Abstract
Hepatocyte nuclear factor 4α (HNF4α) is a highly conserved member of the nuclear receptor superfamily of ligand-dependent transcription factors. It is best known as a master regulator of liver-specific gene expression, especially those genes involved in lipid transport and glucose metabolism. However, there is also a growing body of work that indicates the importance of HNF4α in the regulation of genes involved in xenobiotic and drug metabolism. A recent study identifying the essential fatty acid linoleic acid (LA, C18:2) as the endogenous, reversible ligand for HNF4α suggests that HNF4α may also be a potential drug target and that its activity may be regulated by diet. This review will discuss the role of HNF4α in drug metabolism, including the genes it regulates, the factors that regulate its activity, and its potential as a drug target.
Introduction
HNF4α (NR2A1), a highly conserved member of the nuclear receptor superfamily (NR) of ligand-dependent transcription factors (TFs), is known as a master regulator of liver-specific gene expression [1]. It was originally identified as an activity in rat liver that bound the APOC3 gene and characterized as an orphan receptor as its ligand status was unknown [2]. It was subsequently identified as the gene mutated in Maturity Onset Diabetes of the Young 1 (MODY1), an inheritable form of non-insulin-dependent diabetes [3]. Therefore, HNF4α was initially known for its role in carbohydrate and lipid metabolism in the liver and its role in insulin signaling in the pancreas. Subsequently, cytochrome P450 (CYP) genes involved in xenobiotic and drug metabolism were identified as targets of HNF4α [4], although its role in those processes was eclipsed by that of ligand receptors. The recent identification of the endogenous ligand for HNF4α, as well as several genome-wide studies that have greatly expanded the repertoire of HNF4α targets, has renewed interest in the role of HNF4α in drug metabolism. In this review, we will discuss the key findings on this topic and how they relate to the notion of targeting HNF4α for drug discovery.
Background on drug metabolism and HNF4α
The detoxification of xenobiotics and metabolism of drugs occurs primarily in the liver and consists of two phases. Phase I is carried out by hundreds of cytochrome P450 enzymes (CYP450s) and a handful of flavin-containing monooxygenases (FMOs) that add or expose a polar functional group to lipophilic compounds. Phase II, which consists of conjugation reactions that help eliminate Phase I products from the body, is carried out by glutathione-S-transferases (GSTs), UDP-glucuronosyltransferase (UGTs) and steroid-and bile acid-sulfotransferases (SULTs). In addition to acting on exogenous compounds, the Phase I and Phase II enzymes also act on endogenous compounds, such as steroids, bile acids and fatty acids, as well as drugs.
The expression of Phase I and Phase II genes is influenced by a wide variety of factors, including species, gender, age, diet, genetics and the environment [5]. Not surprisingly, many of the proteins responsible for turning on the expression of the Phase I and Phase II genes are TFs that are highly enriched in the liver. Some of those factors were originally identified as hepatocyte nuclear factors (HNFs) (e.g., HNF1, HNF2 (C/EBPα), HNF3 (now FOXA), HNF6) while several other factors belong to the NR superfamily. HNF4α belongs to both groups.
The human HNF4α gene consists of 12+ exons and two promoters and gives rise to many splice variants/isoforms [1]. The proximal P1 promoter is active in the adult liver, kidney and intestine, where most of the xenobiotic/drug metabolism occurs, and gives rise to two major isoforms -- HNF4α1 and HNF4α2. Recent genome-wide location analyses of HNF4α and other liver-enriched TFs (HNF1α, HNF6 and C/EBPα), showed that in hepatocytes HNF4α is much more often found bound to the promoters of 1iver-specific genes than other HNFs [6,7]. Therefore, since one of the main functions of the liver is detoxification of foreign compounds, the notion that HNF4α plays a major role in drug metabolism is a reasonable one.
Phase I and Phase II genes regulated by HNF4α
Phase I and Phase II target genes – identification by classical means
Several CYP450 genes have been identified as HNF4α targets using classical means (e.g., promoter cloning, gel shift analysis, reporter assays, etc.) and have been reviewed previously [8] (Figure 1). Among the Phase I enzymes, CYP450 3A4 (CYP3A4) is arguably one of the most important as it is involved in the metabolism of nearly half of all drugs currently used. Early studies on CYP3A4 gene regulation focused on ligand-activated NRs, pregnane X receptor (PXR; NR1I2) and constitutive androstane receptor (CAR; NR1I3) [9,10]. Subsequently, it was found that HNF4α is required for both PXR-and CAR-mediated transcriptional activation of CYP3A4 [11]. Tirona et al. examined a variety of NRs, including farnesoid X receptor (FXR, NR1H4), liver receptor homolog-1 (LRH1, NR5A2), liver X receptor α (LXR1, NR1H3), retinoid X receptor α (RXRα, NR2B1), and found HNF4α to have the greatest effect on the CYP3A4 promoter. Furthermore, when an HNF4α binding site in the CYP3A4 enhancer region was mutated, the ability of PXR and CAR to activate the CYP3A4 promoter was eliminated [11]. Thus, HNF4α plays a critical and central role in the regulation of this key Phase I gene.
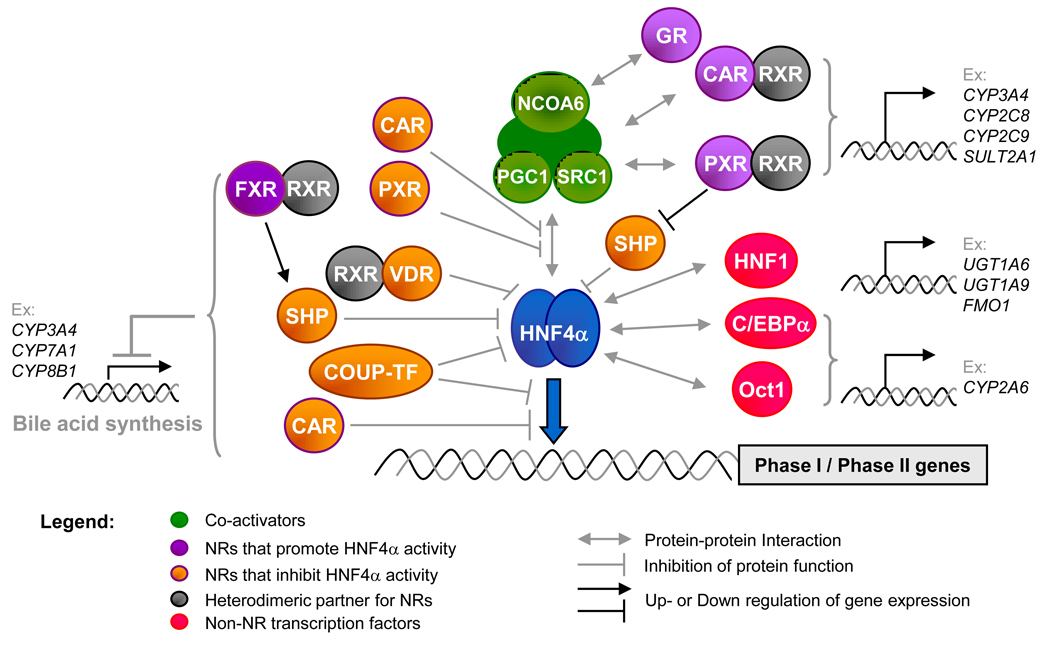
Figure 1. Central role of HNF4α in the regulation of Phase I and Phase II genes.
Depicted are the NRs and liver-enriched transcription factors known to interact with HNF4α protein (an obligate homodimer) either physically and/or functionally to regulate the various Phase I/II genes shown. See Table 1 for a complete list of potential Phase I/II targets of HNF4α. See Figure 2 for additional interactions on the level of gene expression. See main text for references and additional detail.
HNF4α has been shown to activate the expression of several other Phase I genes, including CYP2C8, CYP2C9, CYP2C19, CYP7A1 and CYP8B1 [8,12]. Knockdown of HNF4α by antisense and siRNA in primary human hepatocytes also identified CYP2A6, CYP2B6, CYP2D6, CYP3A4 and CYP3A5 as targets of HNF4α [13,14]. Classical techniques have also shown that HNF4α can bind and activate FMO1 promoter activity in concert with HNF1α [15].
In terms of Phase II genes, HNF4α also plays an important role in regulating the basal SULT2A1 promoter, as well as synergizing with PXR and CAR [16]. The expression of the UGT1A6 and UGT1A9 genes were found to be positively correlated with HNF4α and HNF1α expression in human liver [17] and functional HNF4α and HNF1α binding sites were identified in the promoter regions of both UGT genes using several classical techniques [17–20]. Several transporters that are critical for the elimination of drugs and toxicants (ABCB1, ABCB11, ABCC2, SLCO1B1, SLC22A1), have also been found to be decreased when HNF4α is knocked down [14].
Phase I and Phase II target genes – identification by genomic approaches
The sequencing of the human genome has led to the use of genome-wide approaches that are more high throughput and less biased than the classical approach to target gene identification. Chief among those techniques are genome-wide location analysis (ChIP-chip/seq) and expression profiling. Another less well known but highly complementary technique is protein binding microarrays (PBMs) [21]. PBMs are a high throughput in vitro DNA binding assay that allows for the identification of 1000’s of distinct DNA binding sequences in a given experiment. We recently modified the PBMs to characterize the DNA binding specificity of native, full length HNF4α in crude nuclear extracts [22]. This led to the identification of >1400 new binding sequences for HNF4α that we then used to search the regulatory regions of human genes to identify potential new targets of HNF4α. The PBM data were also used to train a Support Vector Machine (SVM) algorithm to predict additional HNF4α binding sites with high accuracy. Finally, cross referencing with expression profiling data from an HNF4α RNAi knockdown in a human liver cancer cell line (HepG2) and published HNF4α ChIP-chip data identified >240 new direct, functional targets of HNF4α [22]. The results for the Phase I and Phase II genes are given in Table 1, along with target genes identified by classical methods. All told there are at least 50 CYP, 7 FMO, 21 GST, 13 SULT and 19 UGT (~110 total) human genes involved in drug metabolism that are predicted or proven targets of HNF4α.
Table 1.
Verified and predicted HNF4α Phase I and Phase II human target genes1
| Family | Gene Symbol |
Family | Gene Symbol |
Family | Gene Symbol |
Family | Gene Symbol |
|---|---|---|---|---|---|---|---|
|
Phase I |
Phase II |
||||||
| CYP450 | CYP1A1* | CYP450 | CYP7A1 | GST | GSTA1 | SULT | SULT1A1 |
| CYP1A2 | (cont) | CYP7B1* | GSTA2* | SULT1A2 | |||
| CYP1B1* | CYP8B1 | GSTA3* | SULT1A3 | ||||
| CYP2A6 | CYP11A1* | GSTA4* | SULT1A4 | ||||
| CYP2A7* | CYP11B1* | GSTA5* | SULT1C1 | ||||
| CYP2A13* | CYP11B2* | GSTCD* | SULT1C2* | ||||
| CYP2B6 | CYP17A1* | GSTK1* | SULT1C3* | ||||
| CYP2C8 | CYP19A1* | GSTM1 | SULT1C4* | ||||
| CYP2C9 | CYP20A1* | GSTM2* | SULT1E1 | ||||
| CYP2C19 | CYP21A2* | GSTM1L* | SULT2A1 | ||||
| CYP2D6 | CYP24A1 | GSTM4 | SULT2B1* | ||||
| CYP2J2* | CYP26A1* | GSTM5 | SULT4A1* | ||||
| CYP2R1* | CYP27A1* | GSTO1* | SULT6B1* | ||||
| CYP2S1* | CYP27B1* | GSTO2* | |||||
| CYP3A4 | CYP27C1* | GSTP1* | UGT | UGT1A1 | |||
| CYP3A5 | CYP27E1* | GSTP2* | UGT1A3 | ||||
| CYP3A7 | CYP27F1* | GSTT1* | UGT1A4 | ||||
| CYP3A43* | CYP39A1* | GSTT2* | UGT1A5 | ||||
| CYP4A11* | CYP46A1* | GSTT2B* | UGT1A6 | ||||
| CYP4A22* | CYP51A1* | GSTTP1* | UGT1A7 | ||||
| CYP4B1* | GSTZ1 | UGT1A8 | |||||
| CYP4F2* | FMO | FMO1 | UGT1A9 | ||||
| CYP4F3* | FMO2* | UGT1A10 | |||||
| CYP4F8* | FMO3* | UGT2A1* | |||||
| CYP4F11* | FMO4* | UGT2A2* | |||||
| CYP4F12 | FMO5 | UGT2A3 | |||||
| CYP4F22* | FMO6P* | UGT2B4* | |||||
| CYP4V2* | UGT2B7* | ||||||
| CYP4X1* | UGT2B10* | ||||||
| CYP4Z1* | UGT2B11* | ||||||
| UGT2B28* | |||||||
| UGT3A1* | |||||||
| UGT8* | |||||||
Target genes identified by both classical promoter analysis (see text for references) and genome-wide techniques [22]: see Supplemental Tables S7A and S7B for PBM and SVM-predicted HNF4α binding sites in −2kb to +1 kb relative to the transcription start site (+1), including location and sequence, and Table S3A for expression profiling data of HNF4α RNAi in HepG2 cells [22]. See HNF4 Motif Finder (http://nrmotif.ucr.edu/fuzzhtmlform.html) to identify additional sites.
Predicted targets for which an HNF4α binding site has been identified in −2kb to +1 kb but for which no functional data (e.g., reporter assays, expression profiling) are currently available. These genes require additional verification. Genes without an asterisk have been verified by expression profiling, RT-PCR [14,22,36] or by classical approaches (see text for references). Not listed are transporters (SLC genes) or pumps (ABC) that also play a role in toxicant/drug elimination (see [14; 22]).
Factors that influence HNF4α-mediated regulation of drug metabolism genes
Transcriptional regulation network
Many NRs such as PXR, CAR, LXR, FXR, small heterodimer partner (SHP, NR0B2), Vitamin D receptor (VDR, NR1I1), chicken ovalbumin upstream promoter transcription factor (COUP-TF, NR2F1) and glucocorticoid receptor (GR, NR3C1) interact with HNF4α to regulate the expression of drug metabolism genes in a complex fashion (Figure 1). For example, HNF4α is involved in crosstalk with PXR and CAR on the promoters of the CYP3A4 [23], CYP2C8 [24], CYP2C9 [25], CYP7A1 [26,27] and SULT2A1 [28] genes. While for several of these genes the crosstalk has not been characterized beyond requirements for specific NRs and their respective binding sites, on two genes – CYP3A4 and CYP7A1 -- the mechanisms have been explored. Ligand (rifampacin)-activated PXR recruits HNF4α and co-activator SRC-1 to the CYP3A4 promoter, thereby increasing its expression [23]. HNF4α is one of the few NRs able to activate the human CYP7A1 gene, a key player in bile acid synthesis (see [26]), but its ability to do so is negatively modulated by other NRs. In contrast to CYP3A4, a direct physical interaction between HNF4α and PXR, that is enhanced by rifampacin, displaces the co-activator PGC1α from HNF4α resulting in down regulation of CYP7A1 (and CYP8B1) gene expression [26,29]. Likewise, on CYP7A1, CAR competes with HNF4α for binding both DNA response elements and co-activators GRIP1 and PGC1α, thereby repressing the ability of HNF4α to activate this gene [27]. The VDR also interacts directly with HNF4α to suppress CYP7A1 expression [30]. High levels of SHP, a NR that lacks a DNA binding domain, represses CYP3A4 and CYP7A1 gene expression by interacting with HNF4α and PXR [23,31]. In contrast, rifampacin-activated PXR suppresses SHP, allowing for the crosstalk between HNF4α, PXR and coactivators (PGC1, SRC1) to maximally induce CYP3A4 [23]. FXR also suppresses CYP7A1 through up regulation of SHP. However, down regulation of CYP7A1 reduces bile acid synthesis and subsequently hepatic JNK activity that in turn increases HNF4α expression [32,33]. Interestingly, HNF4α not only co-regulates target genes with PXR, CAR and SHP but it also regulates their expression [14,22,23,34–36]. FXR gene is also direct target of HNF4α while FXR protein can indirectly up regulate the expression of the HNF4A gene [22,32] (Figure 2A). Finally, on CYP2C9, HNF4α works with GR and NR coactivator 6 (NCOA6) to mediate maximum induction of the gene [37]. It is anticipated that similar complex networks involving multiple NRs will be found on one or more of the other HNF4α-regulated Phase I and Phase I genes in Table 1.
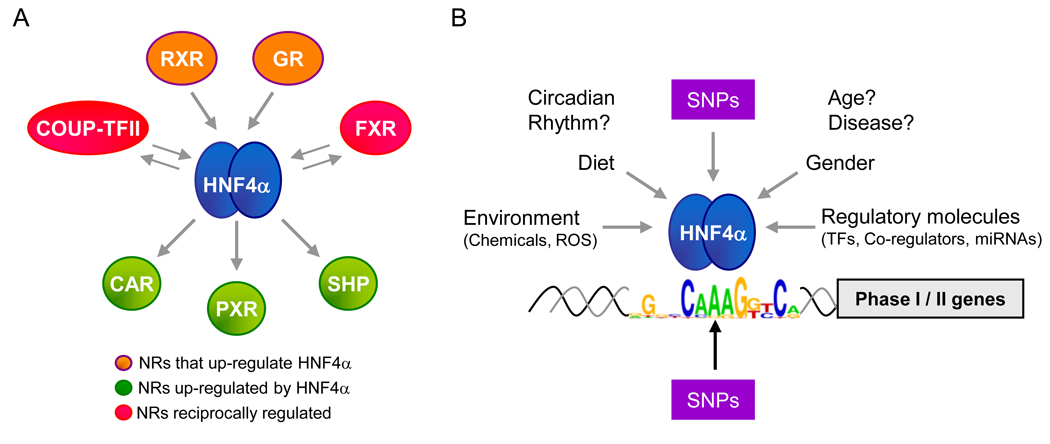
Figure 2. Factors affecting HNF4α expression and function.
A. The HNF4A gene is at the center of a complex transcriptional network of NR genes that are involved in xenobiotic and drug metabolism. All interactions are on the HNF4α P1 promoter except COUP-TF2 that acts on both the P1 and P2 promoter. See main text and [1] for references.
B. Shown are broad categories of factors known to regulate the activity and level of HNF4α that could ultimately affect the expression of Phase I and Phase II genes and hence alter one’s ability to metabolize drugs. SNPs in the HNF4A gene as well as in the response elements bound by HNF4α protein could also account for some of the variability in drug metabolism among individuals. Shown is the position weight matrix for strong binders from [22]. ROS, reactive oxygen species. ?, effects not proven but are likely. See main text for references and additional detail.
In addition to protein-protein and protein-promoter interactions between HNF4α and other NRs, there is also direct competition for binding to common response elements between HNF4α and the COUP-TFs, orphan NRs that tend to repress transcription. Both COUP-TFs and HNF4α bind DNA via direct repeats of a half site AGGTCA with a spacing of 1–2 nucleotides (DR1s and DR2s, respectively) and thus compete for regulation of the rat Cyp2C and human CYP2D6 genes. COUP-TFs also repress the strong transactivation function of HNF4α by binding additional response elements on the rat Cyp3a1 and Cyp3a23 promoters. In humans, COUP-TF2 inhibits HNF4α transactivation activity on the CYP3A4 promoter via both the DNA binding-dependent and –independent mechanisms [8] (Figure 1). Interestingly, HNF4α and COUP-TF2 can transactivate the expression of each others’ expression [38] (Figure 2A). Crosstalk between HNF4α and other TFs, such as C/EBPα and Oct-1, has also been observed on CYP2A6 [8]. More recently, two microRNAs have been identified that down regulate the expression of HNF4α (miR-24 and miR-34a). These miRNAs are up-regulated by reactive oxygen species (ROS) and phorbol ester (PMA), both of which down regulate HNF4α protein levels [39] (Figure 2B).
Genetics
Mutations in HNF4α response elements contribute to diseases such as hemophilia and MODY3 [4], while mutations in the HNF4α P2 promoter result in MODY1 [3]. Likewise an altered HNF4α binding site in a drug metabolism gene promoter could result in dysregulation of that gene and nucleotide changes in the HNF4α P1 promoter could result in altered levels of HNF4α1/2 protein in the liver, kidney and intestine, which would in turn affect Phase I and Phase II gene expression (Figure 2B). Indeed, a single nucleotide polymorphism (SNP) at a putative HNF4α binding site in the CYP2B6 promoter is able to alter the level of expression of that gene [40]. A SNP has also been identified in the HNF4α coding region that affects expression of the CYP2D6 gene that metabolizes a wide variety of drugs including beta blockers for heart disease and Prozacs and other drugs for mental disorders [41]. Numerous genetic variations resulting in altered levels of Cyp2D6 have also been associated with altered metabolism of 4-hydroxy tamoxifen and responsiveness to treatment for breast cancer [42]. While it is not known if HNF4α is directly involved in those effects, it is possible that HNF4α activity in the liver could affect one’s response to chemotherapeutic agents such as tamoxifen via Cyp2D6. Therefore, it will be of great interest to identify SNPs in HNF4α response elements in other Phase I and Phase II promoters, and to further characterize SNPs in the promoter and coding region of the HNF4A gene itself (Figure 2B).
Gender
In humans, it is estimated that there is a 40% difference in pharmacokinetics between men and women [43]. Much of this difference could be due to differential expression of drug metabolism genes between genders. CYP3A4, for example, is expressed at a higher level in women than in men [44], resulting in higher clearance rates of CYP3A4-targeted xenobiotics in women [45]. While these differences in human have not yet been attributed to HNF4α, in rodents HNF4α has been found to be a key player in gender-specific gene expression in liver and to be involved in crosstalk with STAT5b, a key intermediary in the growth hormone-mediated sexual dimorphism seen in hepatic gene expression [46,47]. An additional layer of complexity is achieved by the fact that STAT5b and HNF4α also co-regulate the expression of TFs that regulate gender-specific Cyp genes. For example, in response to growth hormone stimulation, STAT5b and HNF4α bind and activate the promoter of HNF6 (Onecut1) [48], a female-predominant transcription factor [49] that positively regulates the female-specific gene Cyp2c12 [50,51].
Environmental factors
The liver is exposed to a wide variety of toxic agents, many of which damage DNA and result in increased levels of the tumor suppressor protein p53. We previously showed that p53 inhibits the transactivation function of HNF4α1 [52] and suppresses the HNF4α P1 promoter in human liver cells exposed to doxorubicin [53]. Other cellular stress conditions known to up regulate p53 expression, such as hypoxia [54] and arsenic trioxide [55], also down regulate the expression of HNF4α, and hence presumably affect Phase I and Phase II gene expression. We have also shown that protein kinase C (PKC) phosphorylates the DNA binding domain (DBD) of HNF4α, thereby inhibiting its ability to bind DNA and increasing its degradation. Importantly, the PKC site in the HNF4α DBD is completely conserved in all non steroid NRs, suggesting that other NRs such as CAR, RXR, FXR and COUP-TF, could also be affected by PKC activators [56]. This suggests that compounds such as phorbol esters that activate PKC could also alter the regulation of drug metabolism enzymes via HNF4α and other NRs. Finally, while HNF4α mRNA levels do not appear to be affected by the circadian clock, as are many other NR mRNAs [57], HNF4α protein does interact with Period (PER), a key component of the circadian clock regulatory pathway [58]. This opens the possibility that HNF4α activity may also be affected by the light/dark cycle.
Diet and the HNF4α Ligand
Diet can influence the levels and hence activity of HNF4α in several ways. HNF4α protein and mRNA levels are up regulated in the liver during fasting [59], as is the potent HNF4α co-activator PGC1α [60]. The effect on HNF4α is due in part to increased levels of insulin after feeding, which up regulates the sterol response element binding proteins SREBPs which in turn down regulate the expression of the HNF4α P1 promoter [59]. The energy/metabolic sensing kinase AMPK also phosphorylates both HNF4α and PGC1α thereby altering their activity [61,62]. Finally, bile acids, as a product of dietary metabolism, alter HNF4α expression in a complex fashion [32,33]. Hence, nutritional status can be expected to affect the transcription of many Phase I and Phase II genes via HNF4α.
One of the most controversial issues regarding HNF4α, and other orphan NRs, has been whether they respond to ligands [63]. HNF4α was known as a constitutively active NR that did not require the addition of exogenous ligand in order to activate transcription [2,64], but the identity of its endogenous ligand remained elusive. The crystallographic structures of bacterially expressed HNF4 showed that it did indeed contain a hydrophobic ligand binding pocket that could accommodate lipophilic compounds [65,66]. However, since those compounds, a mixture of fatty acids, bound HNF4 in an irreversible fashion, the ligand status and potential for HNF4 as a drug target remained in doubt. These issues were resolved once the endogenous HNF4α ligand expressed in the appropriate physiological context – mammalian tissues and cells – was identified. Using an unbiased and highly sensitive approach (affinity isolation followed by mass spectrometry, AIMS), linoleic acid (LA, C18:2\ω6) was found bound to HNF4α in the livers of fed but not fasted mice [36]. This made physiological sense as LA is an essential polyunsaturated fatty acid that must be obtained from the diet. Furthermore, it was shown that LA binds mammalian-expressed HNF4α in a reversible fashion, suggesting that it could be available for drug targeting. While a specific transactivation function of LA on HNF4α, was not observed, a modest but consistent decrease in HNF4α target gene expression in the presence of LA that was associated with a decrease in HNF4α protein levels was noted [36]. These findings raise the intriguing possibility that different diets containing different amounts of LA could affect HNF4α levels, and hence drug metabolism, although this remains to be proven in vivo.
HNF4α as a drug target?
While the function of the endogenous HNF4α ligand remains uncertain, what is clear is that HNF4α contains a ligand binding pocket that is occupied in a reversible fashion dependent upon the feeding state of the host. Hence, it is possible that one could design a drug that could bind in the pocket of HNF4α and alter its ability to activate transcription. However, since HNF4α regulates so many Phase I/II and other genes in the liver, there is a high likelihood of side effects, unless one can develop a drug that is specific to a given HNF4α target. But this is not a new problem, nor is it unique to HNF4α. Any NR or TF with multiple targets faces similar problems. The trick will be to develop drugs that are specific to HNF4α on one or at most a few genes. This specificity could be found in the DNA sequence of the response elements that we now know can vary greatly between different target genes [22].
Perspectives
It is now evident that HNF4α is a key player in the regulation of genes involved in drug metabolism. Indeed, since HNF4α is one of the most ancient of the NRs and since Cyp genes have equally ancient evolutionary origins [63], it is possible that HNF4α was the first NR to control the expression of this critical gene family in animals. The regulation of these genes, however, has evolved, just as the genes themselves. In addition to HNF4α, that regulation now involves many additional NR, and other TFs, in a complex network of synergy, inhibition and mutual regulation that is affected by a variety of internal factors such as SNPs and external factors such as diet and environmental stress. Detailed knowledge of this network and the factors that influence it is essential for understanding individual responses to drug treatment.
Acknowledgements
We apologize to all of our colleagues whose work we were not able to cite due to space limitations. WH-V is funded by a postdoctoral fellowship from Academia Sinica. The Sladek lab is funded by grants from the National Institutes of Health (DK053892 and MH087397).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1. Bolotin E, Schnabl J, Sladek FM. HNF4A (Homo sapiens) [Accessed August 14, 2010];Transcription Factor Encyclopedia. (Updated August 14, 2010) http://www.cisreg.ca/tfe. This continually updated, on-line review provides an overview of the most important aspects of HNF4α as well as comprehensive lists of target genes and interacting proteins with direct links to the literature.
- 2.Sladek FM, Zhong WM, Lai E, Darnell JE., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 3.Ellard S, Colclough K. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Hum Mutat. 2006;27:854–869. doi: 10.1002/humu.20357. [DOI] [PubMed] [Google Scholar]
- 4.Sladek FM, Seidel SD. Hepatocyte Nuclear Factor 4α. In: Burris TP, McCabe E, editors. Nuclear Receptors and Genetic Diseases. Academic Press; 2001. pp. 309–361. [Google Scholar]
- 5.Stine KE, Brown TM. Principles of Toxicology. edn 2nd. Boca Raton: CRC Press, Taylor & Francis Group, LLC; 2006. [Google Scholar]
- 6.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. In this study the authors follow up on their previous ChIP-chip work with ChIP-seq for HNF4α and C/EBPα in the livers of five vertebrates, including man. The results provide a global view of potential HNF4α and C/EBPα target genes and is an invaluable resource.
- 8. Jover R, Moya M, Gomez-Lechon MJ. Transcriptional regulation of cytochrome p450 genes by the nuclear receptor hepatocyte nuclear factor 4-alpha. Curr Drug Metab. 2009;10:508–519. doi: 10.2174/138920009788898000. An excellent, very detailed review of the role of HNF4α and other NRs in the regulation of several key CYP450 genes.
- 9.Goodwin B, Hodgson E, D'Costa DJ, Robertson GR, Liddle C. Transcriptional regulation of the human CYP3A4 gene by the constitutive androstane receptor. Mol Pharmacol. 2002;62:359–365. doi: 10.1124/mol.62.2.359. [DOI] [PubMed] [Google Scholar]
- 10.Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- 11.Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 12.Kawashima S, Kobayashi K, Takama K, Higuchi T, Furihata T, Hosokawa M, Chiba K. Involvement of hepatocyte nuclear factor 4alpha in the different expression level between CYP2C9 and CYP2C19 in the human liver. Drug Metab Dispos. 2006;34:1012–1018. doi: 10.1124/dmd.106.009365. [DOI] [PubMed] [Google Scholar]
- 13.Jover R, Bort R, Gomez-Lechon MJ, Castell JV. Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology. 2001;33:668–675. doi: 10.1053/jhep.2001.22176. [DOI] [PubMed] [Google Scholar]
- 14.Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. Role of human hepatocyte nuclear factor 4alpha in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab Pharmacokinet. 2007;22:287–298. doi: 10.2133/dmpk.22.287. [DOI] [PubMed] [Google Scholar]
- 15.Luo Z, Hines RN. Regulation of flavin-containing monooxygenase 1 expression by ying yang 1 and hepatic nuclear factors 1 and 4. Mol Pharmacol. 2001;60:1421–1430. doi: 10.1124/mol.60.6.1421. [DOI] [PubMed] [Google Scholar]
- 16.Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B. The xenobiotic-sensing nuclear receptors pregnane X receptor, constitutive androstane receptor, and orphan nuclear receptor hepatocyte nuclear factor 4alpha in the regulation of human steroid-/bile acid-sulfotransferase. Mol Endocrinol. 2007;21:2099–2111. doi: 10.1210/me.2007-0002. [DOI] [PubMed] [Google Scholar]
- 17.Aueviriyavit S, Furihata T, Morimoto K, Kobayashi K, Chiba K. Hepatocyte nuclear factor 1 alpha and 4 alpha are factors involved in interindividual variability in the expression of UGT1A6 and UGT1A9 but not UGT1A1, UGT1A3 and UGT1A4 mRNA in human livers. Drug Metab Pharmacokinet. 2007;22:391–398. doi: 10.2133/dmpk.22.391. [DOI] [PubMed] [Google Scholar]
- 18.Auyeung DJ, Kessler FK, Ritter JK. Differential regulation of alternate UDP-glucuronosyltransferase 1A6 gene promoters by hepatic nuclear factor-1. Toxicol Appl Pharmacol. 2003;191:156–166. doi: 10.1016/s0041-008x(03)00230-8. [DOI] [PubMed] [Google Scholar]
- 19.Barbier O, Girard H, Inoue Y, Duez H, Villeneuve L, Kamiya A, Fruchart JC, Guillemette C, Gonzalez FJ, Staels B. Hepatic expression of the UGT1A9 gene is governed by hepatocyte nuclear factor 4alpha. Mol Pharmacol. 2005;67:241–249. doi: 10.1124/mol.104.003863. [DOI] [PubMed] [Google Scholar]
- 20.Gardner-Stephen DA, Mackenzie PI. Hepatocyte nuclear factor1 transcription factors are essential for the UDP-glucuronosyltransferase 1A9 promoter response to hepatocyte nuclear factor 4alpha. Pharmacogenet Genomics. 2007;17:25–36. doi: 10.1097/FPC.0b013e32801112b5. [DOI] [PubMed] [Google Scholar]
- 21.Bulyk ML. Analysis of sequence specificities of DNA-binding proteins with protein binding microarrays. Methods Enzymol. 2006;410:279–299. doi: 10.1016/S0076-6879(06)10013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. This is the first application of PBMs to any TF under physiologically relevant conditions, and the first integration of PBM data with expression profiling and ChIP-chip data. Extremely useful are the extensive supplementary data and a web-based tool (HNF4 Motif Finder) to identify potential HNF4α binding sites in any DNA sequence (http://nrmotif.ucr.edu/fuzzhtmlform.html).
- 23.Li T, Chiang JY. Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4alpha and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab Dispos. 2006;34:756–764. doi: 10.1124/dmd.105.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4alpha. Mol Pharmacol. 2005;68:747–757. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Kissling G, Negishi M, Goldstein JA. The nuclear receptors constitutive androstane receptor and pregnane X receptor cross-talk with hepatic nuclear factor 4alpha to synergistically activate the human CYP2C9 promoter. J Pharmacol Exp Ther. 2005;314:1125–1133. doi: 10.1124/jpet.105.087072. [DOI] [PubMed] [Google Scholar]
- 26.Li T, Chiang JY. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol. 2005;288:G74–G84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- 27.Miao J, Fang S, Bae Y, Kemper JK. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J Biol Chem. 2006;281:14537–14546. doi: 10.1074/jbc.M510713200. [DOI] [PubMed] [Google Scholar]
- 28.Fang HL, Strom SC, Ellis E, Duanmu Z, Fu J, Duniec-Dmuchowski Z, Falany CN, Falany JL, Kocarek TA, Runge-Morris M. Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4alpha and pregnane X receptor. J Pharmacol Exp Ther. 2007;323:586–598. doi: 10.1124/jpet.107.124610. [DOI] [PubMed] [Google Scholar]
- 29.Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem. 2004;279:45139–45147. doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- 30.Han S, Chiang JY. Mechanism of vitamin D receptor inhibition of cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes. Drug Metab Dispos. 2009;37:469–478. doi: 10.1124/dmd.108.025155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ourlin JC, Lasserre F, Pineau T, Fabre JM, Sa-Cunha A, Maurel P, Vilarem MJ, Pascussi JM. The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol Endocrinol. 2003;17:1693–1703. doi: 10.1210/me.2002-0383. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Yin L, Anderson J, Ma H, Gonzalez FJ, Willson TM, Edwards PA. Identification of novel pathways that control farnesoid X receptor-mediated hypocholesterolemia. J Biol Chem. 2010;285:3035–3043. doi: 10.1074/jbc.M109.083899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiang JY. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J Hepatol. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Ding X, Lichti K, Kim I, Gonzalez FJ, Staudinger JL. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Biol Chem. 2006;281:26540–26551. doi: 10.1074/jbc.M600931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamiya A, Inoue Y, Gonzalez FJ. Role of the hepatocyte nuclear factor 4alpha in control of the pregnane X receptor during fetal liver development. Hepatology. 2003;37:1375–1384. doi: 10.1053/jhep.2003.50212. [DOI] [PubMed] [Google Scholar]
- 36. Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. This is the first identification of an endogenous ligand bound to any endogenous NR in a mammalian tissue. It also documents that functional apoHNF4α exists under physiological conditions and that ligand binding is reversible, indicating that HNF4α could be a potential drug target.
- 37.Surapureddi S, Rana R, Reddy JK, Goldstein JA. Nuclear receptor coactivator 6 mediates the synergistic activation of human cytochrome P-450 2C9 by the constitutive androstane receptor and hepatic nuclear factor-4alpha. Mol Pharmacol. 2008;74:913–923. doi: 10.1124/mol.108.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perilhou A, Tourrel-Cuzin C, Zhang P, Kharroubi I, Wang H, Fauveau V, Scott DK, Wollheim CB, Vasseur-Cognet M. The MODY1 gene for hepatocyte nuclear factor 4alpha and a feedback loop control COUP-TFII expression in pancreatic beta cells. Mol Cell Biol. 2008;28:4588–4597. doi: 10.1128/MCB.01191-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285:4415–4422. doi: 10.1074/jbc.M109.085431. This is the first report of miRNAs regulating HNF4α expression and sets the stage for these important regulatory molecules to play a role in drug metabolism.
- 40.Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, Fackenthal JD, Rogan PK, Ring B, Wrighton SA, et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther. 2003;307:906–922. doi: 10.1124/jpet.103.054866. [DOI] [PubMed] [Google Scholar]
- 41. Lee SS, Cha EY, Jung HJ, Shon JH, Kim EY, Yeo CW, Shin JG. Genetic polymorphism of hepatocyte nuclear factor-4alpha influences human cytochrome P450 2D6 activity. Hepatology. 2008;48:635–645. doi: 10.1002/hep.22396. This work documents for the first time that a SNP in HNF4α affects its ability to regulate the expression of a Phase I/II gene.
- 42. Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer. 2009;9:576–586. doi: 10.1038/nrc2683. This excellent review on tamoxifen metabolism includes many of the CYP genes mentioned here as well as information about functional SNPs in the CYP2D6 gene. It provides strong support for the notion that SNPs play a role in an individual's response to drugs.
- 43.Anderson GD. Sex and racial differences in pharmacological response: where is the evidence? Pharmacogenetics, pharmacokinetics, and pharmacodynamics. J Womens Health (Larchmt) 2005;14:19–29. doi: 10.1089/jwh.2005.14.19. [DOI] [PubMed] [Google Scholar]
- 44.Diczfalusy U, Miura J, Roh HK, Mirghani RA, Sayi J, Larsson H, Bodin KG, Allqvist A, Jande M, Kim JW, et al. 4Beta-hydroxycholesterol is a new endogenous CYP3A marker: relationship to CYP3A5 genotype, quinine 3-hydroxylation and sex in Koreans, Swedes and Tanzanians. Pharmacogenet Genomics. 2008;18:201–208. doi: 10.1097/FPC.0b013e3282f50ee9. [DOI] [PubMed] [Google Scholar]
- 45.Krecic-Shepard ME, Park K, Barnas C, Slimko J, Kerwin DR, Schwartz JB. Race and sex influence clearance of nifedipine: results of a population study. Clin Pharmacol Ther. 2000;68:130–142. doi: 10.1067/mcp.2000.108678. [DOI] [PubMed] [Google Scholar]
- 46. Holloway MG, Miles GD, Dombkowski AA, Waxman DJ. Liver-specific hepatocyte nuclear factor-4alpha deficiency: greater impact on gene expression in male than in female mouse liver. Mol Endocrinol. 2008;22:1274–1286. doi: 10.1210/me.2007-0564. The authors use expression profiling to document for the first time differences in liver gene expression in male and female rodents as well as the role of HNF4α in those differences.
- 47. Waxman DJ, Holloway MG. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol. 2009;76:215–228. doi: 10.1124/mol.109.056705. This comprehensive review covers the molecular basis of sex differences in pharmacology with special emphasis on growth hormone, STAT5b and HNF4α.
- 48.Lahuna O, Rastegar M, Maiter D, Thissen JP, Lemaigre FP, Rousseau GG. Involvement of STAT5 (signal transducer and activator of transcription 5) and HNF-4 (hepatocyte nuclear factor 4) in the transcriptional control of the hnf6 gene by growth hormone. Mol Endocrinol. 2000;14:285–294. doi: 10.1210/mend.14.2.0423. [DOI] [PubMed] [Google Scholar]
- 49.Lahuna O, Fernandez L, Karlsson H, Maiter D, Lemaigre FP, Rousseau GG, Gustafsson J, Mode A. Expression of hepatocyte nuclear factor 6 in rat liver is sex-dependent and regulated by growth hormone. Proc Natl Acad Sci U S A. 1997;94:12309–12313. doi: 10.1073/pnas.94.23.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delesque-Touchard N, Park SH, Waxman DJ. Synergistic action of hepatocyte nuclear factors 3 and 6 on CYP2C12 gene expression and suppression by growth hormone-activated STAT5b. Proposed model for female specific expression of CYP2C12 in adult rat liver. J Biol Chem. 2000;275:34173–34182. doi: 10.1074/jbc.M004027200. [DOI] [PubMed] [Google Scholar]
- 51.Gardmo C, Mode A. In vivo transfection of rat liver discloses binding sites conveying GH-dependent and female-specific gene expression. J Mol Endocrinol. 2006;37:433–441. doi: 10.1677/jme.1.02116. [DOI] [PubMed] [Google Scholar]
- 52.Maeda Y, Seidel SD, Wei G, Liu X, Sladek FM. Repression of hepatocyte nuclear factor 4alpha tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol Endocrinol. 2002;16:402–410. doi: 10.1210/mend.16.2.0769. [DOI] [PubMed] [Google Scholar]
- 53.Maeda Y, Hwang-Verslues WW, Wei G, Fukazawa T, Durbin ML, Owen LB, Liu X, Sladek FM. Tumour suppressor p53 down-regulates the expression of the human hepatocyte nuclear factor 4alpha (HNF4alpha) gene. Biochem J. 2006;400:303–313. doi: 10.1042/BJ20060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazure NM, Nguyen TL, Danan JL. Severe hypoxia specifically downregulates hepatocyte nuclear factor-4 gene expression in HepG2 human hepatoma cells. Tumour Biol. 2001;22:310–317. doi: 10.1159/000050632. [DOI] [PubMed] [Google Scholar]
- 55.Yu D, Wang ZH, Cheng SB, Li HK, Chan HB, Chew EC. The effect of arsenic trioxide on the expression of Hsc and HNF4 in nuclear matrix proteins in HepG2 cells. Anticancer Res. 2001;21:2553–2559. [PubMed] [Google Scholar]
- 56.Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, Parpura V, Paschal BM, Sladek FM. Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol. 2007;21:1297–1311. doi: 10.1210/me.2006-0300. [DOI] [PubMed] [Google Scholar]
- 57.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 58. Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. This is one of the first indications that the activity of HNF4α may be regulated in a circadian fashion and has important implications for drug treatment.
- 59.Xie X, Liao H, Dang H, Pang W, Guan Y, Wang X, Shyy JY, Zhu Y, Sladek FM. Down-regulation of hepatic HNF4alpha gene expression during hyperinsulinemia via SREBPs. Mol Endocrinol. 2009;23:434–443. doi: 10.1210/me.2007-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 61.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong YH, Varanasi US, Yang W, Leff T. AMP-activated protein kinase regulates HNF4alpha transcriptional activity by inhibiting dimer formation and decreasing protein stability. J Biol Chem. 2003;278:27495–27501. doi: 10.1074/jbc.M304112200. [DOI] [PubMed] [Google Scholar]
- 63.Sladek FM. What are nuclear receptor ligands? Mol Cell Endo. 2010 doi: 10.1016/j.mce.2010.06.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruse MD, Jr, Privalsky ML, Sladek FM. Competitive cofactor recruitment by orphan receptor hepatocyte nuclear factor 4alpha1: modulation by the F domain. Mol Cell Biol. 2002;22:1626–1638. doi: 10.1128/MCB.22.6.1626-1638.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 2002;277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- 66.Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]