Abstract
SH-SY5Y neuroblastoma cells were examined to determine changes in protein expression following exposure to the organophosphate paraoxon (O,O-diethyl-p-nitrophenoxy phosphate). Exposure of SH-SY5Y cells to paraoxon (20 µM) for 48 h showed no significant change in cell viability as established using an MTT assay. Protein expression changes from the paraoxon-treated SH-SY5Y cells were determined using a comparative, sub-proteome approach by fractionation into cytosolic, membrane, nuclear, and cytoskeletal fractions. The fractionated proteins were separated by 2D-PAGE, identified by MALDI-TOF mass spectrometry, and expression changes determined by densitometry. Over four-hundred proteins were separated from the four fractions and sixteen proteins were identified with altered expression ≥ 1.3-fold including heat shock protein 90 (−1.3 fold), heterogeneous nuclear ribonucleoprotein C (+2.8 fold), and H+ transporting ATPase beta chain (−3.1 fold). Western blot analysis conducted on total protein isolates confirmed expression changes in these three proteins.
Introduction
Organophosphate insecticides (OPs) are widely used in agriculture and domestic applications but their use continues to be a public safety concern because OPs share chemical traits, properties, structure and biological mechanisms with nerve gas agents. These concerns have persisted for decades with reports of neurotoxicity and non-neurologic toxicities associated with acute and chronic exposure to OPs. The neurotoxicity observed following exposure to OP compounds is typically initiated by the covalent modification of acetylcholinesterase (AChE) forming OP-AChE adducts. The OP-AChE adduct is relatively stable and unable to hydrolyze its substrate acetylcholine resulting in an increase in this neurotransmitter followed by overstimulation of acetylcholine receptors (AChR’s) (1–3).
Recently, certain reactive OP compounds have been found to covalently and non-covalently interact with protein targets other than AChE and may be the causative step in other toxic responses. The involvement of OPs at nicotinic and muscarinic AChRs and non-cholinergic protein targets has been reported with increasing frequency (4–12). However, no unified or integrative approach has been conducted to identify possible alterations in protein expression following exposure to OPs despite mounting evidence that OPs interact with a wide range of proteins in addition to AChE (5, 7, 13–14) and may modify many cellular processes. This study seeks to preliminarily identify the protein expression changes resulting from exposure to the representative OP, paraoxon.
Paraoxon (O,O-diethyl p-nitrophenylphosphate; Figure 1), the bioactive metabolite of parathion, is well-known to covalently modify and inhibit AChE, and has been used to investigate and identify toxicologically relevant targets of OPs (11, 14–15). In neuronal and glial cells, alterations in signaling molecules and pathways (12, 16–18), mitochondrial integrity and ATP production (18–21), and stress responses (16, 22) were observed following OP exposure.
Figure 1.
Chemical structure of paraoxon.
In order to better understand how cells respond to OP exposure and gain insight into the mechanisms involved in OP toxicity, the SH-SY5Y human neuroblastoma cell line was selected for study since they express AChE, the nicotinic AChR (23) and also many neuron-specific enzymes including dopamine hydroxylase, tyrosine hydroxylase, and aromatic L-amino acid decarboxylase. The in vitro toxicity of paraoxon has been demonstrated in a variety of systems including the SH-SY5Y cell line (19, 24–26) and AChE present in SH-SY5Y cell culture is inhibited within minutes at sub-micromolar levels. However, loss of SH-SY5Y cell viability required incubations with millimolar concentrations for 24–48 h suggesting cellular toxicity occurs by non-cholinergic events well after AChE inhibition (25). In this study, SH-SY5Y cells were treated with paraoxon and a comparative, sub-cellular fractionation proteomic approach was used to identify changes in protein expression after 48 h.
Material and Methods
Paraoxon Synthesis
Equimolar amounts of p-nitrophenol and diethyl chlorophosphate were dissolved in diethyl ether at 0 °C to which triethylamine (Et3N, 1.1 equiv) was added. The reaction mixture was stirred at 25 °C for 8 h whereupon the mixture was filtered, and the residue chromatographed on silica gel (EtOAc:hex, 1:1). Paraoxon was isolated as a pale-yellow oil. The structure was confirmed by comparison with authentic material and characterized by NMR and MS. The chemical purity was determined to be greater than 98% by LC-MS. Paraoxon was stored neat at 5 °C or as a solution in acetonitrile at 5 °C prior to use.
Culture of SH-SY5Y Cells
Human neuroblastoma cells (SH-SY5Y) were obtained from American Type Culture Collection (Rockville, MD) and cultured in Dulbecco's Modified Eagle's medium and Ham's F-12 nutrient mixture (DMEM/F12) (GIBCO BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) (Hyclone; Thermo Fisher Scientific Inc., Waltham, MA), 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM L-glutamine in a CO2 incubator maintained at 5% CO2 and 37 °C. The media was changed every two days. Cells were allowed to reach 80% confluence before exposure to paraoxon.
SH-SY5Y Exposure to Paraoxon
A 10 mM stock solution of paraoxon was prepared in 100% acetonitrile. Experimental concentrations were prepared by dilution of stock solution in DMEM/F12 medium, 1% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM L-glutamine. The FBS in the SH-SY5Y cell culture media was reduced from 10% to 1% 24 h prior to paraoxon treatment. Original culture media was replaced with paraoxon culture media and allowed to incubate at 37 °C for the various time points described below. Cells were thoroughly rinsed with ice cold PBS prior to cell collection. Cells were then transferred to 1.5 mL storage tubes and stored at −80 °C prior to protein extraction.
Cell Viability Assay
Cell viability was determined using an MTT assay kit (Roche Applied Science, Indianapolis, IN) with ~ 0.25 × 105 cells/mL per well in 96-well culture plates using eight replicates per treatment group. Cells were exposed to paraoxon concentrations ranging from 10 nM – 100 µM for 24, 48, or 72 h. As a positive control for cell death, wells of untreated cells were separately exposed to 2% Triton X-100 solution in assay medium. Culture media was removed and 100 µL of fresh medium containing paraoxon (various concentrations) or Triton-X-100 was added to each well (n = 8 for each paraoxon concentration and Triton-X-100).
After incubation for appropriate time points, 10 µl of the supplied MTT labeling reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was added to each well. Plates were incubated with MTT for 4 h and 100 µl of solubilizing solution was added to each well. Plates were incubated overnight before being measured at 575 nm (formazan) using a microplate reader. Viability was determined by comparing the absorbance readings of the wells containing paraoxon-treated cells with those of the vehicle (0.1% acetonitrile) treated cells. Statistical significance was determined by analysis of variance (ANOVA) followed by a Dunnet's Post Hoc test, p < 0.05 (n = 8 samples per group).
Subcellular Protein Fractionation
For the proteomic analysis, SH-SY5Y cells were cultured and exposed to 20 µM paraoxon in culture media for 48 h as described. To overcome the low abundance of certain proteins found in the 2-D gels an increase in extraction efficiency and sensitivity was accomplished in the SH-SY5Y proteome by extraction according to their subcellular localization (cytosolic, membrane bound, nuclear, and cytoskeletal) using a detergent based proteome extraction kit (Calbiochem ProteoExtract Subcellular Proteome Extraction Kit; EMD Chemicals Inc., Gibbstown, NJ). The kit contains four different extraction buffers that exploit the differential solubility of certain cell compartments to separate proteins into differing subcellular fractions. Western Blot analyses and 1-D SDS PAGE were used to estimate the extraction efficiency of the subproteome extraction (Supporting Information, Figure S-1). After extraction, the protein concentration was determined for each fraction using a Bradford protein assay and samples were stored at −80 °C. To concentrate proteins and remove any impurities such as salts and detergents that may interfere with 2-D SDS PAGE separation, proteins were precipitated (Calbiochem ProteoExtract Protein Precipitation Kit; EMD Chemicals Inc., Gibbstown, NJ), and 200 µg of the precipitated proteins were resuspended in 185 µL of 7 M urea, 2 M thiourea, 1% (w/v) ASB-14 detergent, 40 mM Tris base, and 0.001% bromophenol blue.
Two-Dimensional (2-D) Electrophoresis and Proteomic Analyses
Proteins were separated using two-dimensional gel electrophoresis and changes in spot density between the control and paraoxon exposure group (n = 3 for each group) were determined using 200 µg of total protein from each treatment group. Total protein from subcellular fractions was loaded onto an 11 cm immobilized pH gradient strip (IPG; pH 3–10; Bio-Rad Laboratories, Hercules, CA) and the samples placed in a 12-well rehydration tray. Protein samples were allowed to enter the strip for 60 min whereupon a layer of mineral oil was then placed over the strips and left in the tray 16 h. The IPG strips were next subjected to isoelectric focusing (250 V for 15 min then ramped to 8,000 V for 2–5 h depending on sample content). The voltage was limited by a 50 µA/gel current restriction. After reaching 8000 V, the samples were focused for an additional 5 h. In preparation for the second dimension, sulfhydryl groups were reduced with dithiothreitol (DTT) and alkylated using iodoacetamide. The IPG strips were placed in an 11 cm rehydration tray containing 1 mL of reducing solution (3.6 g urea, 0.2 mg DTT, 2.0 mL 10% SDS, 2.5 mL 1.5 M Tris (pH 8.8), 2.0 mL glycerol, and 1.0 mL H2O) for 30 min whereupon the reducing solution was decanted and the channels were refilled with alkylating solution (3.6 g urea, 0.25 g iodoacetamide, 2.0 mL 10% SDS, 2.5 mL 1.5 M Tris (pH 8.8), 2.0 mL glycerol, 1.0 mL H2O, and 5 mg bromophenol blue) for 30 min. After the IPG strips were equilibrated, they were positioned in a 12% bis-Tris Criterion gel (Criterion precast gel; Bio-Rad Laboratories, Hercules, CA) and overlaid with molten agarose. Gels for all treatment groups were run simultaneously in a Bio-Rad Criterion dodeca cell (Bio-Rad Laboratories, Hercules, CA) for 1.5 h at 20 °C at 150 V.
2-D Gel Staining and Image Analysis
2-D gels were removed from the gel cassette, stained with Bio-Safe Coomassie G-250 stain (Bio-Rad Laboratories, Hercules, CA) for 1 h and destained using distilled water. Coomassie stained gel images were recorded, digitized by densitometry, and analyzed using Bio-Rad PDQuest 2-D Gel Analysis Software (v 6.2). The comparative gels from each cellular fraction (n = 3) for the control and paraoxon treatment sets were grouped together. The average spot density for each protein spot was then used to compare the spot density among control and paraoxon treatment groups for each cellular fraction. Protein spots that demonstrated a statistically significant change of ±1.3 fold or greater between control and paraoxon treatment groups were excised and identified by mass spectrometry. Significance was determined by t-test using Bio-Rad PDQuest 2-D Gel Analysis software (Bio-Rad Laboratories, Hercules, CA).
In-gel Trypsin Digestion and Sample Preparation for Mass Spectrometry
Protein spots were excised from Coomassie stained gels using a ProteomeWorksTM Spot Cutter (Bio-Rad Laboratories, Hercules, CA) and gel plugs transferred to a 96-well plate. Gel plugs were first destained by washing for 30 min in 200 µl of 50% H2O/50% acetonitrile, dried in a non-humidified oven (37 °C) for approximately 2 h, and then digested with 50 µl trypsin solution (12.5 ng trypsin/25 mM NH4HCO3 in 50% acetonitrile). The sample was incubated at 4 °C in the trypsin solution for 20 min, excess trypsin was removed and the sample overlaid with 30 µL of 25 mM NH4HCO3 and allowed to incubate overnight at 37 °C. The supernatant containing peptides was removed, placed in a fresh 0.5 mL Eppendorf tube and extracted twice with 0.1% trifluoroacetic acid/60% acetonitrile (2 × 200 µL) and concentrated to 10 µL. The extracts were applied to ZipTips (ZipTip® Pipette Tips, P10 C18; Millipore, Billerica, MA) and following the manufacturer’s instructions, the tryptic peptides were eluted using 60% acetonitrile/0.2% formic acid (4 µL). For MALDI-TOF analysis, 10 µg-cyano hydroxycinnamic acid was dissolved in 1 mL of acetonitrile-water (1:1), spiked with 2 µL of Bruker peptide calibration standard (#206195 Bruker Daltonics Inc., Billerica, MA) and mixed 1:1 with tryptic peptide before samples were analyzed.
Protein Identification by Mass Spectrometry
Peptide mass spectra were recorded in positive ion mode on a Voyager-DE Pro MALDI time-of-flight mass spectrometer (Applied Biosystems, Foster City, CA). Mass spectra were obtained by averaging 500 individual laser shots. A calibration standard mixture (#206195 Bruker Daltonics Inc., Billerica, MA) containing seven different peptide standards was used as internal calibration standards. The selected proteins were identified by MALDI-TOF mass spectrometry and Mascot protein database search. Acceptable Mascot database criteria used in this study were: (a), the protein match must have a MOWSE (Molecular Weight Search) score of at least 56 to be considered significant (p < 0.05), (b) average sequence coverage of approximately 25%, and (c) experimental molecular weights and isoelectric points (pIs) must be in close approximation to the protein’s actual molecular weight and/or pI.
Database Search
The peptide mass data was used to identify proteins using a Mascot peptide mass fingerprint database search (Matrix Science; www.matrixscience.com) against the SwissProt/Uniprot database (database is developed by the SWISS-PROT groups at the Swiss Institute of Bioinformatics and The European Bioinformatics Institute). The following search parameters were used: SwissProt database; taxonomy, homo sapiens (human); maximum missed cleavages, 1; variable modifications, carbamidomethyl (C) and oxidation (M); and peptide tolerance, ± 0.2 Da. The Mascot search engine was used to calculate a theoretical molecular weight, isoelectric point, and search (MOWSE) score for each experimental peptide mass spectra (protein score in −10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 56 were considered significant (p < 0.05) (Supporting Information, Table S-4).
Western Blot Analysis
For the western blot analysis, SH-SY5Y cells were treated with 20 µM paraoxon and collected at 12–72 h. Cells were homogenized in lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM ethylenediamine-tetraacetic acid (Na2EDTA), 1 mM ethylene glycol-O, O'-bis(2-aminoethyl)-N, N, N', N'-tetraacetic acid (EGTA), 1% Triton-X, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate and 1 mM Na3VO4. Additions were made to give final concentrations of 0.5% Na-deoxycholate, 0.5% SDS, 1 µM okadaic acid, 1 mM phenylmethylsulfonyl (PMSF), 0.1 mg/mL benzamidine, 8 µg/mL calpain inhibitors I and II and 1 µg/mL each leupeptin, pepstatin A and aprotinin. Homogenates were sonicated and vortexed for 5 min before centrifugation at 10,000 × g for 20 min. The remaining insoluble pellet was discarded and the total protein concentration of the supernatant was then determined by Bradford protein assay (27). Proteins were first separated by SDS-PAGE using 4–12% gradient bis-Tris Criterion precast gel (Bio-Rad Laboratories, Hercules, CA) and transferred to PVDF membranes for immunoblotting. Gels were transferred to Immun-Blot PVDF membrane (Bio-Rad Laboratories, USA) using Criterion transfer cells with plate electrodes (Bio-Rad Laboratories, Hercules, CA) for 60 min at 100 V and blocked in 5% milk in TBST (Tris-buffered saline, 0.1% Tween 20) overnight at 4 °C. Membranes were washed in TBST and then incubated overnight at 4 °C with the specific primary antibody. The primary antibodies (Abcam Inc. Cambridge, MA) were: (a) mouse monoclonal antibody for HSP 90 (antibody S88; ab1429 @ dilution 1:200); (b) mouse monoclonal antibody for hnRNP (C1/C2) (antibody 4F4; ab10294 @ dilution 1:1000); (c) mouse monoclonal antibody for ATP synthase beta subunit (antibody 3D5; ab14730 @ dilution 1:1000) and; (d) rabbit polyclonal antibody against GAPDH (ab9485 @ dilution 1:2000) (Supporting Information, Figure S-2). Membranes were washed in TBST and incubated with the appropriate secondary antibody for 2 h. Secondary antibodies were purchased from Vector Laboratories (Vector Laboratories, Inc., Burlingame, CA) HRP-conjugated anti-rabbit, PI-1000 (dilution 1:2000) and HRP-conjugated anti-mouse, PI-1000 (dilution 1:2000). Membranes were then washed in TBST before imaging with the Fuji LAS-3000 CCD based imaging system (FujiFilm Life Science, Valhalla, NY). Membranes were re-probed with GAPDH to confirm equal protein loading. Band pixel intensity was measured using the Fuji LAS-3000 software. Band intensities from control and treated groups were normalized to GAPDH. Statistical significance was determined by one-way ANOVA followed by a Dunnets Post Hoc, p < 0.05 was considered significant (n = 3 samples per group), using GraphPad Prism 4.0 software (GraphPad Software, La Jolla, CA).
Results
SH-SY5Y Cell Viability
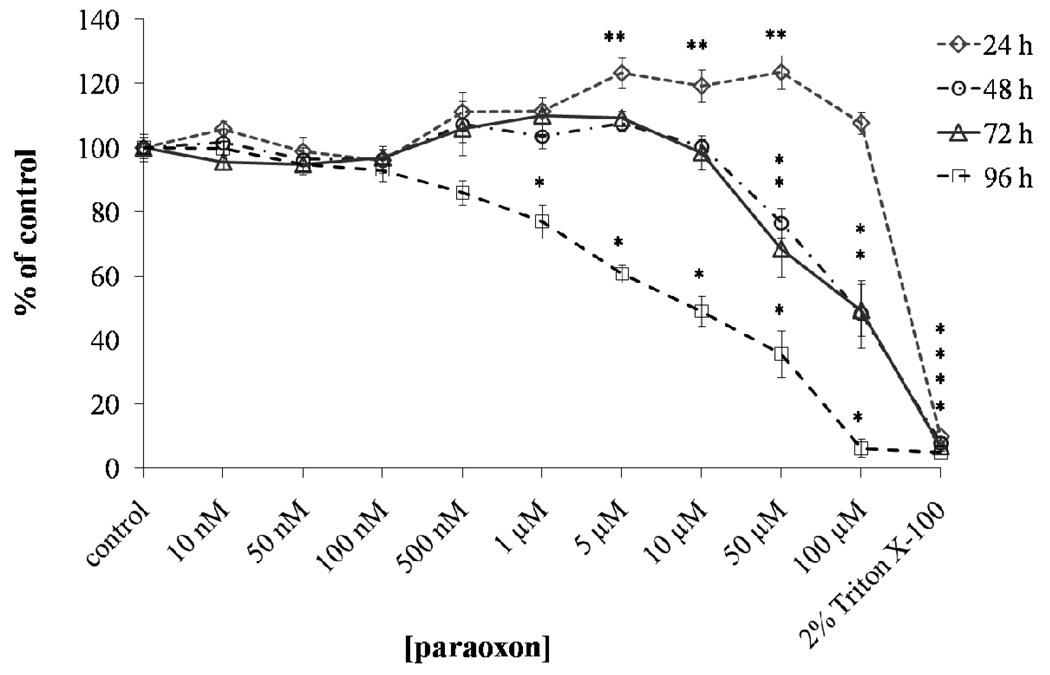
To obtain the optimum conditions for the proteome analysis, the tolerance of the SH-SY5Y cell line to variable paraoxon concentrations was determined using a MTT colorimetric assay at several time points. MTT analysis at 24 h showed that exposure to paraoxon results in no significant decrease in cell viability over the concentration range tested from 10 nM to 100 µM. At the 48 h and 72 h timepoints, SH-SY5Y cell viability dropped to ≤ 80% of control when the paraoxon concentration exceeded 10 µM. At 96 h exposure, significant cytotoxicity (approx. 70% of control) occurred as low as 1 µM and only 5% of control cell viability remained at 100 µM paraoxon exposure (Figure 2). Based on the MTT assay, an exposure of 20 µM paraoxon for 48 h was used for our proteomic studies since this was below the threshold where reduction in cell viability was observed. No loss in cell viability at 20 µM paraoxon was observed at 48 h of exposure as determined by trypan blue exclusion (See: Supporting Information, Figure S-3).
Figure 2.
Cytotoxicity of paraoxon to SH-SY5Y cells. Cells were treated with paraoxon (10 nM – 100µM) for 24, 48, 72 and 96 h or no paraoxon (control) and the cell viability estimated by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) colorimetric assay. Results are presented as % of vehicle control, determined by comparing the absorbance readings of the wells containing the paraoxon treated cells with those of the vehicle (0.1% acetonitrile) treated cells. Triton X-100 was used as a positive control (= 100% cell death). Significance determined by one-way ANOVA followed by Dunnet’s post hoc test (* significant decrease; ** significant increase) (p < 0.05; n = 8).
Proteomic Analysis
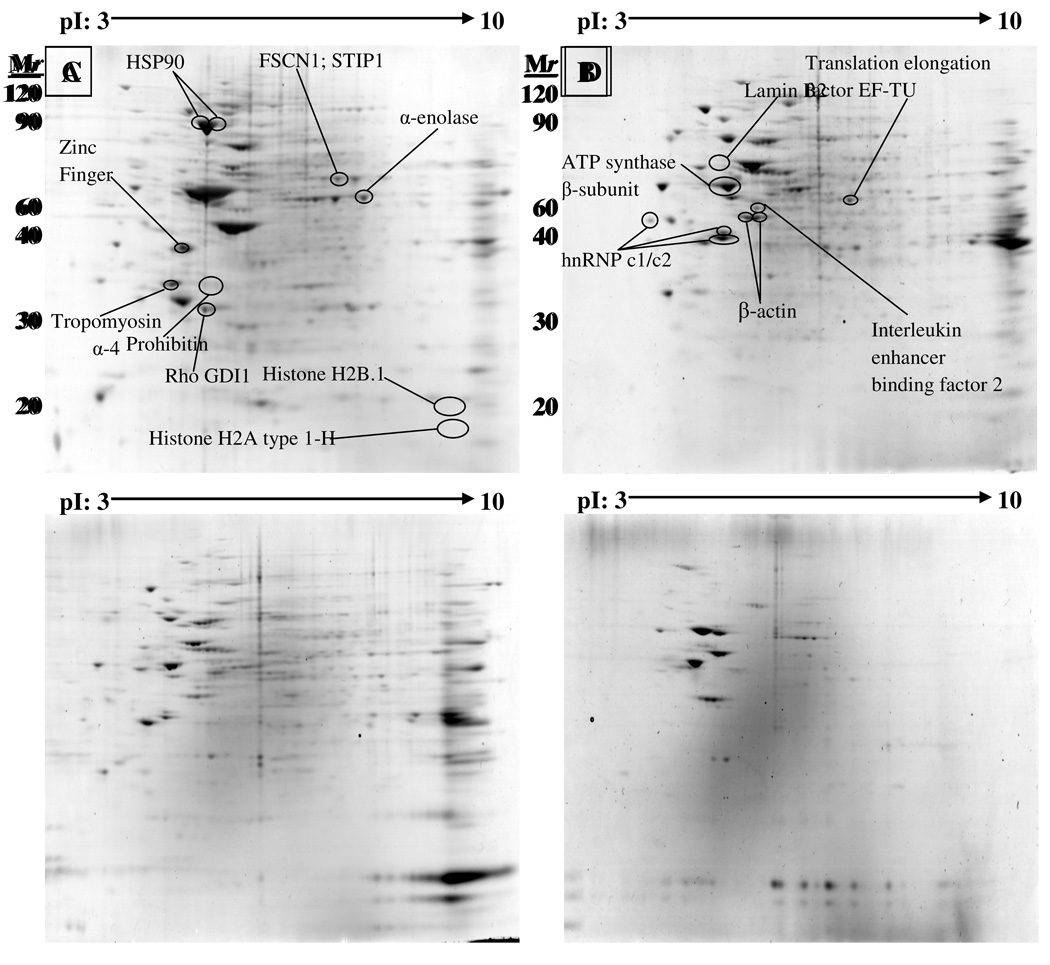
Changes in protein expression levels in SH-SY5Y cell cultures were examined following exposure to 20 µM paraoxon for 48 h. A comparative, sub-fractionation proteomic analysis was utilized to identify changes in protein expression yielding a number of identifiable proteins in both control and paraoxon exposed cells. Representative 2-D gel maps of overall protein expression from the cytosolic, membrane, nuclear and cytoskeletal fractions from SH-SY5Y cells (Figure 3) showed that proteins were widely distributed across the pI range between 20 and 120 kDa. Protein spots were analyzed from three pairs of gels from independent experiments using Bio-Rad PDQuest 2D analysis software. Some proteins were represented by multiple spots distributed across a range of isoelectric points (pI), and certain proteins exist in different cellular compartments and therefore, may be found in one or more cellular fractions. The 2-D gels from paraoxon-treated samples were compared to control gels and the protein spots displaying an expression change of ±1.3 fold or greater were excised from the gel and identified by MALDI-TOF mass spectrometry using peptide mass fingerprint searches (Table 1). Mascot database search scores (values > 67 represent 95% confidence or higher), calculated isoelectric points (pI), molecular weight (Mr), and sequence coverage for each protein is included for each protein (Table 1).
Figure 3.
Representative two-dimensional gel maps of overall protein expression in SH-SY5Ycells. Protein expression from (A) cytosolic fraction, (B) membrane fraction, (C) Nuclear fraction, and (D) cytoskeletal fractions are shown. The majority of the proteins are located between 20 and 120 kDa, and are widely distributed across the entire pI range (pI: 3–10). Many proteins are identified as multiple spots in a single fraction, suggesting possible post-translational modifications such as glycosylation or phosphorylation. Circled spots displayed an expression change of ±1.3 fold or greater. Protein spots were excised from the gel and identified by MALDI-TOF mass spectrometry and a peptide mass fingerprint search.
Table 1.
Proteins differentially expressed ≥ 1.3-fold in SH-SY5Y cells, search values and the identification measures obtained from 2D-PAGE.
| Spot ID |
Accession no. | Protein Name | Mascot Score |
Sequence Coverage |
Calculated Mr/pI |
Relative Fold Change |
|---|---|---|---|---|---|---|
| Cytosolic Fraction | ||||||
| 1101 | P67936 | tropomyosin alpha-4 chain | 76 | 27% | 32093/4.86 | 2.5 |
| 1105 | BC021092 | zinc finger, FYVE domain containing 19 | 73 | 24% | 37170/4.88 | 2.6 |
| 2001 | P52565 | rho GDI 1 | 71 | 28% | 29470/5.01 | 1.5 |
| 2703 | P08238 | heat shock protein 90-beta | 89 | 22% | 84415/5.07 | −1.7 |
| 2705 | BC000987 | heat shock protein 90kDa alpha (cytosolic) | 104 | 30% | 85430/4.95 | −1.3 |
| 7501 | Q96IH1; Q3ZCU9 | FSCN1 protein; STIP1 protein (mix) | 130 | 18%, 18% | 55975/7.05 | −1.5 |
| 8301 | Q8WU71 | alpha-enolase | 80 | 26% | 57339/7.84 | −2.3 |
| Membrane Fraction | ||||||
| 104 | BC089438 | heterogeneous nuclear ribonucleoprotein C (C1/C2) (hnRNP c1/c2) | 89 | 38% | 37775/4.97 | 2.8 |
| 1101 | BC089438 | heterogeneous nuclear ribonucleoprotein C (C1/C2) (hnRNP c1/c2) | 113 | 38% | 38828/4.98 | 2.8 |
| 201 | BC089438 | heterogeneous nuclear ribonucleoprotein C (C1/C2) (hnRNP c1/c2) | 93 | 37% | 41614/4.04 | 1.6 |
| 1402 | A33370 | ATPase synthase beta chain, mitochondrial | 211 | 51% | 48974/5.04 | −3.1 |
| 2201 | Q53FG3 | interleukin enhancer binding factor 2 variant (Fragment) | 80 | 30% | 44445/5.59 | 2.1 |
| 1204 | BC012854 | actin, beta | 116 | 38% | 42084/5.41 | 1.7 |
| 2204 | BC012854 | actin, beta | 74 | 29% | 42458/5.79 | −3.2 |
| 6301 | S62767 | translation elongation factor EF-Tu | 122 | 35% | 47300/7.70 | 1.5 |
| Nuclear Fraction | ||||||
| 2101 | I52690 | prohibitin | 76 | 30% | 31765/5.80 | −1.6 |
| 9004 | M60756 | histone H2B.1 | 70 | 53% | 13983/9.85 | −3.3 |
| 9005 | Q96KK5 | histone H2A type 1-H | 69 | 33% | 12120/9.46 | −2.2 |
| Cytoskeletal Fraction | ||||||
| 4702 | BC006551 | lamin B2 | 77 | 24% | 67443/5.51 | 1.4 |
The cellular subfractionation approach led to the separation of distinct proteins into four groups: (a) cytosolic, (b) membrane, (c) nuclear, and (d) cytoskeletal. (a) The cytosolic fraction afforded 117 total spots that were resolved of which 21 were identified by MALDI and 13 were found to be non-redundant proteins. Spot density analysis showed differential expression for 7 of the 13 total proteins identified in the cytosol, 3 proteins were up-regulated and 4 proteins were down-regulated (including the α- and β-HSP 90 subunits) (Table 1). (b) The membrane fraction yielded 132 total resolved spots of which 21 were identified by MALDI and 15 were found to be non-redundant. Spot density analysis resulted in 8 differentially-expressed proteins of which 6 increased in expression and 2 decreased in expression. Three of the up-regulated proteins were hnRNP (C1/C2), which varied in expression change from 1.6 to 2.8-fold. Two forms of β-actin showing disparate increase (1.7-fold) and decrease (3.2-fold) in expression were identified in response to paraoxon treatment. (c) The nuclear fraction led to 132 resolved protein spots with 8 protein identifications by MALDI. Seven of the nuclear proteins were non-redundant and 3 of these proteins showed significant decrease in expression from 1.6 to 3.3-fold (prohibitin). (d) The cytoskeletal fraction was resolved into 33 protein spots of which 11 proteins were identified (9 non-redundant proteins) including lamin-B2 that increased by 1.4-fold in response to paraoxon exposure.
Western Blot Analysis
Western blot analysis was conducted for three proteins that changed significantly in expression following exposure to paraoxon: ATP synthase β-subunit (membrane fraction), hnRNP c1/c2 (membrane fraction), and HSP 90 (cytosolic fraction). For western blot analyses (See: Supporting Information, Figure S-2), SH-SY5Y cell cultures were exposed to 20 µM paraoxon in culture media for 12–72 h, but were not fractionated prior to blot analysis. A significant increase in expression for all three proteins was observed at 24 h post-exposure. The expression of ATP synthase β-subunit was increased in the first 24 h by 1.66-fold, however, this value dropped to 1.35-fold at 72 h relative to control. Likewise, HSP 90 protein levels increased to 2.6-fold at 24 h but dropped to 2.0-fold from 48–72 h. For hnRNP c1/c2, the expression level sustained a 1.2 to 1.3-fold increase in the 24–72 h time period following exposure to paraoxon.
Discussion
The current study was undertaken in order to gain further insight into the mechanisms involved in paraoxon cytotoxicity through alterations in protein regulation. For our proteomic analysis, the cells were treated with 20 µM paraoxon for 48 h since the MTT assays indicated there was no significant decrease in cell viability at 72 h. Cell viability was adversely affected at paraoxon concentrations greater than 500 nM at the 96 h time point, however, SH-SY5Y cells showed normal viability with paraoxon concentrations as high as 100 µM at 24 h, which is consistent with earlier reports with undifferentiated and differentiated cells (21, 25, 28). Although cell viability was unaffected, treatment with 20 µM paraoxon inhibits acetylcholinesterase in SH-SY5Y cell lines within one hour (25, 28). Interestingly, 24 h exposure to 5, 10, and 50 µM paraoxon appeared to have a modest proliferative effect on cell growth as indicated by an increase in absorbance at these time points. This observation is contrary to that found for human hepatoma HepG2 cells that show a reduction in cell proliferation after exposure to 100 µM methyl paraoxon (29).
A sub-proteome fractionation approach was undertaken to facilitate the identification of low abundance proteins and aid separation of individual proteins by their locus of compartment. These advantages can be offset, in part, by carryover of certain proteins that are distributed into more than one cellular compartment thereby complicating measures in the expression changes. Sixteen proteins changed in expression level > 1.3-fold following exposure of SH-SY5Y cells to 20 µM paraoxon at 48 h (Table 1) and these proteins were found to be located in the four different cellular compartments (Figure 3). This is a significant finding indicating that paraoxon exposure results in cell-wide changes in protein expression. The total number of proteins that changed in expression roughly correlates with the fifteen genes that were identified by gene array in SH-SY5Y cells treated with paraoxon (3 µM and 30 µM) for 24 h (17).
Based on our sub-proteome analysis, our results indicate that paraoxon exposure leads to protein expression changes throughout cell compartments and not just AChE and serine hydrolase-type proteins. Good to excellent (18–53%) sequence coverage was determined by MALDI MS analyses and all protein spots were in good agreement with the theoretical molecular weight and pI. As such, the proteomic analysis identified a number of proteins that may be sensitive to paraoxon exposure, only one of which had been previously uncovered during a gene expression study (17). A few proteins changed in expression three-fold or greater including ATPase (membrane fraction), beta actin (membrane fraction) and histone H2B.1 (nuclear fraction). During the tryptic peptide analysis by mass spectrometry, no evidence was obtained that proteins were directly phosphorylated by paraoxon although the possibility that covalent modification may have occurred cannot be excluded.
To address the possibility that overall protein expression changes due to paraoxon exposure may differ from those determined using a subproteome fractionation approach, western blot analysis was conducted for three of the identified proteins using a total protein extraction. SH-SY5Y cells were exposed to 20 µM paraoxon and the cells collected at several different time points to investigate dynamic expression patterns for HSP 90, hnRNP c1/c2 and the ATP synthase β-subunit, which displayed large changes in expression as determined by the 2-D gel analysis.
HSP 90 showed a decrease in expression of 1.3 to 1.7-fold in the cytosolic fraction following paraoxon exposure but when analyzed by Western blot analysis the expression change showed an increase 2.0 to 2.6-fold. This discrepancy has not been resolved although it is possible that isolated cytosolic HSP 90 expression differs from protein located throughout the cell. HSP 90 has been reported to play a role in various cellular processes including signal transduction, protein folding, and protein degradation (30–31). The results observed by Western blot analysis agrees with previous studies using cultured PC-12 cells, which found that HSP90 was induced by a variety of different pesticides, including the organophosphate chlorpyrifos, and believed to be a cellular protection response to oxidative stress (32).
Following paraoxon exposure, the heterogeneous nuclear ribonucleoprotein C (C1/C2) (hnRNP c1/c2) was up-regulated from 1.6 to 2.8-fold when analyzed from the membrane fraction and validated by Western blot by a 1.2 to 1.3-fold increase from total protein. HnRNP c1/c2 belongs to the RNA-binding protein family (33) and is primarily located in the nucleus but may also be found in the cytoplasm. It was therefore surprising that hnRNP c1/c2 was found in the membrane fraction although the detergents used to sub-fractionate the proteome may have played a role in distribution to this compartment. HnRNP c1/c2 may be associated with pre-mRNA processing, metabolism and transport (34), cellular homeostasis (34), plays a role in cell cycle regulation (35), and hypothesized to be associated with disease pathways such as cancer (36–37) and schizophrenia (38). Down-regulation of hnRNP c1/c2 by siRNA in HeLa cells resulted in cells hypersensitivity to a variety of cellular stresses including H2O2 (oxidative stress) (34).
Results from our proteomic analysis indicated that the ATPase beta chain demonstrated a 3.1-fold decrease in spot density, which was contrary to the 1.66-fold increase found using Western blot analysis based on total protein at the same time point. Although ATPase expression is clearly altered by paraoxon exposure, it is difficult to account for the reversed responses. The ATP synthase β-subunit is part of a multi-subunit complex found in mitochondria that catalyzes ATP synthesis utilizing an electrochemical gradient of protons across the inner membrane during oxidative phosphorylation (39) and some early reports suggested that mitochondria may be a target for reactive OPs resulting in a decrease in ATP synthesis (21).
The proteomics study was designed to obtain information about changes in protein expression levels in SH-SY5Y cells following exposure to paraoxon. To more accurately identify those changes, a sub-proteome fractionation approach was employed to separate paraoxon-affected proteins based on their cellular compartment. In addition to improved identification accuracy, the fractionation was also envisioned to assist with the analysis of low abundance proteins and/or better understand how paraoxon might alter protein expression changes based on the cellular localization. However, certain inherent problems in the sub-proteome extraction led to difficulties in correlating and validating protein regulation changes as noted for the three proteins selected for Western blot analysis. For example, many proteins likely appear in more than one of the cell fractions, extraction efficiency can be poor, and some proteins may exist in different post-translationally modified forms in different cellular compartments. Therefore, measures of sub-proteome expression changes may not correlate well with whole cell expression changes, and further interpretation should take the limitations of each experimental approach into account.
Although reactive OPs like paraoxon are known to inhibit AChE to initiate organism level toxicity, it is well known that other individual proteins and cellular pathways are affected by interaction with OPs and may or may not be related to AChE inhibition (13, 28). Further, the concentration of OP required to inhibit AChE is typically much less than the lethal dose of OP indicating that other cellular pathways are affected by OP exposure (25). It is likely that, as AChE inhibition occurs, cells respond by initiating responses such as oxidative stress. Many environmental toxicants induce stress responses in affected cells and these stress responses can be broken down into several broad categories, such as oxidative stress, DNA damage, heat shock (chaperone), and inflammation, among others (40). Certain OPs have been shown to cause oxidative damage in cell culture and animal model systems, as well as oxidative stress-like symptoms in human populations (41). Protein expression changes uncovered in this study suggest that exposure to OPs may induce a cellular stress response. Whether the oxidative stress response elicited by OPs is due to downstream effects of AChE inhibition or to other direct targets of OP phosphorylation is not presently known.
Tropomyosin and actin both changed in regulation following exposure to paraoxon, and have been previously shown to have altered regulation in response to several toxicants, including the herbicide atrazine (42). Likewise, exposure of rat aortic smooth muscle cells to homocysteine, which causes oxidative stress, affected protein expression of tropomyosin, actin, and prohibitin (43), although prohibitin has also been shown to be up-regulated following oxidative stress (44). In the proteomic analysis, FSCN1, actin, and tropomyosin all changed in expression in response to paraoxon exposure and are all implicated in cytoskeletal rearrangement pathways (45) and although this pathway was not observed in the course of this exposure. A decrease in the expression of FSCN1 has been shown to be associated with the suppression of cellular proliferation, possibly indicative of slowing down of cell division (46). Alpha enolase, which was down-regulated 2.3-fold in SH-SY5Y at 48 h following exposure, was briefly examined to see if this enzyme interacts directly with paraoxon. However, the activity of recombinant alpha enolase was largely unaffected by paraoxon at concentrations up to 10 mM (data not shown).
In order to fully elucidate the molecular mechanisms involved in OP toxicity, a better understanding is needed of the molecular events following OP exposure. The results from this study indicate that exposure to paraoxon resulted in the altered expression of a number of proteins involved in a diverse array of cellular processes. Further, it was found that protein expression changes may vary between sub-proteome analysis and gene expression analysis (17) and conclusions using either method should be independently validated. The significance of these protein expression changes to the overall paraoxon cytotoxicity remains to be determined and future studies need to be aimed at determining whether these expression changes are a specific result of paraoxon exposure or due to a general cellular stress response.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant UO1-ES016102 (CMT) and R43 ES016392 and U44 NS058229 (ATERIS Technologies LLC). We also received support from the Core Laboratory for Neuromolecular Production (NIH P30-NS055022) and the Center for Structural and Functional Neuroscience (NIH P20-RR015583). We thank Sarah Ulatowski and Saskia Jacobson for experimental assistance and Dr Sandip Bharate for synthesizing and purifying paraoxon.
Footnotes
Supporting Information Available: Efficiency of the Calbiochem Subproteome extraction kit on SH-SY5Y cells using 1D and Western blot analysis (Figure S-1), time-dependent Western blot analyses of ATP synthases, hnRNP, and HSP90 following exposure to 2µM and 20 µM paraoxon (Figure S-2), trypan exclusion data (Figure S-3), and a table of the proteins found in sub-cellular fractions (Table S-4). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ballantyne B, Marrs TC. Clinical and Experimental Toxicology of Organophosphates and Carbamates. Oxford ; Boston: Butterworth Heinemann; 1992. [Google Scholar]
- 2.Broomfield CA, Millard CB, Lockridge O, Caviston TL. Enzymes of the Cholinesterase Family. New York: Plenum Press; 1995. [Google Scholar]
- 3.Fukuto TR. Mechanism of action of organophosphorus and carbamate insecticides. Environ. Health Perspect. 1990;87:245–254. doi: 10.1289/ehp.9087245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albuquerque EX, Aracava Y, Cintra WM, Brossi A, Schonenberger B, Deshpande SS. Structure-activity relationship of reversible cholinesterase inhibitors: activation, channel blockade and stereospecificity of the nicotinic acetylcholine receptor-ion channel complex. Braz J Med Biol Res. 1988;21:1173–1196. [PubMed] [Google Scholar]
- 5.Bomser J, Casida JE. Activation of extracellular signal-regulated kinases (ERK 44/42) by chlorpyrifos oxon in Chinese hamster ovary cells. J Biochem Mol Toxicol. 2000;14:346–353. doi: 10.1002/1099-0461(2000)14:6<346::AID-JBT7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 6.Bomser JA, Casida JE. Diethylphosphorylation of rat cardiac M2 muscarinic receptor by chlorpyrifos oxon in vitro. Toxicol Lett. 2001;119:21–26. doi: 10.1016/s0378-4274(00)00294-0. [DOI] [PubMed] [Google Scholar]
- 7.Bomser JA, Quistad GB, Casida JE. Chlorpyrifos oxon potentiates diacylglycerol-induced extracellular signal-regulated kinase (ERK 44/42) activation, possibly by diacylglycerol lipase inhibition. Toxicol Appl Pharmacol. 2002;178:29–36. doi: 10.1006/taap.2001.9324. [DOI] [PubMed] [Google Scholar]
- 8.Ehrich M, Intropido L, Costa LG. Interaction of organophosphorus compounds with muscarinic receptors in SH-SY5Y human neuroblastoma cells. J Toxicol Environ Health. 1994;43:51–63. doi: 10.1080/15287399409531903. [DOI] [PubMed] [Google Scholar]
- 9.Eldefrawi ME, Eldefrawi AT. Neurotransmitter receptors as targets for pesticides. J Environ Sci Health B. 1983;18:65–88. doi: 10.1080/03601238309372358. [DOI] [PubMed] [Google Scholar]
- 10.Quistad GB, Sparks SE, Casida JE. Fatty acid amide hydrolase inhibition by neurotoxic organophosphorus pesticides. Toxicol Appl Pharmacol. 2001;173:48–55. doi: 10.1006/taap.2001.9175. [DOI] [PubMed] [Google Scholar]
- 11.Richards P, Johnson M, Ray D, Walker C. Novel protein targets for organophosphorus compounds. Chem Biol Interact. 1999;119–120:503–511. doi: 10.1016/s0009-2797(99)00064-2. [DOI] [PubMed] [Google Scholar]
- 12.Schuh RA, Lein PJ, Beckles RA, Jett DA. Noncholinesterase mechanisms of chlorpyrifos neurotoxicity: altered phosphorylation of Ca2+/cAMP response element binding protein in cultured neurons. Toxicol Appl Pharmacol. 2002;182:176–185. doi: 10.1006/taap.2002.9445. [DOI] [PubMed] [Google Scholar]
- 13.Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem Res Toxicol. 2004;17:983–998. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- 14.Casida JE, Quistad GB. Serine hydrolase targets of organophosphorus toxicants. Chem Biol Interact. 2005;157–158:277–283. doi: 10.1016/j.cbi.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 15.Pope CN. Organophosphorus pesticides: do they all have the same mechanism of toxicity? J Toxicol Environ Health B Crit Rev. 1999;2:161–181. doi: 10.1080/109374099281205. [DOI] [PubMed] [Google Scholar]
- 16.Garcia SJ, Seidler FJ, Crumpton TL, Slotkin TA. Does the developmental neurotoxicity of chlorpyrifos involve glial targets? Macromolecule synthesis, adenylyl cyclase signaling, nuclear transcription factors, and formation of reactive oxygen in C6 glioma cells. Brain Res. 2001;891:54–68. doi: 10.1016/s0006-8993(00)03189-9. [DOI] [PubMed] [Google Scholar]
- 17.Qian Y, Venkatraj J, Barhoumi R, Pal R, Datta A, Wild JR, Tiffany-Castiglioni E. Comparative non-cholinergic neurotoxic effects of paraoxon and diisopropyl fluorophosphate (DFP) on human neuroblastoma and astrocytoma cell lines. Toxicol Appl Pharmacol. 2007;219:162–171. doi: 10.1016/j.taap.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Hong MS, Hong SJ, Barhoumi R, Burghardt RC, Donnelly KC, Wild JR, Venkatraj V, Tiffany-Castiglioni E. Neurotoxicity induced in differentiated SK-N-SH-SY5Y human neuroblastoma cells by organophosphorus compounds. Toxicol Appl Pharmacol. 2003;186:110–118. doi: 10.1016/s0041-008x(02)00016-9. [DOI] [PubMed] [Google Scholar]
- 19.Carlson K, Ehrich M. Human neuroblastoma cell viability and growth are affected by altered culture conditions. In Vitr Mol Toxicol. 2000;13:249–262. [PubMed] [Google Scholar]
- 20.Knoth-Anderson J, Veronesi B, Jones K, Lapadula DM, Abou-Donia MB. Triphenyl phosphite-induced ultrastructural changes in bovine adrenomedullary chromaffin cells. Toxicol Appl Pharmacol. 1992;112:110–119. doi: 10.1016/0041-008x(92)90286-2. [DOI] [PubMed] [Google Scholar]
- 21.Massicotte C, Knight K, Van der Schyf CJ, Jortner BS, Ehrich M. Effects of organophosphorus compounds on ATP production and mitochondrial integrity in cultured cells. Neurotox Res. 2005;7:203–217. doi: 10.1007/BF03036450. [DOI] [PubMed] [Google Scholar]
- 22.Sachana M, Flaskos J, Alexaki E, Glynn P, Hargreaves AJ. The toxicity of chlorpyrifos towards differentiating mouse N2a neuroblastoma cells. Toxicol In Vitro. 2001;15:369–372. doi: 10.1016/s0887-2333(01)00038-8. [DOI] [PubMed] [Google Scholar]
- 23.Ross RA, Spengler BA, Biedler JL. Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J Natl Cancer Inst. 1983;71:741–747. [PubMed] [Google Scholar]
- 24.Carlson K, Ehrich M. Organophosphorus compound-induced modification of SH-SY5Y human neuroblastoma mitochondrial transmembrane potential. Toxicol Appl Pharmacol. 1999;160:33–42. doi: 10.1006/taap.1999.8741. [DOI] [PubMed] [Google Scholar]
- 25.Ehrich M, Correll L, Veronesi B. Acetylcholinesterase and neuropathy target esterase inhibitions in neuroblastoma cells to distinguish organophosphorus compounds causing acute and delayed neurotoxicity. Fundam Appl Toxicol. 1997;38:55–63. doi: 10.1006/faat.1997.2330. [DOI] [PubMed] [Google Scholar]
- 26.Saleh AM, Vijayasarathy C, Fernandez-Cabezudo M, Taleb M, Petroianu G. Influence of paraoxon (POX) and parathion (PAT) on apoptosis: a possible mechanism for toxicity in low-dose exposure. J Appl Toxicol. 2003;23:23–29. doi: 10.1002/jat.880. [DOI] [PubMed] [Google Scholar]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Cho T, Tiffany-Castiglioni E. Neurofilament 200 as an indicator of differences between mipafox and paraoxon sensitivity in Sy5Y neuroblastoma cells. J Toxicol Environ Health A. 2004;67:987–1000. doi: 10.1080/15287390490447287. [DOI] [PubMed] [Google Scholar]
- 29.Hreljac I, Zajc I, Lah T, Filipic M. Effects of model organophosphorous pesticides on DNA damage and proliferation of HepG2 cells. Environ Mol Mutagen. 2008;49:360–367. doi: 10.1002/em.20392. [DOI] [PubMed] [Google Scholar]
- 30.Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 31.Grenert JP, Sullivan WP, Fadden P, Haystead TA, Clark J, Mimnaugh E, Krutzsch H, Ochel HJ, Schulte TW, Sausville E, Neckers LM, Toft DO. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem. 1997;272:23843–23850. doi: 10.1074/jbc.272.38.23843. [DOI] [PubMed] [Google Scholar]
- 32.Bagchi D, Bhattacharya G, Stohs SJ. In vitro and in vivo induction of heat shock (stress) protein (Hsp) gene expression by selected pesticides. Toxicology. 1996;112:57–68. doi: 10.1016/0300-483x(96)03350-1. [DOI] [PubMed] [Google Scholar]
- 33.Gorlach M, Burd CG, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 34.Hossain MN, Fuji M, Miki K, Endoh M, Ayusawa D. Downregulation of hnRNP C1/C2 by siRNA sensitizes HeLa cells to various stresses. Mol Cell Biochem. 2007;296:151–157. doi: 10.1007/s11010-006-9308-2. [DOI] [PubMed] [Google Scholar]
- 35.Schepens B, Tinton SA, Bruynooghe Y, Parthoens E, Haegman M, Beyaert R, Cornelis S. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007;26:158–169. doi: 10.1038/sj.emboj.7601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fay J, Kelehan P, Lambkin H, Schwartz S. Increased expression of cellular RNA-binding proteins in HPV-induced neoplasia and cervical cancer. J Med Virol. 2009;81:897–907. doi: 10.1002/jmv.21406. [DOI] [PubMed] [Google Scholar]
- 37.Rahman-Roblick R, Roblick UJ, Hellman U, Conrotto P, Liu T, Becker S, Hirschberg D, Jornvall H, Auer G, Wiman KG. p53 targets identified by protein expression profiling. Proc Natl Acad Sci U S A. 2007;104:5401–5406. doi: 10.1073/pnas.0700794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martins-de-Souza D, Gattaz WF, Schmitt A, Novello JC, Marangoni S, Turck CW, Dias-Neto E. Proteome analysis of schizophrenia patients Wernicke's area reveals an energy metabolism dysregulation. BMC Psychiatry. 2009;9:17. doi: 10.1186/1471-244X-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vonck J, Schafer E. Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim Biophys Acta. 2009;1793:117–124. doi: 10.1016/j.bbamcr.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Simmons SO, Fan CY, Ramabhadran R. Cellular stress response pathway system as a sentinel ensemble in toxicological screening. Toxicol Sci. 2009;111:202–225. doi: 10.1093/toxsci/kfp140. [DOI] [PubMed] [Google Scholar]
- 41.Soltaninejad K, Abdollahi M. Current opinion on the science of organophosphate pesticides and toxic stress: a systematic review. Med Sci Monit. 2009;15:RA75–RA90. [PubMed] [Google Scholar]
- 42.Lasserre JP, Fack F, Revets D, Planchon S, Renaut J, Hoffmann L, Gutleb AC, Muller CP, Bohn T. Effects of the endocrine disruptors atrazine and PCB 153 on the protein expression of MCF-7 human cells. J Proteome Res. 2009;8:5485–5496. doi: 10.1021/pr900480f. [DOI] [PubMed] [Google Scholar]
- 43.Hung YC, Wang PW, Pan TL, Bazylak G, Leu YL. Proteomic screening of antioxidant effects exhibited by radix Salvia miltiorrhiza aqueous extract in cultured rat aortic smooth muscle cells under homocysteine treatment. J Ethnopharmacol. 2009;124:463–474. doi: 10.1016/j.jep.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Lee JH, Nguyen KH, Mishra S, Nyomba BL. Prohibitin is expressed in pancreatic beta-cells and protects against oxidative and proapoptotic effects of ethanol. FEBS J. 2010;277:488–500. doi: 10.1111/j.1742-4658.2009.07505.x. [DOI] [PubMed] [Google Scholar]
- 45.Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Fu H, Wen JF, Hu ZL, Luo GQ, Ren HZ. Knockdown of fascin1 expression suppresses the proliferation and metastasis of gastric cancer cells. Pathology. 2009;41:655–660. doi: 10.3109/00313020903273100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.