic side, most (85%) of the residue positions that had an increase in Cys reactivity11; 14 also had an increase in the contact surface area for the EX-r1 and -r4 (Table S2). On the cytoplasmic side, some residues that had a decreased Cys reactivity did not show a decrease in the contact surface area. This may be a result of incomplete cytoplasmic closure observed during the simulations.
Site specific labeling with methanethiosulfonate ethylsulfonate (MTSES), a small hydrophilic, membrane-impermeant thiol reagent was used to determine the change in water accessibility at various positions on sugar binding.29; 30; 31; 32; 33; 34; 35 The positions that showed a change in water accessibility were determined. To compare with the MTSES accessibility studies, water accessible surface areas of various residues were calculated from our simulations and were compared with the corresponding surface area values for 1PV7 (Table 4). For EX-r1 and EX-r4, there is near perfect agreement for the positions that measured an increase or decrease in accessibility on sugar binding.
Table 4.
Changes in water accessible surface areas compared to the crystal structure (1PV7) for positions that showed change in MTSES accessibility on sugar binding.
| Increased surface area compared to 1PV7 |
Decreased surface area compared to 1PV7 |
Increased | Decreased | ||
|---|---|---|---|---|---|
| MTSES | MTSES | ||||
| EX-r1 | EX-r4 | EX-r1 | EX-r4 | accessibility | accessibility |
| 18, 32, 38, 42–44, 241- 246, 248, 249, 265, 327 |
18, 32, 38, 42–44, 241- 246, 248, 249, 326, 265, 327 |
60,326 | 60 | 18, 32, 38, 42- 44, 241–246, 248, 249, 265, 327 |
60, 326 |
Sugar binding
Since conformational changes of the protein are coupled to sugar binding (Figure 1),11; 13 in the final section of the result we briefly describe sugar binding. Although these results are based on several explicit membrane simulations, more detailed and comprehensive studies on binding are needed to obtain a complete picture of sugar binding in the periplasmic open state and presented here may not be the entire story. The interactions between the disaccharide, ββ-(Galp)2, with LacY are similar for all EX runs. The disaccharide binding pocket in EX-r1 and -r4 (outward-facing LacY) is similar to the IM simulations. The disaccharide formed hydrogen bonds with Glu126, Arg144, Asp237, Glu269, Lys358, Phe118, Asn119, Asp240, His322 and Tyr350, which is consistent with our previous disaccharide binding studies.18 Separate from these well-known sugar-protein interactions, the disaccharide formed hydrogen bonds with Gln60 and Phe354 (Figure S7). Both these residues have been shown to be important in sugar transport.29; 32; 36; 37 For EX-r1, ββ-(Galp)2 escaped the binding pocket during the last 5 ns of simulation. It migrated ~7 Å towards the periplasm after the protein started opening on the periplasmic side and formed hydrogen bonds with Ser53 and Asn119 in this binding conformation (Figure S8) with the importance of the former in the transport cycle confirmed by experiments.32 Similarly for Ex-r2, where LacY started opening on the cytoplasmic side, the disaccharide escaped the binding pocket and migrated towards the cytoplasm. The disaccharide was found to be undergoing a change in its orientation in order to escape on either sides of the membrane. In the binding pocket, the two sugar rings are aligned nearly parallel to the membrane, whereas the rings become perpendicular to the membrane when the disaccharide starts to escape the protein lumen (Figure S9) as observed by Jensen et al.23 Despite the differences in sugar binding when comparing the cytoplasmic- and periplasmic-open states, our simulations agree with the alternating access model11 in which the same binding site is believed to be accessible to the sugar molecules in inward and outward-facing states. Moreover, in the outward-facing state, the protein lumen is closed on the cytoplasmic side preventing the sugar from escaping to the cytoplasm.
DISCUSSION
LacY undergoes widespread conformational changes during substrate transport based on various biochemical experiments.6 Previous MD simulations have resulted in significant conformational changes on the cytoplasmic side compared to the crystal structure. A decrease in pore radius was observed in the cytoplasmic half for LacY with Glu269 protonated in a 10-ns simulation19 and with Glu325 protonated in 50-ns simulation. Our previous EX simulations with Glu325 protonated18 also resulted in similar decrease in the cytoplasmic pore radius by ~2 Å. However, negligible pore radius changes in the periplasmic half were observed during these simulations.
Several control simulations were performed to verify that our methods open the protein on the periplasmic side only when a sugar is bound and the proper protonation of a titratable residue. Only simulations with Glu269 protonated and a sugar bound resulted in conformational changes in the periplasmic half. Simulations without the sugar (apo), Glu325 protonated, or His322 protonated, all maintained a conformation prevent sugar transport to the periplasm. Although this was expected for simulations without the sugar or Glu325 protonated (Figure 1),6 our simulations suggest that protonation of His322 does not trigger a conformational change.
All IM simulations with Glu269 protonated and a sugar bound resulted in greater periplasmic conformational changes in LacY compared to previous EX simulations because of enhanced sampling. The structural changes were enhanced by the use of SGLD instead of MD and an implicit bilayer. The enhancement alone appears to allow for structural changes and sugar binding is a trigger for protein structural changes because LacY-apo remained closed on the periplasmic side. Structures were screened from the IM simulations and inserted into an explicit bilayer so that a more realistic environment can be used to obtain a final set of conformations (Figure 7, bottom). Essentially, it was our aim to use the implicit membrane simulations with SGLD to overcome a transition state or energy barrier between the two open states in a reasonable amount of time. If the screened structures lie in the metastable transition state region in the free energy profile, then there would be an equal probability to go back to the inward-facing state or further open to the outward-facing state assuming a single transitional barrier. For each of the screened structures, one simulation resulted in a fully periplasmic-open state, while another started closing on the periplasmic side and opening on the cytoplasmic side. Certainly more runs would be required to determine if the screened structures are at or near a transition state, but these results do indicate the ability of this approach to lead to a periplasmic-open state and also return to a conformation closed to the periplasm.
The pore radius profile of LacY’s lumen (Figures 2 and 3) and helix-helix separation distance maps (Figure 5, S2, and S3) clearly demonstrate a protein structural change. An interesting point to be noted is the average pore radius profile for EX-r4 (Figure 4, top). The cytoplasmic half of this profile resembles greatly with the periplasmic half of the pore radius profile of the crystal structure and vice versa. Besides the similarities in the two halves, the two profiles are analogous in the sugar binding region (~z =0 Å).
The helix-helix maps (Figures 5 and S3) indicate that on the periplasmic side structure changes involve significant intradomain as well as interdomain helix movements, whereas only the interdomain displacements are dominant on the cytoplasmic side. The periplasmic conformational changes indicate a more complex mechanism involving significant intradomain changes to yield a final outward-facing state. However, if the initial and final structure of LacY is only considered, then the rigid body rotation of LacY is consistent with the helix map. Therefore, intradomain changes are important as the protein changes between open states, but in the end these collapse back to similar helix packing. More specifically, H-V showed the largest change in the separation on the periplasmic and cytoplasmic sides. The C154G mutant of LacY, which is trapped in an inward-facing conformation, has Gly154 (H-V) at the domain interface. Gly154 is believed to cause tighter packing of H-V near the interface hence hindering the movement of H-V which was found to be significant during the conformational transitions from our simulations.
The Cα-Cα distances calculated from EX-r1 and EX-r4 agree favorably with the DEER experiments (Figure 8 and Table S1).13 On the periplasmic side, the distance changes for pairs V105/T210 and I164/S375 are in agreement with the experimental values. On the cytoplasmic side, the distance changes differ from some of the experimental values indicating a possible partial incomplete cytoplasmic closure. However, the cytoplasmic half is closed to the extent that it prevents and transport of sugar to the cytoplasm (Figure 6, right). The largest discrepancy between the simulations and the experiments is for the cytoplasmic pair R73/S401 suggesting that H-II/XII is the last helix pair to close. Moreover, the distances calculated from the simulations are Cα-Cα distances while those measured in the experiments are the distances between the spin labels (MTSL). The approximate distance between the label and the Cα atom is 8 Å, which may result in the maximum deviation of ± 16 Å. For example, the distance changes reported from the DEER experiments differ considerably for the pairs S136/S401 and N137/S401 even though there is little distance separating the two residues (Ser136 and Asn137). Similar difference is present between the pairs S136/Q340 and N137/Q340. It was suggested that these striking differences may be a result of significant rotation of the helices including winding and unwinding at the ends.13 No such conformational changes were observed during our simulations. The distances calculated from EX-r4 are close to the mean of the DEER measurements for the S136(N137)/Q340 and S136(N137)/S401 pairs. This suggests that the orientation of the spin label may play a role in the DEER measurements. To explicitly investigate the effect of spin labels, simulations with labeled residue pairs will be required and are planned as future work.
In addition to DEER measurements, our simulations are in excellent agreement with the amount of the periplasmic opening measured using cross-linking experiments15 (Table 3) and site-directed alkylation with alkylating agents of various sizes.38 Based on these helix distance measurements, it can be concluded that the periplasmic opening observed from EX-r1 and -r4 closely matches the outward-facing state.
The periplasmic opening of LacY obtained from our simulations is also in compliance with NEM,11 TMRM,14 and MTSES29; 30; 31; 32; 33; 34 labeling experiments. 85% of residues that had a significant increase in NEM and TMRM activity on the periplasmic side showed an increase contact surface area in EX-r1 and -r4. On the cytoplasmic side, the closure obtained from our simulations may not be complete thus preventing the expected decrease in the surface area values for some residues, especially those on H-III and -IX (Table S2). However, we suggest that the steric effect of sugar binding at Gln60 may be a reason behind the reduced NEM32 and TMRM14 reactivity observed with the Cys replacement at that position. Although the comparisons with DEER and NEM/TMRM accessibility studies indicate a possible incomplete cytoplasmic closure for our outward-facing LacY model, the pore radius profiles indicate that the closure is sufficient to block the access of sugar to the cytoplasm as per the alternating access model.11 Inaccessibility of the disaccharide from the cytoplasmic end in the outward-facing state can be clearly seen in Figure 6 (right) where the protein lumen is closed on the cytoplasmic side.
The interdomain hydrogen bonds N245/P31, S41/E374 and K42/Q242 are destabilized in EX-r1 and -r4 while they are maintained in EX-r2 and -r3 (Table 2). Moreover, these hydrogen bonds stabilized the helix-packing in previous EX simulations18 as well as in the initial parts of our IM simulations. These interdomain hydrogen bond interactions appear to be crucial in stabilizing the inward-facing state. Side chains of Asn245, Ser41, Glu374, Lys42 and Gln242 are involved in stabilizing the inward-facing structure and mutations in these residues may destabilize the inward-facing state. Moreover, breaking of these hydrogen bonding interactions in the outward-facing state explains the increase in NEM11 and TMRM14 labeling at residues 31, 42 and 245 on sugar binding. In addition, any change in the interactions between H-VII and the surrounding helices at residue 242 was found to inhibit the transport activity which is consistent with our findings.39
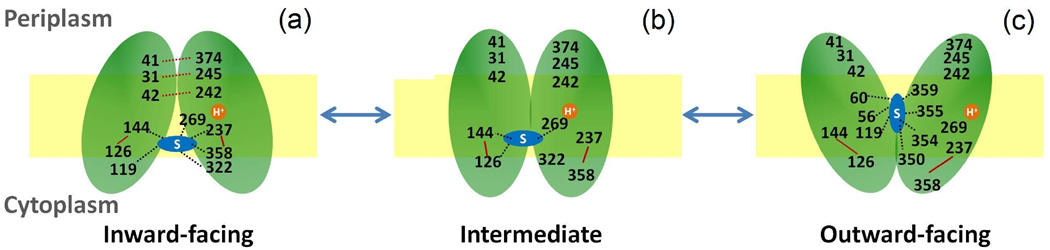
The findings from our simulations are summarized below to give mechanistic insights into the sugar transport cycle and conformational changes of LacY from the outward to inward facing state (Fig. 1, step 3). This is also shown as a cartoon in Figure 9.
Binding of a disaccharide and translocation of proton to Glu269 is necessary to trigger the conformational transformation of LacY. The interdomain hydrogen bonds N245/P31, S41/E374 and K42/Q242 stabilize the inward-facing structure of LacY. In the binding pocket, two rings of the disaccharide are oriented nearly parallel to the membrane.
In the intermediate state, the disaccharide lies in the binding pocket but is weakly bound to the protein at Glu126, Arg144 and Glu269. The structure stabilizing interdomain hydrogen interactions are broken. The salt bridges E126/R144 and D237/K358 remain intact during the transition
The disaccharide remains in the binding pocket during the conformational change which is in accordance with the alternating access model11 but undergoes an orientational change such that the two rings are nearly perpendicular to the membrane. It interacts with Ser56, Gln60, Asn119, Tyr350, Phe354, Cys355, and Gln359 in this binding conformation.
Figure 9.
A proposed mechanism for the conformational transition to the outward-facing state starting with the inward-facing crystal structure (1PV7).5 Important residues are labeled. Interactions between protein residues are represented in red and protein-sugar interactions are represented in black. Salt bridges and hydrogen bonds are represented as solid and dotted lines respectively.
In summary, we were able to obtain significant structural changes on the periplasmic as well as the cytoplasmic side of LacY. The periplasmic changes resulted in the opening of the protein lumen to outside the cell. The outward-facing state from our simulations agrees favorably with a multitude of experimental data. The inward-facing state is stabilized by certain interhelical and interdomain interactions and destabilizing these interactions may help crystallize the outward-facing state. Our simulations are in compliance with the alternating access model11 for the sugar binding but some additional residues that are important in sugar binding are determined. The importance of these residues in sugar binding and conformational change was confirmed by experiments. Ultimately, this IM-EX method offers a possible route to obtain structures of other secondary active transporters based on crystal structures open to the periplasm or cytoplasm in an efficient manner. However, a more systematic study on better characterized proteins will be needed to further test the accuracy and limitations of this method.
METHODS
The CHARMM simulation package40; 41 was used with a simulation time step of 2 fs. The CHARMM family of force fields are used to describe the atomic interactions of the lipids,42; 43 protein (C22-CMAP),44; 45; 46 and disaccharide.47 The TIP3P water model was used (the version consistent with CHARMM parameters48; 49). Long range electrostatic interactions were calculated with the particle-mesh Ewald (PME)50 method. The screening parameter (κ) was set at 0.45 Å−1 and a fast Fourier transform grid density of ~1 Å−1. Lennard-Jones interactions were switched smoothly to zero between 8–10 Å.40 The SHAKE algorithm51 was used to constrain hydrogen atoms. Extended system formalism is used to maintain the pressure with a barostat52; 53 and the temperature constant (EX simulations only) with the Nosé-Hoover method54 at 310.15 K. Glu269 protonated for most simulations, which has been suggested to be the preferred protonated state open to the periplasm7; 19, but other protonated states were also used (see below). All simulation coordinates were saved every picosecond for analysis purposes.
A new two step hybrid simulation method, referred to here as IM-EX (implicit-explicit membrane), was used to probe conformational changes in LacY because fully explicit membrane MD simulations are limited to timescales approaching 1 µs.55 For the first step (IM) in our hybrid simulation procedure, an implicit membrane is used reduce computational demand and enhance conformational changes of the protein. Generalized Born solvation models would be the ideal choice for an implicit membrane, such as those developed by Feig et al.56; 57; 58 However, our benchmark simulations with LacY and the heterogeneous dielectric generalized Born method57 required simulation times that were equal to or greater than a fully explicit bilayer. Instead, an external hydrophobic potential based on the method of Edholm and Jähnig59 is used to represent the lipid/water interface. This potential is assumed to vary exponentially across the membrane surfaces,
where the membrane interface is at ±z0 (assumed to be 15Å), the decay length (λ) is 2, and hi is the hydrophobic energy of atoms taken from reference.59 An explicit water interface is included in all implicit lipid membrane simulations (Figure S1). The lumen of LacY is filled with water18; 19; 22 and not including explicit water may result in an unphysical closing of the cytoplasmic opening. Consequently, the implicit membrane only acts on the protein. At the membrane interface, a thermodynamically stable vapor-liquid interface is formed which prevents water from flooding into the membrane region.
SGLD is used to further enhance protein motions.25 This method has been shown to enhance low-frequency motions and conformational searching of proteins.25; 26; 27 The following SGLD parameters are used in the implicit simulations: local averaging time tL= 0.25 ps, guiding temperature of 1 K, and a friction factor γ = 1 ps−1. The local averaging time enhances all motions with periods larger than tL, while the guiding temperature limits the perturbation to the system. As mentioned in the introduction, this method maintains the correct ensemble averages and fluctuations.25
The implicit bilayer simulations (IM) were performed with three residues (Glu269, His322 and Glu325) separately protonated because of their involvement in the proton translocation mechanism. The initial protein structures for the IM simulations are from certain snapshots of previous explicit membrane (EX) simulations.18 The 5-ns snapshot of EX-wt/ββ-(Galp)2 (run 2) in Klauda and Brooks18 was used for the initial conformations of IM-r1 (run 1 with SGLD), IM-apo (simulation without sugar but using SGLD), IM-322H (simulations with His322 protonated), IM-325H (simulations with Glu325 protonated) and IM-MD (simulation with MD instead of SGLD). The 5-ns snapshot was chosen because it has a slight conformational change in the periplasmic half of LacY compared to the x-ray crystal structure.5 A single disaccharide, ββ-(Galp)2, is used for all but the apo simulation because sugar binding is known to induce structural changes toward the outward-facing state.13 Two different protein coordinates were used as initial structures for the IM-r2 and IM-r3 runs. IM-r2 used a protein snapshot from the EX-wt/ββ-(Galp)2 (run 2) in reference 18 at 12 ns (some conformational change in the periplasmic half) and IM-r3 used the final coordinate of this previous run (no changes in the periplasmic half). Four IM simulations with SGLD were run for 10 ns each and the IM-MD simulation was run for 20 ns. Each simulation consisted of 35,370 total atoms with 9,563 water molecules.
For the second step (EX), two independent simulations were run for each of the two screened conformations from the previous IM step with different initial velocities. All EX simulations were run with a time step of 2 fs. An explicit palmitoyloleoylphosphati-dylethonolamine (POPE) bilayer was used to model the plasma membrane, which is known to be important for the function and topology of LacY.60; 61; 62 Simulations were carried out in the NPAT ensemble at physiological temperature of 310.15 K and an experimentally suggested area of 65.2 Å2/ lipid.63 The details about the simulation time and system size for each run are given in Table 1. The P21 periodic boundary condition64 was used which allows for the redistribution of lipids between the leaflets, which is important for any significant structural change in LacY. For EX-r1, the average number of lipids in the periplasmic leaflet reduced from 142.0±1.7 in the first ns to 133.1±1.9 in the last ns.
Pore radius analysis calculations were done using the program HOLE65 based on 0.5-ns blocks. The distances between all the helices on the periplasmic and cytoplasmic halves of the protein are used as a measure of protein structural change and helix packing. Helix-helix distance maps were generated by calculating average distances between the helix centers on periplasmic and cytoplasmic sides every nanosecond. The helix centers are calculated based on the center of mass of backbone atoms between z=10±1 and −10±1 Å for periplasmic and cytoplasmic sides respectively, as used previously.22 The molecular surface areas are calculated using Lee and Richards algorithm66 that uses a sphere of radius 1.6 Å to probe the molecular surface. Visual molecular dynamics (VMD) was used to make all molecular figures.67
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by University of Maryland startup funds (J.B.K and P.Y.P.) and the Intramural Research Program of the NIH, National Heart, Lung and Blood Institute (B.R.B.). Simulations were run on the High Performance Computational Cluster (HPCC) at the University of Maryland. We also thank Ron Kaback and Vladamir Kosho at UCLA for supplying us with the raw experimental DEER data to compare with our simulations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberts B, Bray D, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Essential Cell Biology: An Introduction to the Molecular Biology of the Cell. New York: Barland Publishing, Inc; 1998. [Google Scholar]
- 2.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobczak I, Lolkema JS. Structural and mechanistic diversity of secondary transporters. Current Opinion in Microbiology. 2005;8:161–167. doi: 10.1016/j.mib.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Milton H, Saier J. Families of transmembrane sugar transport proteins. Molecular Microbiology. 2000;35:699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 5.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 6.Guan L, Kaback HR. Lessons from Lactose Permease. Annual Review of Biophysics and Biomolecular Structure. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abramson J, Iwata S, Kaback HR. Lactose permease as a paradigm for membrane transport proteins - (Review) Molecular Membrane Biology. 2004;21:227–236. doi: 10.1080/09687680410001716862. [DOI] [PubMed] [Google Scholar]
- 8.Huang YF, Lemieux JM, Song MA, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proceedings of the National Academy of Sciences. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaback HR, Dunten R, Frillingos S, Venkatesan P, Kwaw I, Zhang W, Ermolova N. Site-directed alkylation and the alternating access model for LacY. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:491–494. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majumdar DS, Smirnova I, Kasho V, Nir E, Kong XX, Weiss S, Kaback HR. Single-molecular FRET reveals sugar-induced conformational dynamics in LacY. Proceedings from the National Academdy of Science. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smirnova I, Kasho V, Choe J-Y, Altenbach C, Hubbell WL, Kaback HR. Sugar binding induces an outward facing conformation of LacY. Proceedings of the National Academy of Sciences. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie Y, Ermolova N, Kaback HR. Site-directed Alkylation of LacY: Effect of the Proton Electrochemical Gradient. Journal of Molecular Biology. 2007;374:356–364. doi: 10.1016/j.jmb.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Guan L, Freites JA, Kaback HR. Opening and closing of the periplasmic gate in lactose permease. Proceedings from the National Academdy of Science. 2008;105:3774–3778. doi: 10.1073/pnas.0800825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franco PJ, Matzke EA, Johnson JL, Wiczer BM, Brooker RJ. A suppressor analysis of residues involved in cation transport in the lactose permease: Identification of a coupling sensor. Journal of Membrane Biology. 2006;211:101–113. doi: 10.1007/s00232-005-7020-x. [DOI] [PubMed] [Google Scholar]
- 17.Bennett M, D’Rozario R, Sansom MSP, Yeagle PL. Asymmetric stability among the transmembrane helices of lactose permease. Biochemistry. 2006;45:8088–8095. doi: 10.1021/bi060355g. [DOI] [PubMed] [Google Scholar]
- 18.Klauda JB, Brooks BR. Sugar binding in lactose permease: Anomeric state of a disaccharide influences binding structure. Journal of Molecular Biology. 2007;367:1523–1534. doi: 10.1016/j.jmb.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin Y, Jensen MO, Tajkhorshid E, Schulten K. Sugar binding and protein conformational changes in lactose permease. Biophysical Journal. 2006;91:3972–3985. doi: 10.1529/biophysj.106.085993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law CJ, Almqvist J, Bernstein A, Goetz RM, Huang Y, Soudant C, Laaksonen A, Hovmöller S, Wang D-N. Salt-bridge Dynamics Control Substrate-induced Conformational Change in the Membrane Transporter GlpT. Journal of Molecular Biology. 2008;378:826–837. doi: 10.1016/j.jmb.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law CJ, Enkavi G, Wang D-N, Tajkhorshid E. Structural Basis of Substrate Selectivity in the Glycerol-3-Phosphate: Phosphate Antiporter GlpT. 2009;97:1346–1353. doi: 10.1016/j.bpj.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holyoake J, Sansom MSP. Conformational change in an MFS protein: MD simulations of LacY. Structure. 2007;15:873–884. doi: 10.1016/j.str.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Jensen MO, Yin Y, Tajkhorshid E, Schulten K. Sugar Transport across Lactose Permease Probed by Steered Molecular Dynamics. Biophysical Journal. 2007;93:92–102. doi: 10.1529/biophysj.107.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enkavi G, Tajkhorshid E. Simulation of Spontaneous Substrate Binding Revealing the Binding Pathway and Mechanism and Initial Conformational Response of GlpT. Biochemistry. 2010;49:1105–1114. doi: 10.1021/bi901412a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu XW, Brooks BR. Self-guided Langevin dynamics simulation method. Chemical Physics Letters. 2003;381:512–518. [Google Scholar]
- 26.Damjanovic A, Wu X, Garcia-Moreno EB, Brooks BR. Backbone Relaxation Coupled to the Ionization of Internal Groups in Proteins: A Self-Guided Langevin Dynamics Study. Biophysical Journal. 2008;95:4091–4101. doi: 10.1529/biophysj.108.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damjanovic A, García-Moreno EB, Brooks BR. Self-guided Langevin dynamics study of regulatory interactions in NtrC. Proteins: Structure, Function, and Bioinformatics. 2009;76:1007–1019. doi: 10.1002/prot.22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Nie Y, Kaback HR. Residues Gating the Periplasmic Pathway of LacY. Journal of Molecular Biology. 2009;394:219–225. doi: 10.1016/j.jmb.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ermolova NVMR, Kaback HR. Site-Directed Alkylation of Cysteine Replacements in the Lactose Permease of Escherichia coli: Helices I, III, VI and XI. Biochemistry. 2006;45:4182–4189. doi: 10.1021/bi052631h. [DOI] [PubMed] [Google Scholar]
- 30.Frillingos S, Kaback HR. The Role of Helix VIII in the Lactose Permease of Escherichia coli: II. Site-Directed Sulfhydryl Modification. Protein Science. 1997;6:438–443. doi: 10.1002/pro.5560060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwaw I, Zen K, Hu Y, Kaback HR. Site-Directed Sulfhydryl Labeling of the Lactose Permease of Escherichia coli: Helices IV and V That Contain the Major Determinants for Substrate Binding. Biochemistry. 2001;40:10491–10499. doi: 10.1021/bi010866x. [DOI] [PubMed] [Google Scholar]
- 32.Venkatesan P, Hu Y, Kaback HR. Site-Directed Sulfhydryl Labeling of the Lactose Permease of Escherichia coli: Helix X. Biochemistry. 2000;39:10656–10661. doi: 10.1021/bi0004403. [DOI] [PubMed] [Google Scholar]
- 33.Venkatesan P, Kwaw I, Hu Y, Kaback HR. Site-Directed Sulfhydryl Labeling of the Lactose Permease of Escherichia coli: Helix VII. Biochemistry. 2000;39:10641–10648. doi: 10.1021/bi000438b. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Hu Y, Kaback HR. Site-Directed Sulfhydryl Labeling of Helix IX in the Lactose Permease of Escherichia coli. Biochemistry. 2003;42:4904–4908. doi: 10.1021/bi034078e. [DOI] [PubMed] [Google Scholar]
- 35.Venkatesan P, Liu Z, Hu Y, Kaback HR. Site-Directed Sulfhydryl Labeling of the Lactose Permease of Escherichia coli: N-Ethylmaleimide-Sensitive Face of Helix II. Biochemistry. 2000;39:10649–10655. doi: 10.1021/bi0004394. [DOI] [PubMed] [Google Scholar]
- 36.Green AL, Anderson EJ, Brooker RJ. A Revised Model for the Structure and Function of the Lactose Permease. The journal of Biological Chemistry. 2000;275:23240–23246. doi: 10.1074/jbc.M909202199. [DOI] [PubMed] [Google Scholar]
- 37.Green AL, Hrodey HA, Brooker RJ. Evidence for Structural Symmetry and Functional Asymmetry in the Lactose Permease of Escherichia coli. Biochemistry. 2003;42:11226–11233. doi: 10.1021/bi034810+. [DOI] [PubMed] [Google Scholar]
- 38.Nie Y, Sabetfard FE, Kaback HR. The Cys154-->Gly mutation in LacY causes constitutive opening of the hydrophilic periplasmic pathway. Journal of Molecular Biology. 2008;379:695–703. doi: 10.1016/j.jmb.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nie Y, Zhou Y, Kaback HR. Clogging the Periplasmic Pathway in LacY. Biochemistry. 2009;48:738–743. doi: 10.1021/bi801976r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM - a Program for Macromolecular Energy, Minimization, and Dynamics Calculations. Journal of Computational Chemistry. 1983;4:187–217. [Google Scholar]
- 41.Brooks BRCL, Brooks IAD, Mackerell J, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The biomolecular simulation program. Journal of Computational Chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klauda JB, Brooks BR, MacKerell AD, Jr, Venable RM, Pastor RW. An Ab Initio Study on the Torsional Surface of Alkanes and its Effect on Molecular Simulations of Alkanes and a DPPC Bilayer. Journal of Physical Chemistry B. 2005;109:5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 43.Klauda JB, Pastor RW, Brooks BR. Adjacent gauche stabilization in linear alkanes: Implications for polymer models and conformational analysis. Journal of Physical Chemistry B. 2005;109:15684–15686. doi: 10.1021/jp0527608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buck M, Bouguet-Bonnet S, Pastor RW, MacKerell AD. Importance of the CMAP correction to the CHARMM22 protein force field: Dynamics of hen lysozyme. Biophysical Journal. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feig M, MacKerell AD, Jr, Brooks CL. Force field influence on the observation of π-helical protein structures in molecular dynamics simulations. Journal of Physical Chemistry B. 2003;107:2831–2836. [Google Scholar]
- 46.MacKerell AD, Feig M, Brooks CL. Improved treatment of the protein backbone in empirical force fields. Journal of the American Chemical Society. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 47.Kuttel M, Brady JW, Naidoo KJ. Carbohydrate solution simulations: Producing a force field with experimentally consistent primary alcohol rotational frequencies and populations. Journal of Computational Chemistry. 2002;23:1236–1243. doi: 10.1002/jcc.10119. [DOI] [PubMed] [Google Scholar]
- 48.Durell SR, Brooks BR, Bennaim A. Solvent-Induced Forces between Two Hydrophilic Groups. Journal of Physical Chemistry. 1994;98:2198–2202. [Google Scholar]
- 49.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 50.Darden T, York D, Pedersen L. Particle Mesh Ewald - an NLog(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics. 1993;98:10089–10092. [Google Scholar]
- 51.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical Integration of the Cartesian Equations of Motion of a System with Contraints: Molecular Dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 52.Nosé S, Klein ML. A Study of Solid and Liquid Carbon Tetrafluoride Using the Constant Pressure Molecular-Dynamics Technique. Journal of Chemical Physics. 1983;78:6928–6939. [Google Scholar]
- 53.Andersen HC. Molecular-Dynamics Simulations at Constant Pressure and/or Temperature. Journal of Chemical Physics. 1980;72:2384–2393. [Google Scholar]
- 54.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Physical Review A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 55.Khalili-Araghi F, Gumbart J, Wen P-C, Sotomayor M, Tajkhorshid E, Schulten K. Molecular dynamics simulations of membrane channels and transporters. Current Opinion in Structural Biology. 2009;19:128–137. doi: 10.1016/j.sbi.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Im W, Feig M, Brooks CL. An implicit membrane generalized born theory for the study of structure, stability, and interactions of membrane proteins. Biophysical Journal. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanizaki S, Feig M. A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes. Journal of Chemical Physics. 2005;122 doi: 10.1063/1.1865992. [DOI] [PubMed] [Google Scholar]
- 58.Tanizaki S, Feig M. Molecular dynamics simulations of large integral membrane proteins with an implicit membrane model. Journal of Physical Chemistry B. 2006;110:548–556. doi: 10.1021/jp054694f. [DOI] [PubMed] [Google Scholar]
- 59.Edholm O, Jahnig F. The Structure of A Membrane-Spanning Polypeptide Studied by Molecular-Dynamics. Biophysical Chemistry. 1988;30:279–292. doi: 10.1016/0301-4622(88)85023-3. [DOI] [PubMed] [Google Scholar]
- 60.Xie J, Bogdanov M, Heacock P, Dowhan W. Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. Journal of Biological Chemistry. 2006;281:19172–19178. doi: 10.1074/jbc.M602565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang XY, Bogdanov M, Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO Journal. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO Journal. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaikh SR, Brzustowicz MR, Gustafson N, Stillwell W, Wassall SR. Monounsaturated PE does not phase-separate from the lipid raft molecules sphingomyelin and cholesterol: Role for polyunsaturation? Biochemistry. 2002;41:10593–10602. doi: 10.1021/bi025712b. [DOI] [PubMed] [Google Scholar]
- 64.Dolan EA, Venable RM, Pastor RW, Brooks BR. Simulations of membranes and other interfacial systems using P21 and pc periodic boundary conditions. Biophysical Journal. 2002;82:2317–2325. doi: 10.1016/S0006-3495(02)75577-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smart OS, Goodfellow JM, Wallace BA. The Pore Dimensions of Gramicidin A. Biophysical Journal. 1999;65:2455. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee B, Richards FM. The interpretation of protein structures: Estimation of static accessibility. Journal of Molecular Biology. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 67.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.