Abstract
The objective of database AsMamDB is to facilitate the systematic study of alternatively spliced genes of mammals. Version 1.0 of AsMamDB contains 1563 alternatively spliced genes of human, mouse and rat, each associated with a cluster of nucleotide sequences. The main information provided by AsMamDB includes gene alternative splicing patterns, gene structures, locations in chromosomes, products of genes and tissues where they express. Alternative splicing patterns are represented by multiple alignments of various gene transcripts and by graphs of their topological structures. Gene structures are illustrated by exon, intron and various regulatory elements distributions. There are 4204 DNAs, 3977 mRNAs, 8989 CDSs and 126 931 ESTs in the current database. More than 130 000 GenBank entries are covered and 4443 MEDLINE records are linked. DNA, mRNA, exon, intron and relevant regulatory element sequences are provided in FASTA format. More information can be obtained by using the web-based multiple alignment tool Asalign and various category lists. AsMamDB can be accessed at http://166.111.30.6 5/ASMAM DB.html.
INTRODUCTION
In recent years, more and more data about alternatively spliced genes have become available. By the end of 1999, ASDB (1) provided 1922 alternatively spliced proteins. Intronerator (2) collected 844 alternatively spliced genes of Caenorhabditis elegans. All these data greatly promoted the study of alternative splicing of eukaryotic genes. However, more relevant and detailed information is needed, including precisely illustrated splicing patterns of alternatively spliced genes, their gene structures and locations in chromosomes, tissues where they express, feature sequences near splice sites and branch sites, as well as various classifications of these genes. This is why we developed AsMamDB.
Version 1.0 of AsMamDB contains information about alternatively spliced genes of human, mouse and rat, due to the importance of these three species in research on disease and pharmaceutics. In the future, the database will collect this kind of information about genes of other species of mammals. There are 4204 DNAs, 3977 mRNAs, 8989 CDSs and 126 931 ESTs in the current database, which cover more than 130 000 GenBank entries (3) and give links to 4443 MEDLINE records. The data are grouped into 1563 clusters, and each cluster is associated with one alternatively spliced gene. Among all the clusters, 899 are genes from human, 431 from mouse and 233 from rat. The alternative splicing patterns of each gene are depicted by an alignment of its various transcripts. Also, a topological structure is derived from the alignment, which facilitates the use of graph theories in the investigation of alternative splicing. By showing the distribution of exons, introns and various regulatory elements in alignment overview, the gene structure is illustrated to help users in understanding the relationships between these segments. Other information related to a gene, such as expression tissues and chromosome locations, is annotated in a flat file. All nucleotide sequences of DNAs, mRNAs, introns, exons and regulatory elements are provided in FASTA format for users’ convenience. As the first step in revealing the mechanism of alternative splicing, the genes are classified according to their cytobands and expression tissues, etc. The database together with all the category lists can be accessed at http://166.111.30.65/ASMA MDB.html.
DATABASE CONSTRUCTION AND CONTENTS
Data collection
Data for AsMamDB were collected as follows. First, the newest versions of GenBank and UniGene (4) were downloaded as the data sources. GenBank entries were classified according to species. All entries containing the keyword ‘Homo sapiens’ were picked out from PRI division to form the human subset. Entries containing ‘Mus musculus’ and ‘Rattus norvegicus’ were selected from ROD division to form the mouse and rat subsets, respectively. For each species, entries containing ‘alternative splice’ were then picked out and recorded in a list. Secondly, in order to gather sequences that belong to the same gene, UniGene was used. Its clusters were used as raw clusters of AsMamDB, if they contained at least one of the entries in the list obtained in the first step. For each cluster, mRNAs in UniGene *.seq.all file, whose annotation contained ‘/cds=’ and did not contain either ‘/clone=’ or ‘/clone_end=’, were retrieved as kernel sequences. Thirdly, GenBank entries mentioned in UniGene *.data were parsed. Any entry, if it appeared in ‘mRNA join()’, ‘mRNA complement()’, ‘CDS join()’ or ‘CDS complement()’ statements but not contained in the raw cluster, was added into the cluster. This procedure was carried out recursively, until no more entries could be added. The purpose of this step was to complete the raw clusters, since some GenBank entries were missed in UniGene. For example, an mRNA of EP3 gene was annotated in the GenBank entry D86096 as ‘join(D86087.1:361..1488, D86088.1:38..304, D86089.1:33..124, D86091.1:26..52, 34..451)’. The corresponding UniGene cluster, however, contained D86096 only and did not contain the other four entries. Fourthly, other aspects of the information not mentioned above were retrieved from UniGene and GenBank to annotate the cluster. In annotations, all sequences including those described in the feature tables of GenBank such as ‘mRNA’ and ‘CDS’ were classified into four subsets, namely, the DNA, mRNA, CDS and EST. Because DNAs, mRNAs and CDSs of other genes might be introduced in the third step, all these sequences were compared with the cluster kernel. A sequence was preserved, if it shared a common region of >30 bp with at least one of the kernel sequences, otherwise it was deleted. The annotations were recorded in flat files as described in the coming ‘Flat Files’ section. Fifthly, all DNA sequences were retrieved from GenBank. Together with kernel sequences, they were written into a file of FASTA format for users’ convenience, and were also used for creating alignment in AsMamDB. Finally, exons, introns and regulatory elements such as promoters and enhancers, which were annotated in feature tables of GenBank, together with their sequences, were retrieved for the further research. Their sequences were also provided in FASTA format to facilitate the study on splice sites and branch sites, etc. Their location information was used for describing gene structure, as described in the following ‘Gene Structures’ section.
It should be pointed out that, after step three, some raw clusters contained members which were not alternatively spliced from the gene of interest. They were introduced mainly in two cases. First, by including all entries mentioned in the ‘join()’ statements, some DNAs might bring mRNAs, which did not belong to the gene of interest, into the cluster. This problem was solved by comparing non-kernel sequences with kernel sequences as mentioned above. Secondly, some members of multigene families were introduced by original UniGene clusters. This problem can be solved by comparing the sequences with the genome. If the optimal alignment between one sequence and the genome is not in the same location as the gene of interest, the sequence is not alternatively spliced transcript, and should be deleted. However, at the time AsMamDB was created, the complete genome sequences were not available. This type of error will be corrected in the next version of AsMamDB.
The distributions of various kinds of data collected from GenBank Release 117 and UniGene (Hs build 117, Mm build 78, Rn build 77) are shown in Table 1.
Table 1. The data distributions of AsMamDB.
| Species |
Cluster |
DNA |
mRNA |
CDS |
EST |
MEDLINE |
GenBank entries |
| Homo sapiens | 899 | 3168 | 2389 | 5552 | 96 949 | 3069 | >100 000 |
| Mus musculus | 431 | 779 | 1424 | 2650 | 27 865 | 898 | >28 000 |
| Rattus norvegicus | 233 | 257 | 164 | 787 | 2117 | 476 | >2400 |
| Total | 1563 | 4204 | 3977 | 8989 | 126 931 | 4443 | >130 000 |
Flat files
The annotations of a cluster are recorded in a flat file. The main contents of a flat file are listed in Table 2.
Table 2. Main contents of a flat file.
| Field Name |
Contents |
| ASMAMACC | Accession number of a cluster in AsMamDB |
| TITLE | Description of the cluster retrieved from UniGene |
| SEEDGENE | Gene name retrieved from UniGene |
| UNIGENEID | Identifier of the raw cluster in UniGene |
| CYTOBAND | Cytological band |
| CHROMOSOME | Chromosomes containing the cluster |
| EXPRESS | Tissues where the gene expresses |
| MEDLINE | Associated journals in MEDLINE |
| GBACC | Related non-EST GenBank entries |
| EST | All related ESTs of the cluster |
| DNA | All related DNAs of the cluster |
| mRNA | All mRNAs of the cluster |
| CDS | All CDSs of the cluster |
Splicing patterns
The splicing patterns of each cluster are illustrated by a multiple alignment, in which the relationships between its various transcripts are visualized. To create the alignment, we have tried to use CLUSTALW (5). The results, unfortunately, are not satisfactory. More than 20 clusters of known alternatively spliced genes from GenBank were selected randomly, and each cluster was aligned by CLUSTALW. Except for some simple patterns, most of the results were different from the splicing patterns given by GenBank, or were obscured by strong noise. This motivated us to develop Asalign, an ad hoc multiple alignment algorithm for revealing the alternative splicing patterns from a group of nucleotide sequences related to a gene. This algorithm can characterize the alternative splicing patterns more precisely and clearly. Its principles and experimental results can be found in the Supplementary Material.
The sequences participating in the aligning are composed of three parts. One is the kernel sequences, which represent various transcripts of a gene. Another is DNAs of the cluster, which help to reduce alignment errors caused by the lack of information, and to reveal the patterns of how various mRNAs are spliced from pre-mRNAs. The third part consists of some auxiliary sequences created by joining DNAs according to the order described in ‘join()’ or ‘complement()’ statements. Although not displayed in the final alignment result, the third part is used in aligning to assure correctness of the spacial order of the result.
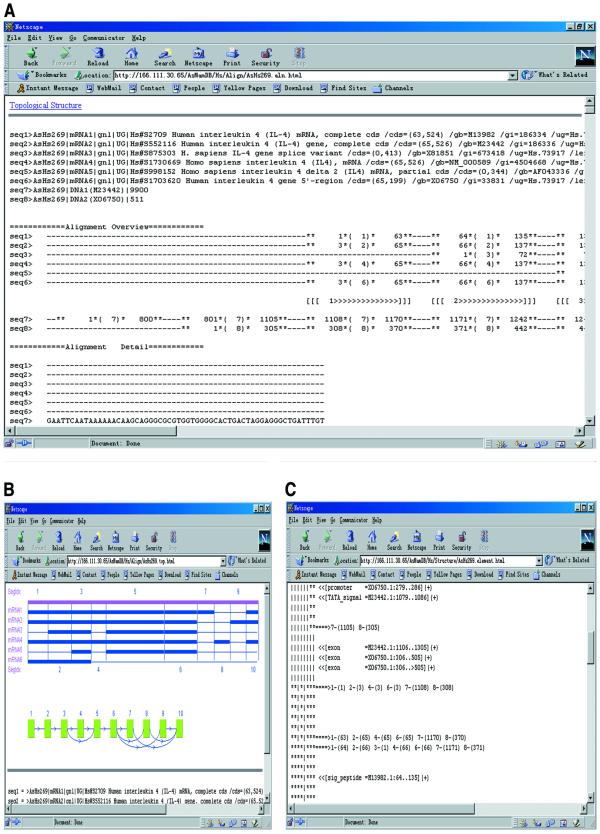
An overview is created for each alignment to illustrate the alternative splicing patterns more clearly. Also, a topological structure graph is extracted from the alignment, to help the use of graph theories in the study of alternative splicing pattern. The overview and the topological structure can be displayed by a Java applet. Figure 1A and B show the alignment of IL4 gene in humans, its overview and the topological structure.
Figure 1.
Splicing patterns and gene structures illustrated by AsMamDB. (A) The alignment of mRNAs and DNAs of IL4 gene of human. (B) Overview of the alignment and the topological structure extracted from the alignment. (C) Gene structure, i.e. the distribution of exons, introns, regulatory elements and other elements.
Gene structures
To help reveal the relationships among introns, regulatory elements and alternative splicing patterns, functional elements annotated in GenBank, such as exons, introns, promoters and enhancers, are designated in the overviews of alignments according to their location information. This results in a gene structure graph, giving a conceptual figure of the distributions of these elements. As an example, the gene structure of human IL4 gene is shown in Figure 1C.
Sequences and classification
All mRNA, DNA, exon, intron, promotor, enhancer and other regulatory element nucleotide sequences are provided in files of FASTA format to facilitate analyses.
Appropriate classifications often serve as a good start point for understanding the mechanism of alternative splicing. All the clusters collected in AsMamDB are classified according to their locations in chromosome and expression tissues, etc. The category lists are part of the database.
USING THE DATABASE
Generally speaking, there are two ways to query the database. One is a keyword query, and the other is a similarity query. In the former case, keywords such as accession number and gene name, etc., can be submitted. The clusters of selected species that contain these keywords will be returned. In the latter case, one nucleotide sequence can be submitted. The submitted sequence will be compared with the representative sequences in the database by BLAST (6). The clusters that contain the most similar sequences will be returned. The query result is linked to topological graphs, alignments and various files of AsMamDB. A number of Internet distributed data sources such as GenBank, UniGene and MEDLINE are linked to provide some other detail information. The human genes can also be accessed through chromosome figures. By clicking a specific region of a chromosome, alternatively spliced genes in that range will be listed.
A simplified version of Asalign, the multiple alignment tool for alternative splicing, can be accessed through a web-interface. When users submit sequences in FASTA format, the tool will give an overview of their alignment. This may be of help in obtaining more information.
Detailed instructions on the database usage can be found at the database homepage.
FUTURE DIRECTIONS
We are planning to classify the introns and retrieve the branch sites of introns. The database will be extended to predict alternatively spliced transcripts. We are also going to classify the clusters according to their gene structures, associate the structure with introns and regulatory elements, and locate all the alternatively spliced genes on the genome. Alternatively spliced genes of other species of mammals will be collected. All errors in the database will be corrected step-by-step. Apart from the errors discussed in Data Collection, another type of error is caused by the greedy heuristic algorithm adopted by Asalign. The correction of this type of error depends on the improvement of Asalign. The ultimate goal of the work is to provide a database with a set of tools. The database not only gives information about known alternatively spliced genes, but also predicts the gene’s splicing and expression patterns.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online, including some specific information on alignment, topological structure of genes and expression of gene structures.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to those who have helped us. In particular we thank Professor Liang Ji, Zhirong Sun, Zhijie Chang and Anming Meng who have discussed the data representation with us and given a lot of useful advice. The work was supported by National Nature Science Foundation of China (Grant No. 69872018, 69935020), the Fundamental and Innovative Research Fund of Tsinghua University and the Technical Project of Beijing.
References
- 1.Dralyuk I., Brudno,M., Gelfand,M.S., Zorn,M. and Dubchak,I. (2000) ASDB: database of alternatively spliced genes. Nucleic Acids Res., 28, 296–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James Kent W. and Zahler,A.M. (2000) The Intronerator: exploring introns and alternative splicing in Caenorhabditis elegans. Nucleic Acids Res., 28, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheeler D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 28, 10–14. Updated article in this issue: Nucleic Acids Res. (2001), 29, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul S.F., Madden,T.L., Schaeffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.