Abstract
Previously, we described an orthotopic cholangiocarcinoma model based on bile duct inoculation of spontaneously-transformed low grade malignant rat BDE1 cholangiocytes (BDEsp cells) compared to high grade malignant erbB-2/neu- transformed BDE1 cholangiocytes (BDEneu cells) into the livers of syngeneic rats, which closely mimics clinical features of early versus advanced stages of the human cancer. We now used gene expression microarray together with quantitative real-time RT-PCR to profile genes differentially expressed in highly tumorigenic BDEneu cells and corresponding tumors compared to less aggressive tumorigenic BDEsp cells and tumors. Genes identified as being commonly overexpressed in parent BDEneu cells, tumors, and in a BDEneu tumor- derived cholangiocarcinoma cell line included Sox17, Krt20, Erbb2, and Sphk1 when respectively compared to BDEsp cells, tumors, and tumor-derived BDEsp cholangiocarcinoma cells. Muc1 was also prominently overexpressed in BDEneu cells and tumor-derived cholangiocarcinoma cells over that expressed in corresponding BDEsp cell lines. Periostin and tenascin-C, which were produced exclusively by cholangiocarcinoma-associated fibroblastic cells, were each significantly overexpressed in BDEneu tumors compared to BDEsp tumors. Interestingly, amphiregulin was representative of a gene found to be significantly underexpressed in vitro in BDEneu cells compared to BDEsp cells, but significantly overexpressed in BDEneu tumors compared to BDEsp tumors, and correlated with BDEneu cholangiocarcinoma progression in vivo. Our data support a unique animal model that recapitulates important molecular features of human cholangiocarcinoma progression, and may serve as a potentially powerful preclinical platform for identifying and rapidly testing novel molecular targeting strategies for cholangiocarcinoma therapy and/or prevention.
Keywords: cholangiocarcinoma, differential gene expression profiling, transformed rat cholangiocytes, orthotopic hepatobiliary cancer model, bile duct cancer cell progression
Introduction
Previously, we described the establishment of a “patient-like” animal model of intrahepatic cholangiocarcinoma based on bile duct inoculation of highly malignant erbB2/neu transformed rat BDE1 cholangiocytes (BDEneu cells) compared to less malignant, spontaneously transformed BDE1 cholangiocytes (BDEsp cells) into the livers of syngeneic rats (Lai et al.,2005; Sirica et al., 2008). More recently, our preclinical syngeneic rat orthotopic cholangiocarcinoma model has been demonstrated to be useful for rapidly assessing the potential therapeutic efficacy of various molecular target-based treatments against intrahepatic cholangiocarcinoma, including those with the tyrosine kinase inhibitor sorafanib (Blechacz et al., 2009), the BH3 mimetic obatoclax (Smoot et al., 2010), and the dual ErbB1/ErbB2 tyrosine kinase inhibitor lapatinib (Zhang et al., 2010).
In an effort to further demonstrate the value of this model as having significance for identifying molecular targets associated with intrahepatic cholangiocarcinoma progression, we have utilized high-density oligonucleotide microarray analysis, selectively validated by quantitative real-time reverse transcriptase polymerase chain reaction (QRT-PCR), to provide a comprehensive gene expression profiling of cultured BDEneu versus BDEsp cell lines, together with that of corresponding tumors. Signature target genes were also assessed by QRT-PCR in novel cholangiocarcinoma cell lines derived from BDEneu and BDEsp liver tumors, respectively, and in a cancer-associated fibroblastic cell line obtained from the BDEsp liver tumor.
Materials and Methods
Cell lines
The parent rat cholangiocarcinoma cell lines, BDEneu and BDEsp, used in this study were each generated in our laboratory from an immortalized rat cholangiocyte cell line initially established in the laboratory of Dr. Douglas C. Hixson at Rhode Island Hospital, Providence, RI (Yang et al., 1993; Rozich et al., 2010) and have been previously described (Lai et al., 2005; Sirica et al., 2008). Cultured BDEneu cells at in vitro passage 14 and BDEsp cells at in vitro passage 11, grown to ≥ 70% confluence in plastic culture dishes, were used in our microarray and QRT-PCR analyses.
Orthotopic syngeneic rat model of Cholangiocarcinoma Progression
The animal experimentation for this study was performed in accordance with criteria outlined in the Guide for the Care and Use of Laboratory Animals, 1996, National Academy Press, Washington, DC, using protocol AM 10149 prepared by A.E.S., and approved by the Institutional Animal Care and Use Committee at Virginia Commonwealth University. The orthotopic syngeneic rat model used to generate rapidly growing, moderately-differentiated invasive and metastatic BDEneu intrahepatic cholangiocarcinomas and slow growing, well-differentiated non-metastatic BDEsp intrahepatic cholangiocarcinomas analyzed in this study has been previously described (Sirica et al., 2008). Briefly, 4 × 106 rat BDEneu cells at in vitro passage 12 or BDEsp cells at in vitro passages 8 to 11, each with ≥ 90% viabilities, were suspended in 0.1ml Hanks’ balanced salt solution and transplanted via bile duct inoculation into the livers of young adult syngeneic Fischer 344 male rats (Harlan, Indianapolis, IN). Rats orthotopically transplanted with BDEneu cells were sacrificed at days 10 (n=3), 15 (n=3), or 25/26 (n=5), respectively, after initial bile duct inoculation of the tumorigenic cells into liver, whereas those inoculated with BDEsp cholangiocytes (n=6) were sacrificed between 47 and 76 days after bile duct inoculation of these tumorigenic cells. For each tumor-bearing rat, total mass-forming intrahepatic tumor tissue was carefully resected from the liver and freed of surrounding liver parenchyma. Gross peritoneal metastases were also resected from the day 25 BDEneu tumor bearing rats (n=3) and separately pooled. None of the rats orthotopically transplanted with BDEsp cells developed extrahepatic metastases. All resected tumor tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until RNA extraction. In addition, corresponding serum samples were also obtained from liver tumor bearing rats by cardiac puncture at sacrifice and assayed for levels of total bilirubin as previously described (Sirica et al., 2008).
Two novel orthotopic tumor-derived neoplastic cholangiocyte cell lines, determined under light microscopy to be epithelial, were separately obtained through selective passages of primary cell outgrowths from 0.1mm tissue fragments isolated from a day 21 BDEneu or from a day 47 BDEsp liver tumor. These two epithelial cell lines, designated as BDEneu-TDE and BDEsp-TDE, respectively, were maintained in culture essentially under the same standard conditions used by us to culture the parent BDEneu and BDEsp cell lines (each at in vitro passage 10) from which the original intrahepatic tumors were generated. A novel cell line highly enriched by ≥ 90% in cancer-associated fibroblastic cells (BDEsp-TDF) was also established by serial passages and selective growth conditions from a primary cell outgrowth of tumor stromal fibroblastic cells from tissue fragments isolated from a day 47 BDEsp liver tumor and then maintained under identical conditions as those used to culture the tumor-derived cholangiocyte cell lines.
Sample handling and RNA extraction
Cultured parent BDEneu and BDEsp cell lines and tumor-derived BDEneu-TDE, BDEsp-TDE, and BDEsp-TDF cell lines were harvested for RNA extraction as previously described (Sirica et al., 2008). In the case of the BDEneu and BDEsp tumor specimens, total RNA was extracted from cryopreserved sections of solid tumor and its quality assessed according to our previously described method (Dumur et al., 2004). Briefly, multiple 10μm thick cryosectioned tissue samples comprised of 100% tumor without necrosis were placed directly in TRIZOL reagent (Invitrogen Corp., Carlsbad, CA) and processed according to the manufacturer’s protocol. A subsequent cleanup step with RNeasy reagents (QIAGEN Inc., Valencia, CA) was then performed according to the manufacturer’s protocol. Total RNA purity was judged by spectrophotometry at 260, 270, and 280nm. Total RNA integrity, as well as cDNA and cRNA synthesis products, were assessed by running 1μL of every sample in RNA 6000 Nano LabChips® on the 2100 Bioanalyzer (Agilent Technologies, Foster City, CA).
Comprehensive gene expression microarray analyses
The Affymetrix® protocol utilized for our microarray analyses has been previously described (Dumur et al., 2004). Starting with 5μg of total RNA from each tissue sample, we performed a single-strand cDNA synthesis primed with a T7-(dT24) oligonucleotide. Second strand cDNA synthesis was performed with the E. coli DNA Polymerase I, using the GeneChip® One-Cycle cDNA Synthesis Kit (Invitrogen Corp.). Resulting cDNA was purified using cDNA cleanup reagents from the GeneChip Sample Cleanup Module (QIAGEN). Biotinylation of the cRNA was achieved by in vitro transcription (IVT), using the GeneChip IVT Labeling Kit. After a 37°C incubation for 16 hours, the labeled cRNA was purified using the cRNA cleanup reagents from the GeneChip Sample Cleanup Module. The cRNA was then fragmented by heating in fragmentation buffer (40mM Tris-Acetate, 100mM KOAc and 30mM MgOAc) at pH 8.1 for 35 minutes at 94°C. As per the Affymetrix® protocol, 10μg of fragmented cRNA were hybridized on the GeneChip® Rat Expression 230 2.0 Array (Affymetrix Inc., Santa Clara, CA) for 16 hours at 60rpm in a 45°C hybridization oven. The GeneChip® Rat Expression 230 2.0 Array provides comprehensive coverage of the transcribed rat genome by including over 31,000 probe sets that analyze the expression level of over 30,000 transcripts and variants from over 28,000 well-substantiated rat genes. The arrays were washed and stained with streptavidin phycoerythrin (SAPE; Molecular Probes, Eugene, OR) in the Affymetrix® fluidics workstation. To amplify the fluorescent signal, SAPE solution was added twice, with an antistreptavidin biotinylated antibody (Vector Laboratories, Burlingame, CA) staining step in between. Every chip was scanned at a high resolution, with pixelations ranging from 2.5μm down to 0.51μm, by the Affymetrix® GeneChip Scanner 3000 according to the GeneChip® Expression Analysis Technical Manual procedures (Affymetrix). After scanning, the raw intensities for every probe were stored in electronic files (in .DAT and .CEL formats) by the GeneChip® Operating Software v1.4 (GCOS) (Affymetrix). Overall quality of each array was assessed by monitoring the 3′/5′ ratios for the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (gapdh), and the percentage of “Present” genes (%P). Arrays exhibiting gapdh 3′/5′ < 3.0 and %P > 40% were considered good quality arrays. Moreover, whisker boxplots were used to assess and compare the intensity distribution across all the arrays included in this study, using the boxplot function from the Bioconductor affy package, run on R 2.4.0.
QRT-PCR
QRT-PCR was used to validate gene expression levels of selected genes using TaqMan® chemistry. Probes and primer sets specific for detection of rat RNA transcripts were purchased from Applied Biosystems, Foster City, CA. These included gene specific probes for the following genes: krt19 (cytokeratin 19), ID# Rn01496870_g1; krt20 (cytokeratin 20), ID# Rn01466577_m1; Sox17 (Sry-related HMG box gene-17), ID# Rn01749232_g1; Erbb2 (neu), ID# Rn00690264_g1; Muc1 (mucin 1), ID # Rn01462585_m1; Areg (amphiregulin), ID# Rn00567471_m1; Mmp7 (metalloproteinase-7; matrilysin), ID# Rn01487001_m1; Acta2 (α-smooth muscle actin), ID# Rn01759928_g1; Postn (periostin), ID# Rn01494627_m1; Tnc (tenascin-C), ID# Rn01454953_m1; Hgf (hepatocyte growth factor), ID# Rn00566673_m1; Ctgf (connective tissue growth factor), ID# Rn01537278_g1; Cxcl12 (C-X-C chemokine ligand-12; stromal-derived factor-1), ID# Rn00573260_m1; Cxcr4 (C-X-C chemokine receptor type 4), ID# Rn01483207_m1; Sphk1 (sphingosine kinase1), ID# Rn00682794_g1; Cav (caveolin-1), ID# R00755834_m1; and Hif1a (hypoxia inducible factor 1α), ID# Rn00694426_m1. Gene-specific probes labeled in the 5′ end with FAM (6-carboxyfluorescein) and in the 3′ end with a dark quencher were used for all the target genes of interest. For each target gene, Gapdh (Applied Biosystems) was used as the endogenous control. The experiments were performed in the ABI Prism 7500 Sequence Detection System (Applied Biosystems) using the TaqMan® Reverse Transcription and Universal PCR Master Mix Reagents. All the samples were tested in triplicate. The cycling conditions were 48ºC for 30 minutes, 95ºC for 10 minutes, 40 cycles of 95ºC for 15 seconds, and 60ºC for 1 minute. The 2−ΔΔCT method was used to calculate fold changes in the expression levels of the genes of interest (Livak et al., 2001).
Western blotting
Western blot analysis of total protein in cell lysates prepared from the various rat cholangiocarcinoma cell lines used in this study was performed as previously described (Lai et al., 2005; Sirica et al., 2008), using the following primary antibodies: Neu (C-18, sc-284) and caveolin-1 (N-20, sc-894) purchased from Santa Cruz Biotechnology, Santa Cruz, CA, and anti-periostin (ab14041) from Abcam Inc., Cambridge, MA. Protein loading was normalized with β-actin control antibody as previously described (Sirica et al., 2008).
Statistical Analysis
For the microarray data analysis, background correction, normalization, and estimation of probe set expression summaries was performed using the log-scale Robust Multi-array Analysis (RMA) method (Irizarry et al., 2003). Hierarchical cluster analyses were performed with the BRB-ArrayTools v3.1.0 (Biometric Research Branch, National Cancer Institute), an Excel add-in that collates microarray data with sample annotations. The resulting expression dataset was filtered to eliminate genes that are presumably not expressed in any of the samples. The latter was assessed by filtering out probe sets with expression summaries lower than the mean value for the limit of quantification (1.5pM) assessed with the lowest hybridization control gene, AFFX-BioB ± 1.96 × standard deviation (S.D.). In order to identify differentially expressed genes between BDEneu and BDEsp cell lines, as well as between samples of BDEneu cell-derived intrahepatic cholangiocarcinoma (BDEneuT) versus BDEsp-cell derived intrahepatic cholangiocarcinoma (BDEspT), we performed t-tests for each probe of the reduced dataset. Statistical significance for multivariate analysis to assess probe set specific false discovery rates (FDR) was performed by estimating the q-values, using the Bioconductor qvalue package (Story, 2002). With the purpose of identifying genes with a significant trend (increase or decrease) associated with BDEneu tumor progressive growth at 10, 15 and 25 days post-bile duct inoculation, we used the Jonckheere-Terpstra test for trend on the normalized gene expression dataset across the three groups of tumors.
Results
Supervised two-dimensional hierarchical cluster analysis of comprehensive gene expression in cultured rat BDEneu versus BDEsp cells and derived tumors
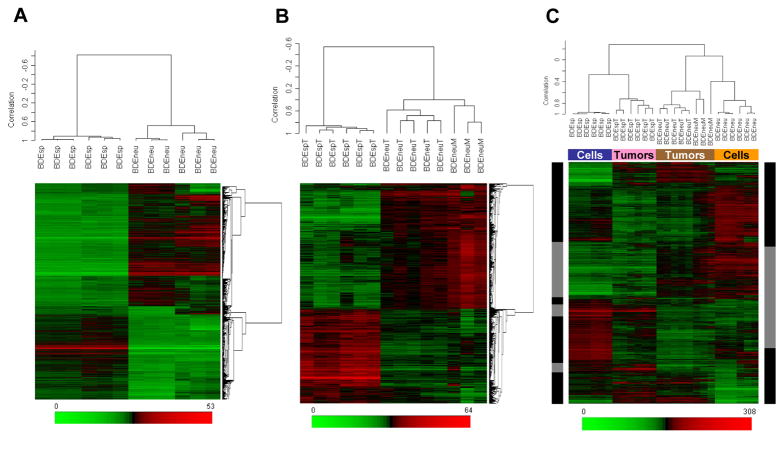
To compare genes differentially expressed in cultured BDEneu versus BDEsp cells and in corresponding BDEneuT, BDEspT, and BDEneu peritoneal metastases (BDEneuM) [see Supplemental Figure 1], we first filtered gene probe sets to eliminate those with very low expression summary values and low variability across all samples. As a result of this filtering step, 15,936 probe sets were retained for further analysis. Univariate parametric two-tailed t-tests were used to identify probe sets that were significantly different at an FDR of 5% (q at < 0.05) and showing a > 1.5-fold change when comparing BDEneu to BDEsp cells; BDEneuT to BDEspT; BDEneuT at day 25 after bile duct inoculation to BDEneuT at day 10; BDEneuM at day 25 to BDEneuT from the same rats, and BDEneuM to BDEspT. The heat maps in Fig. 1 represent the relative expression levels of significantly up-regulated (in red) and down-regulated (in green) genes differentially expressed between cultured BDEneu versus BDEsp cells (Fig. 1A) and between intrahepatic BDEneuT and BDEspT, and also with BDEneuM (Fig. 1B). Among the 4,711 probe sets identified as showing a > 1.5-fold change between cultured BDEneu and BDEsp cells, 2,670 were determined to be significantly overexpressed and 2,041 were significantly underexpressed. Of 3,090 probe sets found to be significantly different between BDEneuT and BDEspT, 1,759 were up and 1,331 were down. Of 4,548 probe sets showing > 1.5-fold change between BDEneuM and BDEspT, 3,184 were overexpressed and 1,364 were underexpressed. Between BDEneuT at day 10 and BDEneuM at day 25, 177 probe sets were found to be up and 18 down, whereas only 4 probe sets were noted as being significantly different between BDEneuT and BDEneuM at day 25. By employing hierarchial clustering analysis based on these datasets, we could clearly distinguish the highly malignant BDEneu cells and their resulting tumors from low grade malignant BDEsp cells and tumors (Fig. 1).
Figure 1.
Two-dimensional hierarchical clustering of samples and genes using Pearson’s (centered) correlation and average linkage, based on (A) 4,711 probe sets that were significantly different (q < 0.05) between BDEneu and BDEsp cultured cell lines, (B) 3,090 probe sets that were significantly different (q < 0.05) between BDEneu liver tumors and peritoneal metastases compared to non-metastatic BDEsp liver tumors, and (C) 4,711 probe sets that were significantly different between BDEneu and BDEsp cell lines used to cluster all of the samples: BDEsp cell line (blue box), BDEneu cell line (orange box), BDEsp tumors (pink box), and BDEneu tumors (brown box). The 1,497 common probe sets for cells in tumors are indicated by gray sidebars, whereas the discordant probe sets are indicated by black sidebars. Each of the colored rows in the individual heat maps show the relative expression for that specific gene in the separate specimen samples (columns). The relative gene expression levels are plotted according to the color scale at the bottom of each heat map, where red and green respectively indicate the relative extents of gene overexpression or underexpression compared to the median intensity across all samples.
Expression profile of selected genes differentiating cultured BDEneu cells and tumors from BDEsp cells and tumors
Table 1 lists selected genes whose expression was found by microarray analysis to be prominently altered in cultured high grade tumorigenic BDEneu cells compared to cultured low grade tumorigenic BDEsp cells. Those genes identified as being differentially overexpressed in BDEneu cells over BDEsp cells included: Rdx (radixin), Sox17, Cdkn2a (cyclin-dependent kinase inhibitor 2A), Mt2A (metallothionine 2A), igfbp7 (insulin-like growth factor binding protein 7), Axl (Axl receptor tyrosine kinase), Krt20, Fn1 (fibronectin), Muc1, Casc4 (cancer susceptibility candidate 4), Tpm2 (tropomyosin 2), Bmp7 (bone morphogenetic protein 7), Erbb2, Sphk1, Wnt7a (wingless-related MMTV integration site 7A), and Smad7 (MAD homolog 7), among others. Select genes whose expression was significantly downregulated in cultured BDEneu cells over that of cultured BDEsp cells included among others: Ocm (oncomodulin), Gja1 (gap junction protein, alpha 1), Cldn18 (claudin 18), Ptgs2 (prostaglandin-endoperoxidase synthase 2), Areg, and Cav.
Table 1.
Selected genes whose expression is significantly altered in rat BDEneu compared to BDEsp cholangiocarcinoma cell lines
| Gene Symbol | Gene Title | Molecular Function | Fold Change | p-value | q-value |
|---|---|---|---|---|---|
| Qk | quaking | RNA binding | 78.3 | 2.12E-11 | 2.63E-08 |
| Rdx | radixin | actin binding | 56.6 | 1.35E-10 | 7.42E-08 |
| Sox17 | SRY-box containing gene 17 | transcription factor | 47.3 | 8.59E-11 | 5.91E-08 |
| Ccl2 | chemokine (C-C motif) ligand 2 | anti-apoptosis | 46.3 | 9.20E-10 | 2.19E-07 |
| Cdkn2a | cyclin-dependent kinase inhibitor 2A | cell cycle checkpoint | 39.8 | 1.21E-09 | 2.37E-07 |
| Mt2A | metallothionein 2A | cellular metal ion homeostasis | 39.5 | 3.97E-06 | 4.53E-05 |
| Lgals9 | lectin, galactose binding, soluble 9 | sugar binding | 36.3 | 5.15E-09 | 5.75E-07 |
| Igfbp7 | insulin-like growth factor binding protein 7 | regulation of cell growth | 36.3 | 6.51E-08 | 2.98E-06 |
| Serpinh1 | serine (or cysteine) peptidase inhibitor, clade H, member 1 | serine-type endopeptidase inhibitor activity | 27.3 | 4.26E-10 | 1.38E-07 |
| Axl | Axl receptor tyrosine kinase | cell proliferation | 25.2 | 2.10E-10 | 9.45E-08 |
| Fn1 | fibronectin 1 | cell-matrix adhesion | 24.5 | 9.01E-08 | 3.68E-06 |
| Krt20 | keratin 20 | intermediate filament | 23.2 | 1.53E-11 | 2.26E-08 |
| Muc1 | mucin 1, transmembrane | cell signaling | 20.4 | 1.18E-07 | 4.29E-06 |
| Casc4 | cancer susceptibility candidate 4 | cell proliferation associated to ErbB2 | 17.6 | 6.46E-10 | 1.71E-07 |
| Tpm2 | tropomyosin 2 | regulation of ATPase activity | 17.1 | 5.78E-10 | 1.66E-07 |
| Cadm1 | cell adhesion molecule 1 | cell adhesion | 15.9 | 4.45E-08 | 2.33E-06 |
| Lgals1 | lectin, galactose binding, soluble 1 | sugar binding | 15.6 | 4.10E-07 | 9.43E-06 |
| Hspb1 | heat shock protein 1 | anti-apoptosis | 11.5 | 1.63E-09 | 2.81E-07 |
| Abcg1 | ATP-binding cassette, sub-family G, member 1 | transport, cholesterol metabolic process | 11.2 | 1.44E-06 | 2.22E-05 |
| Ela1 | elastase 1, pancreatic | proteolysis | 10.7 | 1.53E-07 | 5.18E-06 |
| Rab17 | RAB17, member RAS oncogene family | small GTPase mediated signal transduction | 10.7 | 1.27E-08 | 1.01E-06 |
| Bmp7 | bone morphogenetic protein 7 | extracellular signaling | 7.7 | 2.91E-08 | 1.78E-06 |
| Mt1a | metallothionein 1a | cellular metal ion homeostasis | 5.2 | 1.48E-08 | 1.10E-06 |
| Tspan5 | tetraspanin 5 | cell adhesion | 4.6 | 4.98E-09 | 5.60E-07 |
| Erbb2 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 | cell proliferation | 4.3 | 6.79E-06 | 6.49E-05 |
| Sphk1 | sphingosine kinase 1 | anti-apoptosis | 3.5 | 4.02E-07 | 9.38E-06 |
| Wnt7a | wingless-related MMTV integration site 7A | Wnt receptor signaling pathway | 3.5 | 2.18E-06 | 2.96E-05 |
| Smad7 | MAD homolog 7 (Drosophila) | TGF-β receptor signaling pathway | 3.0 | 2.30E-05 | 1.54E-04 |
| Itga6 | integrin, alpha 6 | cell adhesion | 2.9 | 4.99E-03 | 9.68E-03 |
| Lgals2 | lectin, galactoside-binding, soluble 2 | sugar binding | 2.5 | 2.57E-04 | 9.25E-04 |
| Cdh3 | cadherin 3, type 1, P-cadherin | cell adhesion | 1.5 | 4.21E-04 | 1.37E-03 |
| Areg | amphiregulin | EGFR signaling pathway | −2.3 | 4.02E-06 | 4.57E-05 |
| Cav | caveolin, caveolae protein 1 | endocytosis, signal transduction | −2.5 | 1.57E-04 | 6.41E-04 |
| Foxp1 | Forkhead box P1 | regulation of transcription | −3.4 | 9.46E-06 | 8.18E-05 |
| Gata3 | GATA binding protein 3 | regulation of transcription | −5.0 | 1.73E-07 | 5.59E-06 |
| Tspan13 | tetraspanin 13 | regulation of cell motility | −5.1 | 4.05E-07 | 9.38E-06 |
| Ctsc | cathepsin C | proteolysis | −6.2 | 8.23E-05 | 3.90E-04 |
| Vim | vimentin | intermediate filament-based process | −8.2 | 6.29E-07 | 1.23E-05 |
| Il33 | interleukin 33 | cytokine activity | −13.4 | 1.49E-11 | 2.26E-08 |
| Klk10 | kallikrein related-peptidase 10 | proteolysis | −16.2 | 9.41E-08 | 3.79E-06 |
| Il10 | interleukin 10 | cytokine activity | −16.2 | 1.11E-07 | 4.10E-06 |
| Ptgs2 | prostaglandin-endoperoxide synthase 2 | regulation of cell proliferation | −18.2 | 1.06E-07 | 4.00E-06 |
| Ctsz | cathepsin Z | proteolysis | −29.7 | 2.08E-10 | 9.45E-08 |
| Tspan7 | tetraspanin 7 | regulation of cell motility | −68.6 | 1.02E-09 | 2.27E-07 |
| Cldn18 | claudin 18 | structural molecule activity | −210.1 | 1.61E-12 | 7.97E-09 |
| Gja1 | gap junction protein, alpha 1 | cell-cell signaling | −233.4 | 2.78E-13 | 2.06E-09 |
| Ocm | oncomodulin | calcium ion-binding protein | −305.3 | 4.33E-15 | 6.43E-11 |
Gene expression profiles differentiating the BDEneu and BDEsp cultured cell lines were seen to be partially maintained in the corresponding tumors. Of the 4,711 probe sets that were significantly different between the BDEneu versus BDEsp cultured cell lines, 1,497 probe sets were determined to be altered in the same direction in the corresponding BDEneuT when paired against BDEspT (Fig. 1C). Supplemental Table 1 lists selected genes, which include most of those listed above for the cultured transformed rat cholangiocyte cell lines, whose expression was also found to be significantly altered in BDEneuT compared to that of BDEspT. Notably, when we overlaid the 1,497 common probe sets differentiating BDEneu cells and tumors from BDEsp cells and tumors onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base (IPKB), the most significant biological functions correlated to Cancer and Cellular Movement and Invasion (p = 1.5 × 106).
Selected gene overexpression associated with progressive BDEneu intrahepatic tumor growth and peritoneal metastases
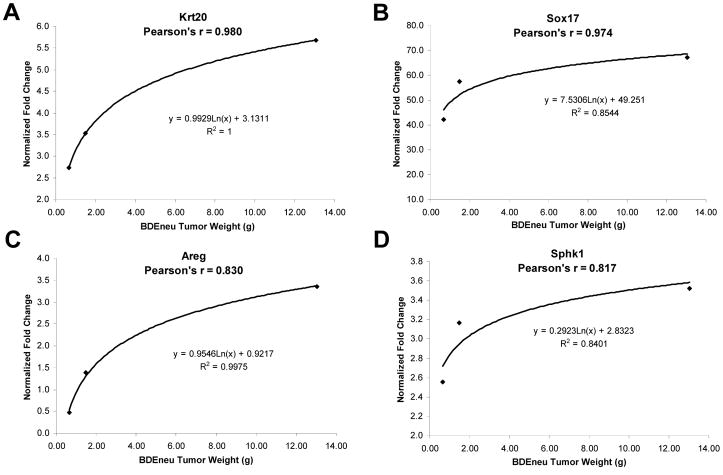
In order to assess a gene expression profile associated with progressive intrahepatic tumor growth and extrahepatic metastases, we looked for genes with a significant trend (increase) in gene expression across the BDEneuT and BDEneuM groups corresponding to tumors analyzed at 10, 15, and 25 days post-BDEneu cell inoculation, using the Jonckheere-Terpstra test for trend. Here, we identified 344 probe sets as having a significant trend across these respective tumor groups. A selection of these probe sets is shown in Supplemental Table 2, with Areg exhibiting the most prominent increase in fold changes in relation to progressive intrahepatic tumor growth at day 15 and 25 and associated peritoneal metastases at day 25. Moreover, in the case of Krt20, Sox17, Areg, and Sphk1, we could demonstrate a strong correlation between increasing gene expression and progressively increased intrahepatic BDEneuT wet weight over time (Fig. 2).
Figure 2.
Regression analysis of selectively analyzed genes, plotting their fold increases relative to expressed levels in BDEsp tumor against BDEneu liver tumor wet weight. Tumors of progressively increased wet weight were harvested from the livers of syngeneic rats at days 10, 15, and 25 after bile duct inoculation of the BDEneu cells. Pearson’s correlations to tumor wet weight are indicated for each analyzed gene. Note that correlation to tumor weight is not linear, but is best-fit by a logarithmic regression curve.
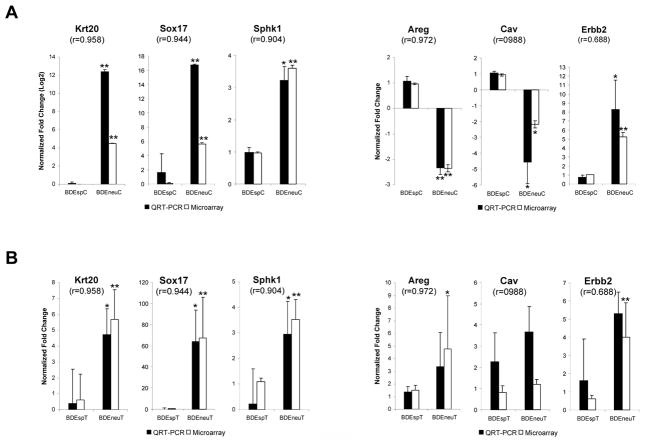
QRT-PCR validation of select differentially expressed genes in cultured BDEneu cells and corresponding tumors compared with BDEsp cells and tumors
QRT-PCR was used to validate the differential expression of 6 selected genes (Krt20, Sox17, Sphk1, Areg, Cav, and Erbb2) in the separate cultured BDEsp and BDEneu cell lines and tumor samples analyzed in this study. For all of the genes validated using this method, we found a significant (p < 0.0001) Pearson’s correlation, thereby proving the robustness of our microarray approach. Our validation was focused on two groups of genes: those significantly affected genes in BDE tumors with an altered expression in the same direction as their corresponding cell lines (Fig. 3A) and those significantly affected genes in BDE tumors with altered expression either in the opposite direction or unchanged from that of their corresponding cell lines (Fig. 3B). The former group appears to be representative of genes (krt20, Sox17, Sphk1, and Erbb2) whose differential expression is an inherently maintained feature of the high grade versus low grade tumorigenic cholangiocarcinoma cell phenotype in vitro and in vivo, whereas the latter group appears to reflect genes (i.e., Areg) whose expression in vitro is increased in vivo as a likely function of tumor progression and a changing tumor microenvironment.
Figure 3.
QRT-PCR validation of selected genes. (A) Significantly affected genes between BDEneu compared to BDEsp cell lines, and (B) between BDEneu compared to BDEsp tumors. Pearson’s correlations between QRT-PCR and microarray measurements are indicated for each gene. Asterisks: unpaired two-tailed t-test BDEneu versus BDEsp samples, * p-value < 0.05; ** p-value < 0.0005. When asterisks are not shown, no significant difference was observed. Error bars represent SD.
Differential expression of genes belonging to the extracellular matrix gene ontogeny
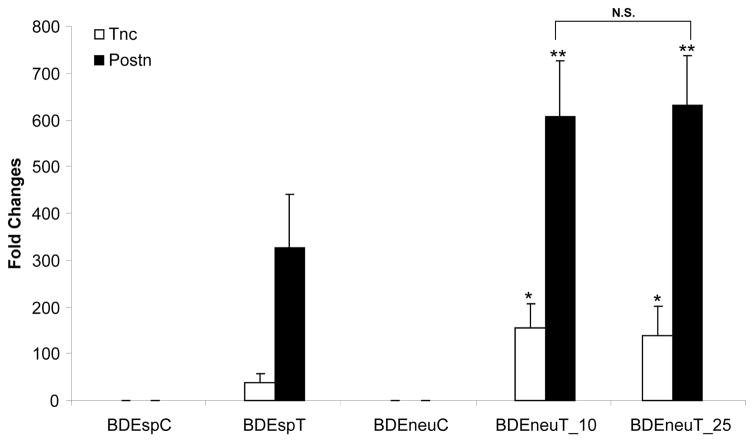
In a preliminary microarray analysis in which we profiled genes aberrantly expressed in BDEneuT samples when compared with pair-matched right liver lobes without evidence of malignant cells obtained from the same rats as the tumors, we observed that the BDEneuT samples exhibited a significant enrichment for genes of the extracellular matrix (ECM) receptor interaction pathway (data not shown). Postn was determined to be the most prominently overexpressed ECM gene in BDEneuT when compared to non-cancerous right liver lobes from the same rats, as well as to normal rat liver and liver from bile duct-ligated rats. Western blotting and quantitative immunohistochemistry confirmed these findings (Sirica et al., 2009). In addition, Tnc was found to be the second highest expressed ECM gene in BDEneuT when compared to non-cancerous right liver lobe tissue from the same rat (Sirica et al., 2009; Supplemental Table 3). Extending these findings, we now demonstrate Postn to be overexpressed by over 600-fold and Tnc to be overexpressed by over 100-fold in BDEneuT analyzed at day 10 and 25 post-BDEneu cell inoculation into liver relative to their levels of expression in cultured BDEneu cells (Fig. 4). As further demonstrated in this figure, both Postn and Tnc were significantly overexpressed in the BDEneuT samples compared to those of BDEspT.
Figure 4.
Bar chart representation of genes (Postn and Tnc) belonging to the extracellular matrix gene ontology classification whose expression is differentially altered in BDEneu liver tumors compared with BDEsp liver tumors. Fold changes for each BDE tumor type (BDEspT, BDEneuT at 10 days and at 25 days) are calculated against their respective parent BDE cell line (BDEspC and BDEneuC). Asterisks: unpaired two-tailed t-test BDEneu tumors versus BDEsp tumors, * p-value < 0.05; ** p-value < 0.005. Error bars represent SD. N.S.=not significant.
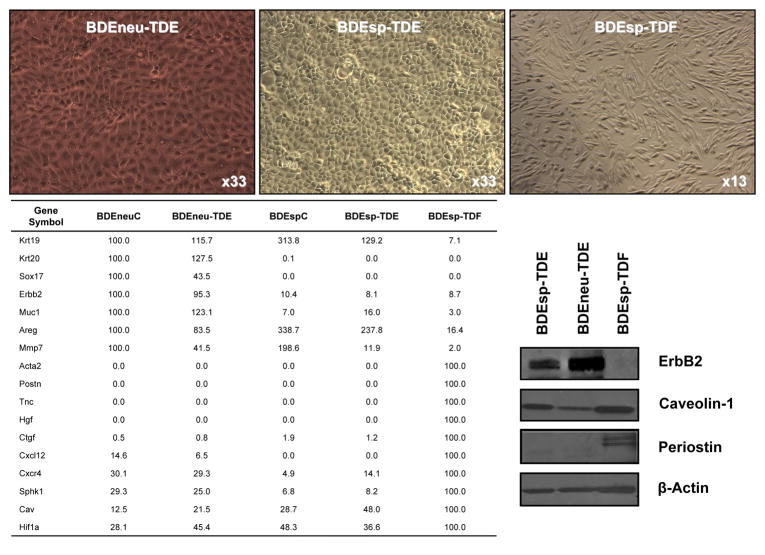
Select differential gene expression profiles differentiating BDEneu-TDE from BDEsp-TDE and BDEsp-TDF
In an effort to further delineate changes in gene expression associated with high grade BDEneuT versus low grade BDEspT, we used QRT-PCR to assess differences in select target genes differentially expressed in derived, newly established BDEneu-TDE versus BDEsp-TDE cultured cell lines, as well as in a novel BDEsp-TDF cell line highly enriched in cancer-associated fibroblasts. Corresponding parent BDEneu (BDEneuC) and BDEsp (BDEspC) cell lines used to generate the respective intrahepatic tumors were also simultaneously analyzed. The results of this analysis are shown in Fig. 5. As indicated, Krt19, a cholangiocyte cytokeratin, was abundantly expressed in all of the BDE cholangiocarcinoma cell lines, but only marginally expressed in the BDEsp-TDF cell line. Conversely, Acta2, which is a gene marker of myofibroblastic cells, together with Postn, Tnc, and Hgf were only detected in the BDEsp-TDF enriched cell line. Ctgf was also marginally expressed in the respective cholangiocarcinoma cell lines and was predominantly a marker gene of the BDEsp-TDF cell line. On the other hand, Krt20, Sox 17, Erbb2, and Muc 1 were abundantly expressed in both the BDEneu-TDE and parent BDEneuC cell lines and only marginally expressed in the BDEsp-TDE, BDEspC and BDEsp-TDF cell lines. Mmp-7 was also primarily expressed in the respective BDE cholangiocarcinoma cell lines, being more highly expressed in the BDEneu-TDE cells compared to the BDEsp-TDE cells, and barely detectable in the BDEsp-TDF cells. Similar to our findings for the parent BDEneu and BDEsp parent cell lines, Areg and Cav1 were determined to be expressed at higher levels in BDEsp-TDE over BDEneu-TDE cells. In comparison, Cav1 was most highly expressed in the BDEsp-TDF cell line, whereas Areg expression was detected only at a low level in this tumor-derived fibroblastic cell culture. Interestingly, while Cxcl12 and the Cxcr4 gene encoding its receptor, as well as Sphk1 were more highly expressed in the BDEneuC and BDEneu-TDE cell lines than in the BDEspC and BDEsp-TDE cell lines, these genes were most abundantly expressed in the BDEsp-TDF cell line. Likewise, HIf1a was more highly expressed in the BDEsp-TDF cell line than in the various cholangiocarcinoma cell lines.
Figure 5.
QRT-PCR analysis of differential gene expression of select genes in parent BDEneuC and tumor-derived BDEneu-TDE cell lines compared to parent BDEspC and tumor-derived BDEsp-TDE and BDEsp-TDF cell lines. Note that representative Western blot banding profiles for ErbB2, caveolin 1, and periostin conform with corresponding QRT-PCR data.
Discussion
Intrahepatic cholangiocarcinoma is an insidious cancer, which in its early stage is usually asymptomatic and slow growing, but which typically presents at an advanced stage where it has progressed to become an aggressive metastatic malignant disease having limited treatment options and a dismal prognosis. As we have now demonstrated, our syngeneic rat model, which closely mimics within a compressed timeframe, key clinical and histopathological features of human intrahepatic cholangiocarcinoma progression (Sirica et al., 2008), also exhibits gene expression profiles which clearly distinguish between the high versus low aggressive malignant rat cholangiocyte phenotype, and which recapitulate a number of critical genes whose overexpression has been shown to correlate with progression and/or poor prognosis in intrahepatic cholangiocarcinoma patients (Sirica et al., 2009; Briggs et al., 2009). Consistent with our results, recent findings from Hixson’s laboratory (Rozich et al., 2010) further suggested that during progression to high passage, cultured rat cholangiocytes undergo preneoplastic changes that enhance their susceptibility to malignant transformation by neu.
Our data demonstrate that Muc1 most notably exemplifed a cholangiocarcinoma cell-specific gene whose overexpression significantly correlated with increased malignant potential of highly tumorigenic BDEneu cells compared to that of less aggressive BDEsp cells, whereas Postn and Tnc best exemplified tumor stromal fibroblastic cell-specific genes (Sirica et al., 2009) whose expression was significantly greater in highly malignant BDEneu tumors than in less malignant BDEsp tumors. Muc1, Postn, and Tnc have each been reported to correlate with tumor progression and/or poor prognosis in human intrahepatic cholangiocarcinoma (Park et al., 2009; Utispan et al., 2010; Aishima et al., 2003).
Our findings demonstrating Sox17 to be uniquely overexpressed in BDEneu cells, corresponding tumors, and in tumor-derived BDEneu-TDE when compared with less tumorigenic BDEsp cells and low grade BDEsp tumor, and tumor-derived BDEsp-TDE is particularly interesting and may have important implications with respect to establishing a relationship between enhanced malignancy and reversion of neoplastically transformed cholangiocytes to a more “endodermal stem cell-like” state. Sox17 has a conserved role in endodermal formation in various vertebrate species (Uemura et al., 2010) and has been shown to function to specify the formation of gallbladder/bile duct progenitors during early foregut morphogenesis in mice (Spence et al., 2009; Uemura et al., 2010). Sox17 function in cholangiocarcinogenesis has not been studied, although it has been proposed that this transcription factor may play a tumor suppressor role in gastrointestinal tumorigenesis (Du et al., 2009). On the other hand, our data would suggest that in cholangiocarcingenesis, Sox17 may be functioning as an oncofetal transcription factor to transcribe endodermal target genes, such as CxCr4 (Katoh et al., 2010), also overexpressed in BDEneu cells compared with BDEsp cells (Fig. 5), and demonstrated to be linked to cholangiocarcinoma cell migration (Ohira et al., 2006).
Differential overexpression of Krt20 in the BDEneu cholangiocarcinoma cells also could reflect a change to a more “stem cell-like” state. In this context, rat “oval” cells or “liver stem cell-like progenitor cells” have been reported to exhibit uniform immunostaining for cytokeratin 20, which is in sharp contrast to the strikingly heterogeneous immunoreactivity for cytokeratin 20 observed in proliferating bile ductules induced in rat liver by bile-duct ligation or treatment with α-naphthylisothiocyanate (Faa et al., 1998). Further studies are now needed to validate if BDEneu cholangiocarcinoma cells have indeed undergone de-differentiation to a hepatobiliary cancer “stem cell-like” state.
Recently, we proposed that novel therapeutic strategies that combine targeting of both malignant cholangiocytes and tumor stromal targets which correlated with human cholangiocarcinoma progression would represent an important development in advancing new therapeutic approaches for this devastating cancer (Sirica et al., 2009). Our results shown in Fig. 5 represent our initial effort to identify select gene targets differentially expressed in cholangiocarcinoma and cancer-associated fibroblastic cells (CAFs) derived from our orthotopic cholangiocarcinoma syngeneic rat model, which are suggested to be playing a role in cholangiocarcinoma progression. We have already noted that Muc1, Postn, and Tnc have been correlated with cholangiocarcinoma progression and poor outcome in humans. MMP7 and CTGF represent prognostic indicators for human intrahepatic cholangiocarcinoma (Gardini et al., 2005; Itatsu K et al., 2008; Sirica et al., 2009). In addition, as also referenced above, signaling between Cxcl12 and Cxcr4 gene products has been shown to regulate cholangiocarcinoma cell migration. Both Areg and Hgf were further demonstrated to be overexpressed in human cholangiocarcinomas (Hansel et al.; 2003; Yotsumoto et al., 2008). Here it is of interest that we determined Areg to be significantly up-regulated in BDEneu cholangiocarcinoma cells in vivo (10) relative to progressive tumor growth, but not in cultured BDEneu cells in basal medium, and was most prominently expressed in both intrahepatic tumors and extrahepatic metastases obtained from rats with malignant biliary obstruction. Our results have also identified for the first time Sphk1 and Cav as putative molecular targets with possible relevance to the progressive human cancer.
Conclusions
We have identified differential gene expression profiles that correlate with high versus low states of malignancy in a unique and important rat model of cholangiocarcinoma progression closely mimicking the human cancer. Moreover, our findings suggest new approaches for cholangiocarcinoma therapy based on combined targeting of identified cholangiocarcinoma and CAF pathways. Based on our findings, we are now devising novel therapeutic strategies to test in our orthotopic cholangiocarcinoma model, which hopefully will validate the concept of combined cholangiocarcinoma cell/CAF targeting as a new approach to cholangiocarcinoma therapy and potentially translate to improving clinical outcome.
Supplementary Material
Acknowledgments
NIH grants R01 CA 39225 and R01 CA 83650 (A.E. Sirica)
Abbreviations
- ECM
extracellular matrix
- FDR
false discovery rates
- IVT
in vitro transcription
- QRT-PCR
quantitative real-time polymerase chain reaction
- RMA
robust multi-array analysis
- SAPE
streptavidin phycoerythrin
- SD
standard deviation
Footnotes
Conflicts of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aishima S, Taguchi K, Terashi T, Matsuura S, Shimada M, Tsueneyoshi M. Tenascin expression at the invasive front is associated with poor prognosis in intrahepatic cholangiocarcinoma. Mod Pathol. 2003;16:1019–1027. doi: 10.1097/01.MP.0000086860.65672.73. [DOI] [PubMed] [Google Scholar]
- Blechacz BRA, Smoot RL, Bronk SF, Werneburg NW, Sirica AE, Gores GJ. Sorafenib inhibits signal transducer and activation of transcription-3 signaling in cholangiocarcinoma cells by activating the phosphatase shatterproof 2. Hepatology. 2009;50:1861–1870. doi: 10.1002/hep.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs CA, Neal CP, Mann CD, Steward WP, Manson MM, Berry DP. Prognostic molecular markers in cholangiocarcinoma: a systematic review. Eur J Cancer. 2009;45:33–47. doi: 10.1016/j.ejca.2008.08.024. [DOI] [PubMed] [Google Scholar]
- Du YC, Oshima H, Oguma K, Kitamura T, Itadani H, Fujimura T, Piao YS, Yoshimoto T, Minamoto T, Kotani H, Taketo MM, Oshima M. Induction and down-regulation of Sox17 and its possible roles during the course of gastrointestinal tumorigenesis. Gastroenterology. 2009;137:1346–1357. doi: 10.1053/j.gastro.2009.06.041. [DOI] [PubMed] [Google Scholar]
- Dumur CI, Nasim S, Best AM, Archer KJ, Ladd AC, Mas VR, Wilkinson DS, Garrett CT, Ferreira-Gonzalez A. Evaluation of quality-control criteria for microarray gene expression analysis. Clin Chem. 2004;50:1994–2002. doi: 10.1373/clinchem.2004.033225. [DOI] [PubMed] [Google Scholar]
- Faa G, Van Eyken P, Roskams T, Miyazaki H, Serreli S, Ambu R, Desmet VJ. Expression of cytokeratin 20 in developing rat liver and in experimental models of ductular and oval cell proliferation. J Hepatology. 1998;29:628–633. doi: 10.1016/s0168-8278(98)80158-x. [DOI] [PubMed] [Google Scholar]
- Gardini A, Corti B, Fiorentino M, Altimari A, Ercolani G, Grazi GL, Pinna AD, Grigioni WF, D’Errico Grigioni A. Expression of connective tissue growth factor is a prognostic marker for patients with intrahepatic cholangiocarcinoma. Dig Liver Dis. 2005;37:269–274. doi: 10.1016/j.dld.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Hansel DE, Rahman A, Hidalgo M, Thuluvath PJ, Lillemoe KD, Shulick R, Ku JL, Park JG, Miyazaki K, Ashfaq R, Wistuba II, Varma R, Hawthorne L, Geradts J, Argani P, Maitra A. Identification of novel cellular targets in biliary tract cancers using global gene expression technology. Am J Pathol. 2003;163:217–229. doi: 10.1016/S0002-9440(10)63645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itatsu K, Zen Y, Yamaguchi J, Ohira S, Ishikawa A, Ikeda H, Sato Y, Harada K, Sasaki M, Sasaki M, Sakamoto H, Nagino M, Nimura Y, Ohta T, Nakanuma Y. Expression of matrix metalloproteinase 7 is an unfavorable postoperative prognostic factor in cholangiocarcinoma of the perihilar, hilar, and extra hepatic bile ducts. Human Pathol. 2008;39:710–719. doi: 10.1016/j.humpath.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Integrative genomic analyses of CXCR4: transcriptional regulation of CXCR4 based on TGFβ, nodal, activin signaling and POU5F1, FOXA2, FOXC2, FOXH1, SOX17, and GFI1 transcriptional factors. Int J Oncology. 2010;36:415–420. doi: 10.3892/ijo_00000514. [DOI] [PubMed] [Google Scholar]
- Lai GH, Zhang Z, Shen XN, Ward DJ, DeWitt JL, Holt SE, Rozich RA, Hixson DC, Sirica AE. erbB-2/neu Transformed rat cholangiocytes recapitulate key cellular and molecular features of human bile duct cancer. Gastroenterology. 2005;129:2047–2057. doi: 10.1053/j.gastro.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ohira S, Sasaki M, Harada K, Sato Y, Zen Y, Isse K, Kozaka K, Ishikawa A, Oda K, Nimura Y, Nakanuma Y. Possible regulation of intrahepatic cholangiocarcinoma cells by interaction of CXCR4 expressed in carcinoma cells with tumor necrosis factor-α and stromal-derived factor-1 released in stroma. Am J Pathol. 2006;168:1155–1168. doi: 10.2353/ajpath.2006.050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Roh SJ, Kim YN, Kim SZ, Park HS, Jang KY, Chung MJ, Kang MJ, Lee DG, Moon WS. Expression of MUC1, MUC2, MUC5AC, and MUC6 in cholangiocarcinoma: prognostic impact. Oncology Rep. 2009;22:649–657. doi: 10.3892/or_00000485. [DOI] [PubMed] [Google Scholar]
- Rozich RA, Mills DR, Brilliant KE, Callanan HM, Yang D, Tantravahi U, Hixon DC. Accumulation of neoplastic traits prior to spontaneous in vitro transformation of rat cholangiocytes determines susceptibility to activated ErbB-2/Neu. Exp Mol Pathol. 2010 doi: 10.1016/j.yexmp.2010.07.005. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirica AE, Zhang Z, Lai GH, Asano T, Shen XN, Ward DJ, Mahatme A, DeWitt JL. A novel “patient-like” model of cholangiocarcinoma progression based on bile duct inoculation of tumorigenic rat cholangiocyte cell lines. Hepatology. 2008;47:1178–1190. doi: 10.1002/hep.22088. [DOI] [PubMed] [Google Scholar]
- Sirica AE, Dumur CI, Campbell DJW, Almenara JA, Ogunwobi OO, DeWitt JL. Intrahepatic cholangiocarcinoma progression: prognostic factors and basic mechanisms. Clin Gastroenterol Hepatol. 2009;7:S68–S78. doi: 10.1016/j.cgh.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot RL, Blechacz BRA, Werneburg NW, Bronk SF, Sinicrope FA, Sirica AE, Gores GJ. A Bax-mediated mechanism for obatoclax-induced apoptosis of cholangiocarcinoma cells. Cancer Res. 2010;70:1960–1969. doi: 10.1158/0008-5472.CAN-09-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Lange AW, Lin S-CJ, Kaestner KH, Lowy AM, Kim I, Whitsett JA, Wells JM. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Developmental Cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. JR Statistic Soc B. 2002;64:479–498. [Google Scholar]
- Uemura M, Hara K, Shitara H, Ishii R, Tsunekawa N, Miura Y, Kurohmaru M, Taya C, Yonekawa H, Kanai-Azuma M, Kanai Y. Expression and function of mouse Sox17 gene in the specification of gallbladder/bile-duct progenitors during early foregut morphogenesis. Biochem Biophys Res Commun. 2010;391:357–363. doi: 10.1016/j.bbrc.2009.11.063. [DOI] [PubMed] [Google Scholar]
- Utispan K, Thuwajit P, Abiko Y, Charngkaew K, Paupairoj A, Chau-in S, Thuwajit C. Gene expression profiling of cholangiocarcinoma-derived fibroblast reveals alterations related to tumor progression and indicates periostin as a poor prognostic marker. Molecular Cancer. 2010;9:13–20. doi: 10.1186/1476-4598-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotsumoto F, Yagi H, Suzuki SO, Oki E, Tsujioka H, Hachisuga T, Sonoda K, Kawarabayashi T, Mekada E, Miyamoto S. Validation of HB-EGF and amphiregulin as targets for human cancer therapy. Biochem Biophys Res Commun. 2008;365:555–561. doi: 10.1016/j.bbrc.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Oyesanya RA, Campbell DJ, Almenara JA, DeWitt JL, Sirica AE. Preclinical assessment of simultaneous targeting of epidermal growth factor receptor (ErbB1) and ErbB2 as a strategy for cholangiocarcinoma therapy. Hepatology. 2010 doi: 10.1002/hep.23773. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.