Abstract
As tumor development relies on a coordination of angiogenesis and tumor growth, an efficient antitumor strategy should target both the tumor and its associated vessels. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces apoptosis in a tumor-selective manner. Additionally, thrombospondin-1, a naturally occurring inhibitor of angiogenesis, and a recombinant protein containing functional domains of thrombospondin-1, 3TSR, have been shown to be necessary and sufficient to inhibit tumor angiogenesis. Here, we show that a combination of a TRAIL receptor 2 agonist antibody, Lexatumumab, and 3TSR results in a significantly enhanced and durable tumor inhibition. We further observed that 3TSR induces apoptosis in primary endothelial cells by up-regulating the expression of TRAIL receptors 1and 2 in a CD36 and Jun NH2-terminal kinase-dependent manner leading to the activation of both intrinsic and extrinsic apoptotic machineries. The modulation of these pathways is critical for 3TSR-induced apoptosis as disrupting either via specific inhibitors reduced apoptosis. Moreover, 3TSR attenuates the Akt survival pathway. These studies indicate that 3TSR plays a critical role in regulating the proapoptotic signaling pathways that control growth and death in endothelial cells and that a combination of TRAIL and 3TSR acts as a double hit against tumor and tumor-associated vessels.
Introduction
Apoptosis of endothelial cells is a prominent feature of blood vessel remodeling (1–3) and may limit the undesired neovascularization of tumors (4). In mammalian systems, there are two major apoptotic pathways. One is the intrinsic pathway, which is triggered by many stress stimuli including chemotherapeutic agents, UV irradiation, and p53 and leads to the release of cytochrome c and other apoptogenic factors from mitochondria causing the activation of caspase-9 and downstream executioner caspases (5). By contrast, the extrinsic apoptotic pathway is activated on binding of members of the tumor necrosis factor family of ligands [e.g., tumor necrosis factor, Fas, and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)] to their cognate receptors (6–8). This interaction results in the recruitment of apical procaspase-8, causing the formation of the death-inducing signaling complex (5, 6, 9). Once formed, the death-inducing signaling complex promotes the release of active caspase-8 to directly activate caspase-3and to trigger apoptosis. However, in most cells (including cancer cells), caspase-8 inefficiently processes caspase-3 (6). In these cells, death signaling bifurcates into two arms, one of which engages mitochondria in a manner equivalent to the intrinsic pathway (6, 10). The death receptor-induced apoptotic pathway can also result in the activation of the stress-activated protein kinases including Jun NH2-terminal kinase (JNK) and p38 (4, 11). JNK and p38 have also been shown to be involved in mediating apoptosis in some cell systems (12) through transcriptional regulation of proapoptotic factors including some death ligands and death receptors (11, 13, 14).
Currently, therapeutic strategies for the selective activation of the intrinsic and/or extrinsic pathways in tumor cells are heavily sought after (6, 9, 15–17). Because one of the physiologic roles of TRAIL is in the immune response against transformed cells (18, 19), and because exogenous TRAIL induces tumor-selective apoptosis, TRAIL has great potential in cancer therapy (18). TRAIL and TRAIL receptor agonist antibodies (Lexatumumab) are currently at various stages of clinical trials (20, 21). These therapeutic agents do not cause complete tumor regression when used as single agents (22).
TSP-1 is one of several naturally occurring inhibitors of angiogenesis (23) and a member of a family of high molecular weight matricellular glycoproteins (24, 25). TSP-1 has been reported to induce apoptosis of endothelial cells through activation of CD36, p59fyn, caspase-3–like activity, and p38 mitogen-activated protein kinase (2, 4, 26, 27). CD36 is the receptor for a functional domain of TSP-1, known as the type 1 repeats (TSR). TSRs, or their synthetic peptides, are the best mimetics of TSP-1, and one peptide, designated ABT-510, is in clinical trials for cancer therapy (26, 28–30). 3TSR is thought to inhibit tumor growth in vivo either by directly targeting endothelial cells, leading to apoptosis of the cells through mechanisms such as inhibition of Bcl-2 and activation of caspase-3, or indirectly via inhibition of vascular endothelial growth factor (VEGF) release from the extracellular matrix. These effects of 3TSR could result in the induction of apoptosis in the tumor-associated endothelial cells, promoting hypoxia and leading to tumor regression. However, in the tumor models that have been studied, 3TSR alone is not sufficient to cause complete inhibition of tumor growth. Additionally, the molecular mechanisms through which TSP-1 and TSRs induce activation of caspase-3 and promote CD36-mediated apoptosis remain to be elucidated.
In this study, we have investigated the effectiveness of inhibiting tumors and tumor-associated vessels by combining 3TSR and Lexatumumab in a tumor model in vivo. We have further analyzed the molecular mechanism by which 3TSR induces CD36-mediated apoptosis of cultured primary human dermal microvascular endothelial cells (HDMEC).
Materials and Methods
Reagents
Lexatumumab, a fully human agonist monoclonal antibody to TRAIL receptor DR5, was a gift from Human Genome Sciences. Matrigel (Matrigel basement membrane matrix) was purchased from BD Biosciences. Each of the following reagents was dissolved in DMSO for use: caspase-8 inhibitor Z-IETD-FMK, caspase-9 inhibitor Z-LEHD-FMK, and caspase-3 inhibitor Z-DEVD-FMK were purchased from R&D Systems; the PI3K inhibitors LY294002 and wortmannin were purchased from Calbiochem and Alexis Biochemicals, respectively; and SP600125 was purchased from Biomol International. Protease inhibitor cocktail tablets were purchased from Roche. Purified VEGF165, expressed in Sf21, was obtained from the National Cancer Institute Preclinical Repository, Biological Resources Branch. The following antibodies were used in our studies: monoclonal antibody to CD36 (clone FA6-152; Immunotech, Coulter); mouse anticytochrome c monoclonal antibody (clone 7H8.2C12; BD Biosciences Pharmingen); mouse IgG, GRB2 (C-23) polyclonal antibody, caspase-8 p20 (H134) polyclonal antibody, human caspase-9 p10 (H-83) polyclonal antibody, Akt1/2 (H136), and anti-DR5/TRAIL receptor 2 (all from Santa Cruz Biotechnology); caspase-3 polyclonal antibody, human-specific caspase-9 polyclonal antibody, phospho-Akt (Ser473), phospho-SAPK/JNK (Thr183/Tyr185), and SAPK/JNK (SC-827) antibodies (all from Cell Signaling Technology); monoclonal anti-β-actin clone AC-15 (Sigma-Aldrich); and anti-DR4/TRAIL receptor 1 (Axxora).
3TSR production
A recombinant 3TSR was prepared using the Drosophila expression vector system, pMT/BiP/V5-HisA (Invitrogen), as described previously (31).
Cell culture
Four colon cancer cell lines, HT29, HCT116, SW480, and COLO205, were obtained from the American Type Culture Collection. HT29 and HCT116 cells are maintained in McCoy’s 5A medium with 1.5 mmol/L l-glutamine and 10% fetal bovine serum (FBS). SW480 and COLO205 cells are maintained in RPMI 1640 with 2 mmol/L l-glutamine, 10 mmol/L HEPES, 4.5 g/L glucose, and 10% FBS. HDMEC were isolated by the procedure of Richard and colleagues (32). The cells were cultured in Vitrogen-precoated dishes and/or 24-well plates, maintained in EBM-2 (Clonetics) containing 20% FBS, 1 mg/mL hydrocortisone acetate, 5 × 105 μmol/L dibutyryl-cAMP, 200 units/mL penicillin, 100 units/mL streptomycin, and 250 mg/mL amphotericin, and grown to ~80% confluency. The cells were then cultured in EBM-2 containing 2% FBS overnight and then switched into EBM-2 containing 0.5% FBS for the experiment. HDMEC of passage 6 or 7 were used in the study. Jurkat cells were obtained from the American Type Culture Collection and maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 50 mg/mL penicillin, 50 units/mL streptomycin, and 2 mmol/L glutamine. The cells were switched into RPMI 1640 containing 0.5% FBS for the apoptosis and kinase assays.
Xenograft tumor model and treatment
The human colon cancer HCT116 or SW480 cells were mixed with Matrigel at 1:1 (v/v), and 100 μL of the mixture (1 × 106 HCT116 cells or 8 × 106 SW480 cells) was injected subcutaneously into the right flank of nu/nu mice (8 weeks old, female; Charles River Laboratories). After ~1 week when tumor volumes reached 100 to 150 mm3, mice were randomized among and regrouped such that tumor volumes of all groups of mice were in the same range. After the randomization, mice began to receive treatment. Lexatumumab was delivered by tail vein (intravenous) injection at 3 mg/kg and given twice weekly. 3TSR was injected intraperitoneally at 1 mg/kg/d. In vehicle control group of mice, 0.9% saline (intravenous) and TSR buffer (intraperitoneal) were used for injection on the same schedule as Lexatumumab and 3TSR treatment groups. Tumor volume was measured using calipers in two dimensions. Tumor volume was calculated using the formula: (width2 × length) / 2. Tumor growth inhibition (TGI) was calculated using the formula: TGI = (100 − treatment /control) × 100. Some of the mice were sacrificed at the indicated time points and the tumors were frozen immediately or fixed in 10% formalin for further pathologic analysis. The rest of the mice continued on the treatment.
Pathologic analysis
Major organs and tumor tissues were preserved in 10% formalin immediately after collection and processed into paraffin embedding samples. Histology examination on major organ tissues was carried out by H&E staining and microscopy. Detection of DR4 and DR5 expression in tumor samples was carried out by immunohistochemistry, and evaluation of apoptosis was done by the terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay as well as by cleaved caspase-3 staining on slides at the Rodent Histopathology Core Laboratory of the Dana-Farber/Harvard Cancer Center. Frozen tumor sections were used for double staining of CD31 and TUNEL as described previously (33).
Apoptosis assay
Primary HDMEC were treated with 3TSR, TSP-1, and camptothecin for 24 h. The cells were then trypsinized, collected, and washed once with PBS. Colon carcinoma cells were treated with TRAIL as indicated in the figure legend. The cells were then resuspended in DNA staining buffer containing propidium iodide (Roche) and incubated for 1 h at 4°C. The cell population was assayed by fluorescence-activated cell sorting (FACSCalibur; Becton Dickinson Immunocytometry System) and CellQuest software was used for measuring the sub-G1 apoptotic cell population, and 10,000 cells were analyzed for each condition. In addition, the morphology of the endothelial cells was observed under a phase-contrast microscope and photos were taken with a CCD digital camera.
In parallel studies, the cells were grown on Lab-Tek chamber slides coated with Vitrogen, and apoptosis was evaluated using a TUNEL assay (Promega). After the cells were treated with 3TSR (2 μmol/L) or 3TSR buffer for 24 h, the slides were washed twice with PBS. The cells were then fixed by immersing the slides in freshly prepared 4% methanol-free formaldehyde solution in PBS (pH7.4) for 25 min at 4°C and processed directly with the apoptosis detection assay as described in the protocol by the manufacturer. Vectashield and 4′,6-diamidino-2-phenylindole (Vector Laboratories) were applied to stain nuclei and internal negative and positive controls were set up for quality control.
Endothelial cell apoptosis was measured using tumor tissues 4 weeks post-tumor inoculation. Frozen tissue sections (5 μm thick) were incubated with rat monoclonal anti-CD31 antibody (BD Pharmingen) in a humidified chamber for 15 to 18 h at 4°C, rinsed with PBS, and incubated with goat anti-rat IgG conjugated to Texas red (Vector Laboratories) for 60 min at ambient temperature in the dark. After sections were washed with PBS containing 0.1% Brij 99 (v/v), TUNEL labeling was done using a commercial kit (Promega) according to the manufacturer’s instructions. Background reactivity was determined by processing slides in the absence of terminal deoxynucleotidyl transferase (negative control); maximum reactivity was observed by preincubating the tissue sections with DNase I to confirm the quality of the specimen. Representative sections from two tumors per group were analyzed with a Bio-Rad 1024 confocal microscope. Endothelial cells were identified by red fluorescence, and DNA fragmentation was detected by localized green and yellow fluorescence within the nuclei of apoptotic cells. Apoptotic endothelial cells were quantified and expressed as an average of the ratio of apoptotic endothelial cells to the total number of endothelial cells.
Assay of caspase activity
Caspase-8 and caspase-9 activities were assayed in lysates from HDMEC treated with 2 μmol/L 3TSR for 24 h following the procedure described by the manufacturer (BD ApoAlert Caspase Fluorescent Assay kits; BD Biosciences Pharmingen). In parallel experiments, the caspase activities were also measured in the presence of Z-IETD-fmk (20 μmol/L) or Z-LEHD-fmk (2 μmol/L) to determine the specificity of caspase-8 or caspase-9 activities. Caspase-3 activity in 3TSR-treated cells (24 h) was also assayed according to the manufacturer’s protocol (BD ApoAlert Caspase Colorimetric Assay kit; BD Biosciences Pharmingen). Caspase-3 activity was also measured in the presence of Z-DEVD-fmk (40 μmol/L).
Detection of cytochrome c release into cytosol
Cell fractionation on HDMEC and Jurkat cells was done as described in the MBL cytochrome c assay protocol (MBL). The supernatant was collected as a cytosolic fraction for Western blot analysis.
Western blot analysis
The cells were washed once with ice-cold PBS, rapidly lysed in lysis buffer [20 mmol/L Tris (pH 7.4), 10% glycerol, 1% Triton X-100, 137 mmol/L NaCl, 2 mmol/L EDTA, 25 mmol/L β-glycerophosphate, 2 mmol/L p-nitrophenyl phosphate, 0.5 mmol/L Pefabloc (4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride; Roche Applied Science), 10 mg/mL aprotinin, 1 mmol/L sodium pervanadate, and protease inhibitor cocktail (phenylmethylsulfonyl fluoride, aprotinin, Pefabloc SC, leupeptin, α2-macroglobulin, EDTA, bestatin, pepstatin, and E64)], and incubated for 10 min on ice. Total cell lysates were then centrifuged at 13,000 × g for 15 min at 4°C. Protein concentration was assayed with the BCA protein assay kit (Pierce). The cytosolic and mitochondrial fractions were prepared as described above. Western blot analyses were done as follows: SDS sample buffer was added to cell lysates (total, cytosol, and mitochondria fractions), samples were boiled, and proteins were resolved on a 12.5% SDS-PAGE followed by transferring the separated proteins to Immobilon transfer membranes (Millipore) and immunoreaction with specific antibodies. The immunoreactive protein was then detected by enhanced chemiluminescence.
Real-time quantitative reverse transcription-PCR
Gene expression in the primary microvascular endothelial cells treated with 3TSR (1 μmol/L) or buffer control was assessed by real-time reverse transcription-PCR following the protocol of Ghaffari and colleagues (34). The primers for DR4 and DR5 as well as GAPDH are as follows: human DR4 forward 5′-CAATGCTGATGAAATGGGTCA-3′ and reverse 5′-GCATCCAGCAGGGTGTGG-3′, human DR5 forward 5′-CTCCTGAGATGTGCCGGAA-3′ and reverse 5′-ATCACCGACCTTGACCATCC-3′, and human GAPDH forward 5′-GGTTTCGGTCGTATCGGACG-3 and reverse 5′-TTGGTCACCAGTGTAAGC-3′. GAPDH transcripts were amplified in a separate tube to normalize variances in input RNA. The relative Ct value was used to compare the fold change of mRNA expression.
Akt activation assay
To determine the effects of 3TSR on Akt phosphorylation, the primary HDMEC were cultured in 2% FBS/EBM-2 overnight. 3TSR buffer control, 3TSR, and LY294002 were added for 9 h to the cells in 0.5% FBS/EBM-2. VEGF was added at the same time or at the last 15, 30, and 60 min of the 9 h incubation. Akt activation was assayed by Western blot with phosphospecific Akt antibody phospho-Akt (Ser473; Cell Signaling Technology). Akt1/2 (H136:sc-8312) (Santa Cruz Biotechnology), and monoclonal anti-β-actin clone AC-15 (Sigma) were used to detect total Akt and β-actin on the same or duplicate membranes, respectively.
JNK activation assay
To determine the effects of 3TSR on JNK phosphorylation, the primary HDMEC were cultured in 2% FBS/EBM-2 overnight. After the cells were pretreated with JNK-specific inhibitor SP600125 (10 μmol/L) and FA6-152 (20 μg/mL) for 30 min, 3TSR buffer control or 3TSR was added for 30 and 60 min. Western blotting was used to detect phosphorylation levels of SAPK/JNK.
Nucleofection
The primary dermal microvascular endothelial cells (5 × 105, passage 6) were grown in freshly prepared 20% FBS/EBM-2 and nucleofected with the pcDNA3.1 plasmid and the plasmid encoding myr-Akt with a protocol optimized for human microvascular endothelial cells by using the HMVEC-L nucleofector kit and Amaxa nucleofector (Amaxa Biosystems). After nucleofection for 36 h, the cells were switched to 2% FBS/EBM-2 overnight and treated with 3TSR or 3TSR buffer for 24 h for an apoptosis assay; parallel cell lysate was obtained for detection of total Akt and phospho-Akt expression.
Xenograft tumor model, treatment, and pathologic analysis
The human colon cancer HCT116 or SW480 cells were used for generation of xenograft tumor model as described previously (35, 36).
Results
Combination treatment with 3TSR and TRAIL leads to cooperative tumor inhibition in vivo
A successful strategy for cancer therapy may involve selective inhibition of tumors in conjunction with inhibition of tumor-associated endothelial cells. To investigate the in vivo effectiveness of Lexatumumab and 3TSR combination, we chose several human colon cancer cell lines, HT29, HCT116, SW480, and COLO205. We first tested these cell lines in growth medium containing vehicle (control) or Lexatumumab (2 μg/mL) for 24 h, and apoptosis was quantified by measuring sub-G1 apoptotic cell population. All four cell lines treated with Lexatumumab showed an increase of 30% to 76% in the level of apoptosis compared with the control cells (Supplementary Fig. S1A) but were not responsive to 3TSR (Supplementary Fig. S1B).
Based on these findings, we chose two cell lines, HCT116 and SW480, which had partial but significant response to Lexatumumab. These cells were subcutaneously injected in nude mice to generate tumor xenografts. One week after tumor injections, the mice with HCT116 or SW480 were divided into four groups receiving vehicle control, Lexatumumab at 3 mg/kg twice a week, 3TSR at 1 mg/kg/d, and a combination of Lexatumumab at 3mg/kg twice a week and 3TSR at 1 mg/kg/d.
In the SW480 xenograft tumor model, treatment with Lexatumumab alone or 3TSR alone started to show tumor inhibition as early as 1 week after treatment. Increased tumor inhibition in the combination group, compared with the Lexatumumab or 3TSR group, was evident as early as day 10 after treatment. As the treatment continued, tumor growth in the combination treatment group was almost completely inhibited: 93% by day 31 after treatment compared with 60% for the Lexatumumab group and 69% for the 3TSR group. The result of inhibition on tumor growth in the combination group can clearly be visualized from the mice bearing SW480 tumors on day 31 after treatment compared with other groups (Fig. 1A).
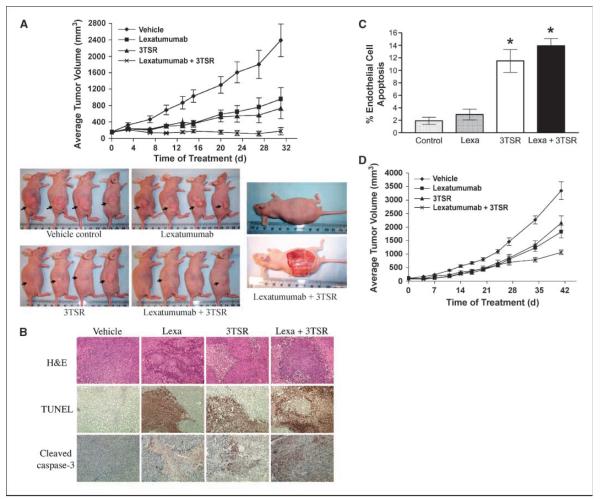
Figure 1.
3TSR and TRAIL cooperate in inhibiting colon cancer. A, human colon cancer SW480 cells were implanted into the right flankof nu/nu mice. Seven days post-implantation, mice were injected daily with Lexatumumab (3 mg/kg intravenously) and 3TSR (1 mg/kg intraperitoneally) or 0.9% saline and TSR buffer. Mean from 10 mice in each group (n = 10). By treatment day 31, ANOVA indicates P < 0.01 for Lexatumumab versus vehicle, P < 0.001 for 3TSR versus vehicle, P < 0.001 for Lexatumumab + 3TSR versus vehicle, and P < 0.05 for Lexatumumab + 3TSR versus Lexatumumab. By day 31, TGI = 60% for Lexatumumab, TGI = 69% for 3TSR, and TGI = 93% for Lexatumumab + 3TSR. B, immunohistochemical staining on SW480 tumor sections for TUNEL and cleaved caspase-3. SW480 xenograft tumors were collected from three mice from each treatment group on day 28 after tumor cell inoculation. Similar results were observed from tumor samples of three mice from each treatment group. Original photos were taken at ×200 magnification. C, analysis of tumor vasculature endothelial cell apoptosis. The apoptosis of tumor-associated vascular endothelial cells was evaluated via double labeling of CD31 and TUNEL. 3TSR significantly induced apoptosis of tumor vascular endothelial cells (P = 0.001 versus vehicle control), whereas Lexatumumab (Lexa) treatment showed little effect on tumor vascular endothelial cell apoptosis. The combination of Lexatumumab and 3TSR resulted in a similar level of endothelial cell apoptosis as with 3TSR alone (*, P < 0.001 versus control). D, growth of HCT116 colon cancer xenograft in nude mice. Tumor implantation and treatments were same as A, except 1 × 106 cells were used for implantation. Points, mean from 12 mice in each group (n = 12). By treatment day 41, statistical analysis by ANOVA indicates P < 0.01 for ×Lexatumumab versus vehicle, P < 0.05 for 3TSR versus vehicle, P < 0.001 for Lexatumumab + 3TSR versus vehicle, and P < 0.05 for Lexatumumab + 3TSR versus 3TSR. By treatment day 41, TGI = 46% for Lexatumumab, TGI = 36% for 3TSR, and TGI = 68% for Lexatumumab + 3TSR.
To investigate whether a combination of Lexatumumab and 3TSR can result in a durable tumor inhibition, we stopped treatment of 6 mice after 31 days. Whereas tumor growth resumed in Lexatumumab- or 3TSR-treated mice, leading to their sacrifice by day 100 due to tumor burden, combined treatment with Lexatumumab and 3TSR led to delayed tumor regrowth in most of the mice and no tumor growth in two mice (n = 6). The latter two mice remained tumor-free for 5 months after treatment was terminated (data not shown).
Pathologic analysis of the SW480 xenograft tumor specimens revealed that there were apoptotic tumor areas in Lexatumumab-treated as well as 3TSR-treated mice as evidenced by positive staining of TUNEL and cleaved caspase-3 (Fig. 1B). However, TUNEL staining and caspase-3 activity was much more prominent in the tumor tissue from the combination Lexatumumab- and 3TSR-treated mice (Fig. 1B). More importantly, H&E staining of lung, kidney, liver, and heart showed normal cellular architecture in normal tissues, indicating that 3TSR does not cause a generalized toxicity to other tissues and organs (Supplementary Fig. S1C). Finally, a costaining of tumors 4 weeks post-tumor inoculation with CD31, which stains endothelial cells, and TUNEL showed that 3TSR but not Lexatumumab induces endothelial cell apoptosis (Fig. 1C).
Combination treatment in HCT116 xenograft mice also showed an enhanced tumor inhibition (Fig. 1D). In these mice, treatment with Lexatumumab alone or 3TSR alone and the combination started to show evidence of tumor inhibition 2 weeks after treatment (Fig. 1D). After the fourth week of treatment, the Lexatumumab or 3TSR group showed significant tumor inhibition compared with the control group, and the combination group showed increased tumor inhibition compared with the Lexatumumab or 3TSR group. When treatment continued, the increased tumor inhibition became significantly stronger in the combination group, and by day 41, the tumor inhibition in the combination group reached 68%, whereas Lexatumumab and 3TSR groups displayed 46% and 36% inhibition, respectively. Statistical analysis showed that combination treatment had significantly enhanced tumor inhibition compared with the 3TSR or Lexatumumab group (P < 0.05).
The notable cooperativity of Lexatumumab and 3TSR in tumor inhibition suggests that this combination therapy could be efficacious in a clinical setting.
3TSR induces apoptosis in microvascular endothelial cells in a CD36-dependent manner
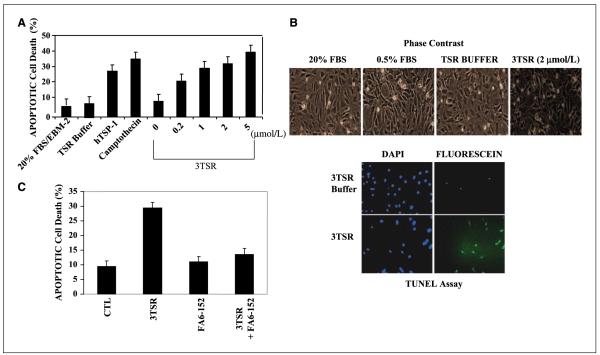
We next determined whether 3TSR could induce apoptosis in HDMEC by exposing these cells to increasing concentrations of 3TSR. HDMEC were treated with various concentrations of 3TSR for 24 h. Flow cytometry of the sub-G1 apoptotic population of 3TSR-treated cells showed that 3TSR could induce apoptosis in a dose-dependent manner (Fig. 2A). After 24 h of treatment with 0.2 μmol/L 3TSR, ~23% of cells were apoptotic, whereas 5 μmol/L 3TSR induced up to 42% apoptosis. Under the same conditions, control cells treated with 3TSR buffer and cells that were grown with 20% serum under normal growth conditions showed 10% basal apoptosis, whereas TSP-1 (30 nmol/L) and camptothecin (0.5 mg/mL) treatment caused 30% and 40% apoptosis, respectively (Fig. 2A). Moreover, the HDMEC that were treated with 3TSR (2 μmol/L) for 24 h showed a more detached and rounded morphology (Fig. 2B) when compared with the untreated controls and were positive for TUNEL staining (Fig. 2B).
Figure 2.
3TSR induces apoptosis of HDMEC. A, primary HDMEC were treated with 3TSR, TSP-1, and camptothecin at the indicated concentrations for 24 h. The apoptotic cell population was assayed by fluorescence-activated cell sorting analysis and indicated that 3TSR-mediated apoptosis increased in a dose-dependent manner. B, phase-contrast microscopy showed that 3TSR-treated cells have a rounded/apoptotic morphology and are positive for TUNEL staining. C, HDMEC were preincubated with anti-CD36 antibody FA6-152 for 1 h before adding 3TSR. Apoptotic cell population was measured as in A and showed that 3TSR-mediated apoptosis is dependent on the activity of the CD36 receptor.
It is known that 3TSR interacts with multiple membrane receptors at the cell surface including CD36 to mediate the apoptotic response of TSP-1 (4, 37, 38). We sought to determine whether the 3TSR-induced apoptosis in HDMEC was also mediated through CD36. HDMEC were preincubated with 30 mg/mL CD36-blocking (FA6-152) antibody followed by treatment with 3TSR (2 μmol/L) for 24 h. As shown in Fig. 2C, monoclonal antibody FA6-152 attenuated 3TSR-induced apoptosis in HDMEC to close to the basal level, indicating that CD36 is required for the apoptotic activity of 3TSR in these cells.
3TSR induces apoptosis of HDMEC through activation of caspase-8 and caspase-3
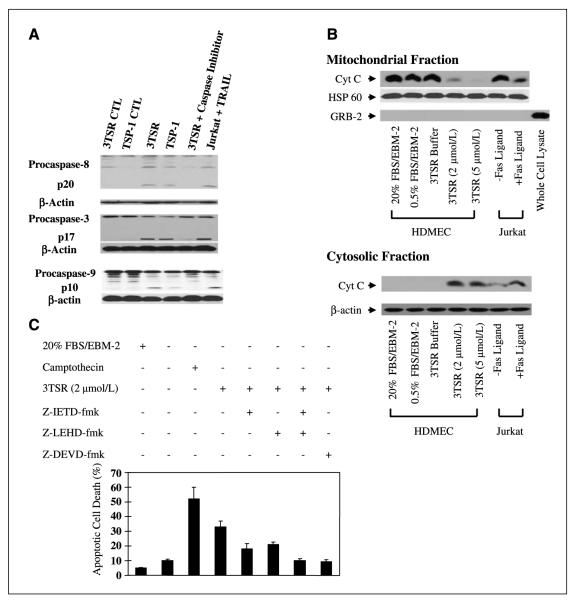
TSP-1–induced apoptosis of endothelial cells has been shown to involve the activation of the apical caspase-8/extrinsic pathway (27). To determine whether 3TSR could also induce apoptosis through the activation of the caspase cascade, we treated HDMEC with recombinant 3TSR (2 μmol/L) for 24 h and harvested cell lysates for Western blot analysis and caspase activity assays. Figure 3A shows that cleaved caspase-8 can be detected following treatment with 3TSR. In addition, we observed that 3TSR induces a 3-fold increase in the activity of caspase-8 compared with the control cells (Supplementary Fig. S2). Furthermore, 3TSR-induced caspase-8 activity was inhibited by IETD inhibitor (Fig. 3A).
Figure 3.
3TSR stimulates caspase-8, caspase-3, and cytochrome c release from mitochondria and promotes activation of caspase-9 in HDMEC. A, HDMEC cells were treated with buffer (CTL), 3TSR, hTSP-1, and caspase-8 (z-IEDT-fmk), caspase-3 (z-DEVD-fmk), or caspase-9 (z-LEHD-fmk) inhibitor. Caspase cleavage was detected by Western blotting with specific caspase-8. Caspase-3 or caspase-9 antibodies. Jurkat cells treated with TRAIL in the presence or absence of z-IETD-fmkwere used as controls. B, cytochrome c (Cyt C) levels in mitochondrial and cytosolic fractions were determined by Western blotting after treatment of cells with 2 or 5 μmol/L 3TSR or buffer. Jurkat cells treated with or without Fas ligand for 6 h were used as controls. Western blot analysis indicated that the mitochondrial preparations were free of GRB2, a protein that is normally excluded from mitochondria. C, HDMEC were treated with 3TSR in the absence or presence of the caspase-8 inhibitor z-IETD-fmk, the caspase-9 inhibitor z-LRHD-fmk, either alone or in combination, and the caspase-3 inhibitor z-DEVD-fmk. Camptothecin was used as a positive control for induction of apoptosis in endothelial cells. Mean ± SD of three separate experiments.
To determine if the principal downstream executioner caspase-3 is also activated, HDMEC cultures were treated with 3TSR (2 μmol/L) for 24 h. Cell lysates were then prepared and the processed caspase-3 signature fragment was detected by Western blot analysis (Fig. 3A). Additionally, in caspase-3 activity assays, we observed a 5- to 6-fold increase in the activity of caspase-3 compared with control cells (Supplementary Fig. S2). This activity was inhibited by caspase-3–specific inhibitor DEVD (Fig. 3A).
3TSR activates the mitochondrial-dependent apoptotic pathways in HDMEC
To determine the involvement of the mitochondrial pathway in mediating 3TSR-induced apoptosis, we evaluated the release of cytochrome c and the activation of caspase-9. HDMEC were treated with buffer control or 3TSR (2 μmol/L) for 24 h. Cell lysates were then prepared and fractionated into cytosolic and mitochondrial fractions. Western blot analysis was done for cytochrome c, HSP-60 (mitochondrial protein), β-actin, and GRB2 (39), a protein that is normally excluded from mitochondria. Figure 3B shows that whereas the majority of cytochrome c is in the mitochondrial fraction in control cells, treatment of HDMEC with 3TSR resulted in the release of cytochrome c into the cytosol. Moreover, the 3TSR-induced release of cytochrome c from the mitochondria also corresponded with the appearance of processed fragments of caspase-9 and a 7-fold increase in the activity of caspase-9 (Fig. 3A; Supplementary Fig. S2). Additionally, stimulation of HDMEC with hTSP-1 resulted in the activation of caspase-9. Finally, the caspase-9-specific inhibitor LEHD prevented the 3TSR-induced activation of caspase-9 (Fig. 3A).
To further show that 3TSR-induced apoptosis is dependent on the activation of caspase-8, caspase-9, and caspase-3, we used specific inhibitors of each of these caspases. The caspase-8-specific inhibitor z-IETD-fmk and the caspase-9-specific inhibitor z-LEHD-fmk both partially inhibited the apoptosis induced by 3TSR (2 μmol/L). Interestingly, a combination of these inhibitors resulted in a complete attenuation of 3TSR-mediated apoptosis in HDMEC (Fig. 3C). Similarly, the inhibition of the executioner caspase-3 by z-DEVD resulted in a complete inhibition of 3TSR-mediated apoptosis (Fig. 3C). These observations suggest that the activities of both caspase-8 and caspase-9 are required for mediating 3TSR-induced apoptosis and their independent activation results in a cooperative effect to promote the induction of apoptosis.
3TSR-induced apoptosis is mediated by JNK activation and induction of DR4 and DR5 expression
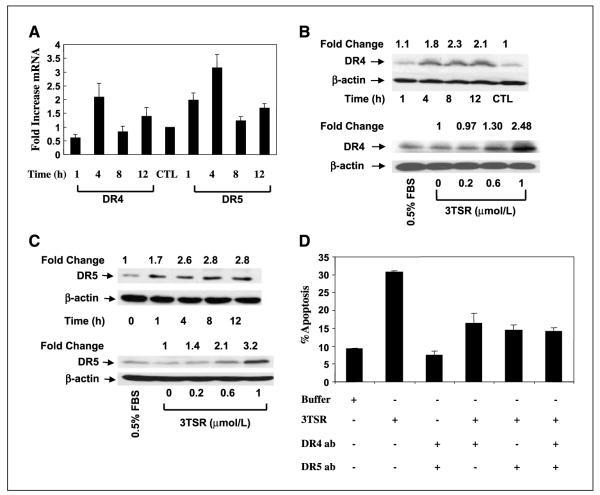
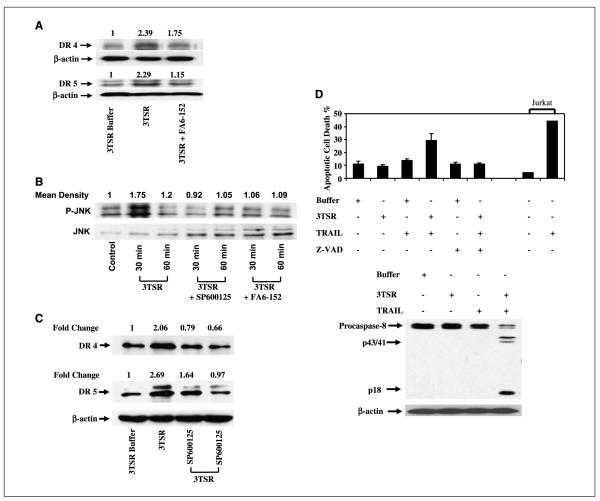
Whereas 3TSR-induced activation of the extrinsic and intrinsic apoptotic pathways is involved in mediating the subsequent apoptosis, the molecular mechanism for interaction and/or cross-talk between 3TSR and death receptor pathways is not well characterized. Low levels of VEGF have been shown previously to induce Fas ligand expression, targeting endothelial cells for TSP-1-induced apoptosis (27). Here, we investigated whether 3TSR can modulate the expression of Fas, TRAIL, and/or TRAIL receptors. Treatment of HDMEC with 1 μmol/L 3TSR does not significantly alter Fas or Fas ligand mRNA expression as measured by real-time PCR analysis (data not shown). However, 1 μmol/L 3TSR resulted in a transient increase in DR4 and DR5 mRNA expression (2-fold increase in DR4 mRNA after 4 h, ~2-fold increase in DR5 after 1 h, and ~3-fold increase after 4 h 3TSR treatment, respectively; Fig. 4A). Additionally, Western blot analysis showed a time- and dose-dependent increase in DR4 and DR5 protein levels (Fig. 4B and C). Notably, 3TSR-induced apoptosis in HDMEC was dependent on the activity of DR4 and DR5 receptors, as DR4- and DR5-blocking antibodies attenuated it (Fig. 4D). As shown in Fig. 5A, a 3TSR-induced increase in DR4 and DR5 is attenuated by CD36-blocking antibody (FA6-152), indicating the involvement of CD36 in this pathway.
Figure 4.
3TSR regulates expression of the proapoptotic death receptors DR4 and DR5. A, 3TSR treatment of HDMEC for 1 to 12 h significantly increased expression of DR4 and DR5 at the transcriptional level. B and C, 3TSR treatment of HDMEC for 1 to 12 h increased both DR4 and DR5 protein expression, whereas 3TSR treatment of HDMEC for 8 h increased both DR4 and DR5 expression in a concentration-dependent manner. D, pretreatment of HDMEC cells with DR4- and/or DR5-blocking antibody for 30 min inhibited 3TSR-mediated apoptosis.
Figure 5.
3TSR-induced DR4 and DR5 expression is mediated by JNK activation. A, 3TSR and IgG treatment of HDMEC for 4 h up-regulated DR4 and DR5 expression and this increase was suppressed with the CD36-blocking antibody FA6-152. B, both 3TSR-stimulated phosphorylation of SAPK/JNK and JNK activity in HDMEC were inhibited by treatment with SP600125, a selective inhibitor of JNK and FA6-152. C, 3TSR treatment of HDMEC for 4 h up-regulated DR4 and DR5 expression. Treatment of these cells with SP600125, however, similarly decreased DR4 and DR5 levels. D, 3TSR sensitizes HDMEC to TRAIL-induced apoptosis. HDMEC were treated with control buffer, 3TSR, and TRAIL alone or in combination. 3TSR and TRAIL combination enhances apoptosis induced in HDMEC. Inhibition of caspase activity by z-VAD suppresses this cooperation. Additionally, combination of 3TSR and TRAIL enhances caspase-8 activity in HDMEC. Processing of caspase-8 was evaluated by Western blot analysis. Arrow, processed p43/p41 signature fragments of caspase-8 processing. Jurkat cells are used as a control to show the activity of TRAIL.
TSP-1 has been shown to regulate the activity of JNK (4), which modulates the expressions of several death receptors including DR5 (13). Therefore, we investigated whether the 3TSR-induced increases in DR4 and DR5 expression are mediated by the JNK pathway. As a basis for these studies, we first confirmed that treatment of HDMEC with 3TSR does result in a transient activation of JNK (Fig. 5B). Following 30 min treatment with 1 μmol/L 3TSR, we observed a 1.75-fold increase in the level of JNK phosphorylation in HDMEC. Additionally, cotreatment of HDMEC with 1 μmol/L 3TSR and 10 μmol/L JNK inhibitor (SP600125) or 10 μmol/L CD36-blocking antibody (FA6-152) inhibited 3TSR-induced JNK phosphorylation, indicating that 3TSR-induced JNK activation is dependent on the interaction between 3TSR and CD36 (Fig. 5B). Finally, to investigate the effect of JNK activation on 3TSR-induced DR4 and DR5 expression, HDMEC were treated with 3TSR (1 μmol/L) alone or in combination with SP600125 (10 μmol/L) for 4 h. As shown in Fig. 5C, inhibition of JNK activity resulted in a significant reduction in the level of 3TSR-induced DR4 and DR5 protein expression. Similarly, 3TSR results in activation of p38 mitogen-activated protein kinase (Supplementary Fig. S3). However, inhibition of p38 with SB203580 does not ablate the 3TSR-induced expression of DR5 (Supplementary Fig. S3).
HDMEC are generally highly resistant to TRAIL-induced apoptosis (Fig. 5D). The 3TSR-induced increase in the levels of the TRAIL receptors, DR4 and DR5, in HDMEC suggests that these cells may become sensitive to TRAIL-induced apoptosis. As shown in Fig. 5D, treatment of HDMEC with TRAIL (10 ng/mL) for 18 h does not result in any significant increase in the rate of apoptosis. However, pretreatment of HDMEC with 3TSR (1 μmol/L) for 6 h followed by 18 h treatment with TRAIL (10 ng/mL) resulted in close to 30% apoptosis in these cells (Fig. 5D). In support of this cooperation between 3TSR and TRAIL, we also observed a synergistic effect in the activation of caspase-8 by 3TSR and TRAIL. Whereas treatment of HDMEC with TRAIL (10 ng/mL) for 18 h or with 3TSR (1 μmol/L) for 6 h did not result in any significant processing of caspase-8 (Fig. 5D), pretreatment of HDMEC with 3TSR for 6 h followed by treatment with TRAIL (10 ng/mL) for 18 h did result in processing of caspase-8 as is evident from the appearance of the signature p43/p41 fragments (Fig. 5D).
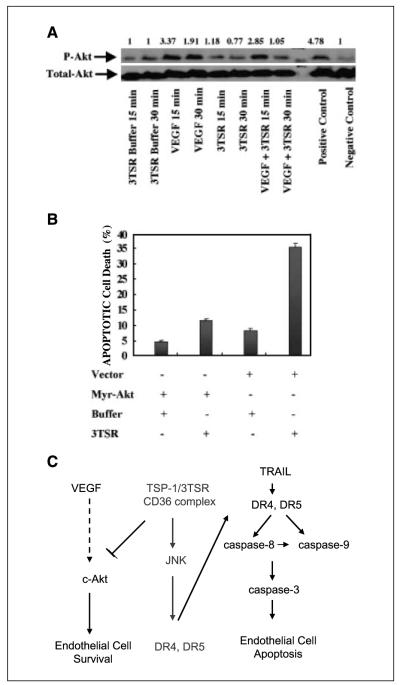
3TSR inhibits VEGF-stimulated survival and promotes down-regulation of c-Akt activity
A delicate balance of negative and positive signals regulates angiogenesis in normal and pathologic conditions. VEGF is a positive regulator of angiogenesis and protects them from apoptosis (40–42). The survival activity of VEGF and other growth factors is thought to promote resistance of endothelial cells to apoptosis (43–45). Recently, it has been shown that the protein kinase c-Akt functions as a critical part of the survival signaling pathway downstream of VEGF (44, 46). Therefore, we investigated whether 3TSR could suppress survival of endothelial cells through suppression of the VEGF-induced activation of c-Akt. To test this hypothesis, HDMEC were preincubated with 3TSR (2 μmol/L for 8 h) followed by 15 or 30 min treatment with 100 ng/mL VEGF (Fig. 6A). We observed that 3TSR reduced the basal level of c-Akt phosphorylation and also significantly inhibited the VEGF-induced phosphorylation of c-Akt.
Figure 6.
VEGF induces Akt activation in HDMEC and 3TSR inhibits basal and VEGF-induced Akt activation of HDMEC. A, HDMEC were treated with control buffer, 3TSR (2 μmol/L), VEGF (100 ng/mL), or a combination of 3TSR and VEGF for the indicated times. B, exogenous expression of Myr-Akt inhibits 3TSR-induced apoptosis in HDMEC. Fluorescence-activated cell sorting analysis indicated that exogenous expression of myr-Akt inhibited 3TSR-mediated apoptosis in microvascular endothelial cells. C, 3TSR acts as a double-edged sword promoting apoptosis via up-regulation of DR4 and DR5 and inhibiting cell survival via attenuation of the Akt pathway.
To further corroborate these observations, HDMEC were transfected with a constitutively active mutant of Akt (Myr-Akt). Thirty-six hours after transfection, the cells were either treated with 1 μmol/L 3TSR for 24 h to analyze the level of apoptosis in these cells (Fig. 6B) or lysed to verify the expression of myr-Akt (data not shown). As shown in Fig. 6B, Myr-Akt protects HDMEC from 3TSR-induced apoptosis.
Discussion
Here, we report that Lexatumumab and 3TSR cooperate in inhibiting tumor growth in vivo. This cooperative action highlights the importance of targeting tumors and their microenvironment for an efficacious therapeutic strategy. Additionally, we report that the mechanism by which 3TSR promotes apoptosis of endothelial cells is through the JNK-dependent induction of DR4 and DR5 expression. This up-regulation of DR4 and DR5 subsequently leads to the activation of the intrinsic and extrinsic apoptotic pathways, culminating in the activation of a cascade of executioner caspases including caspase-3. These studies establish that the death receptors are integrally involved in the mechanism by which 3TSR promotes apoptosis. As TRAIL and TRAIL receptors have been shown previously to play a key physiologic role in the immune response against tumor cells, here we show that TRAIL receptors can also be involved in regulating the angiogenic response in endothelial cells. More importantly, we show that whereas HDMEC are highly resistant to TRAIL-induced apoptosis, 3TSR and TRAIL synergistically cooperate to induce apoptosis of these cells. Thus, the 3TSR-induced increase in DR4 and DR5 sensitizes these cells to TRAIL-induced apoptosis.
The induction of DR4 and DR5 receptors by 3TSR on endothelial cells suggests that the cooperative action of 3TSR and Lexatumumab could be due to Lexatumumab-induced apoptosis of both the tumor and tumor-associated endothelial cells. It is likely that future clinical trials may reveal that Lexatumumab combined with 3TSR can indeed be more beneficial in humans, as it may be able to induce apoptosis of 3TSR-primed endothelial cells. However, reagents for investigating the direct effect of TRAIL (TRAIL or TRAIL receptor agonist antibodies directed against mouse cells) on tumor-associated endothelial cells are not currently available. We are currently investigating the molecular mechanism for cooper-ative action of Lexatumumab and 3TSR. One possible mechanism for this cooperative action is that 3TSR, through its apoptotic and antiangiogenic effects, can induce hypoxia in the tumor microenvironment, which could sensitize these tumors to the action of Lexatumumab (47).
The intrinsic apoptotic pathway can be triggered by death receptors as well as by a variety of stress conditions including chemotherapeutic agents, leading to the induction of mitochondrial perturbation and the release of cytochrome c. Our results indicate that the mitochondrial pathway activation is necessary for 3TSR-induced apoptosis in endothelial cells. As mitochondria are potent integrators/coordinators of apoptotic signaling pathways (5, 48), the induction of the intrinsic pathway by TSP-1 and 3TSR may play a critical role in determining the fate of endothelial cells as well as determining their response to death receptor antibody therapy and chemotherapy.
In summary, we show that 3TSR sensitizes endothelial cells by up-regulating death receptor expression and down-regulating survival signaling, leading to an enhanced antiangiogenic effect (Fig. 6C). Furthermore, these studies suggest that therapeutic approaches that use a combination of 3TSR and TRAIL may prove to be more efficacious in cancer therapy than 3TSR or TRAIL alone by providing a double hit, targeting both tumor and tumor-associated endothelial compartments.
Supplementary Material
Acknowledgments
Grant support: National Cancer Institute grant CA92644; National Heart, Lung, and Blood Institute grants HL68003 and HL80192; and Medical Foundation/Dolphin Trust Grant, American College of Surgeons Faculty Research Fellowship, and National Cancer Institute K08 award CA88965 (S. Parangi). R. Khosravi-Far is an American Cancer Society Scholar.
We thank Carole Perrozzi and Eric Galardi for technical assistance and Susan Glueck for editing of this article.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Armstrong LC, Bornstein P. Thrombospondins 1 and 2 function as inhibitors of angiogenesis. Matrix Biol. 2003;22:63–71. doi: 10.1016/s0945-053x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 2.Nor JE, Mitra RS, Sutorik MM, Mooney DJ, Castle VP, Polverini PJ. Thrombospondin-1 induces endothelial cell apoptosis and inhibits angiogenesis by activating the caspase death pathway. J Vasc Res. 2000;37:209–18. doi: 10.1159/000025733. [DOI] [PubMed] [Google Scholar]
- 3.Stupack DG, Cheresh DA. Apoptotic cues from the extracellular matrix: regulators of angiogenesis. Oncogene. 2003;22:9022–9. doi: 10.1038/sj.onc.1207110. [DOI] [PubMed] [Google Scholar]
- 4.Jimenez B, Volpert OV, Crawford SE, Febbraio M, Silverstein RL, Bouck N. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin 1. Nat Med. 2000;6:41–8. doi: 10.1038/71517. [DOI] [PubMed] [Google Scholar]
- 5.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 6.Ozoren N, El-Deiry WS. Cell surface death receptor signaling in normal and cancer cells. Semin Cancer Biol. 2003;13:135–47. doi: 10.1016/s1044-579x(02)00131-1. [DOI] [PubMed] [Google Scholar]
- 7.Peter ME, Krammer PH. Mechanisms of CD95 (APO-1/Fas)-mediated apoptosis. Curr Opin Immunol. 1998;10:545–51. doi: 10.1016/s0952-7915(98)80222-7. [DOI] [PubMed] [Google Scholar]
- 8.Abe K, Kurakin A, Mohseni-Maybodi M, Kay B, Khosravi-Far R. The complexity of TNF-related apoptosis-inducing ligand. Ann N Y Acad Sci. 2000;926:52–63. doi: 10.1111/j.1749-6632.2000.tb05598.x. [DOI] [PubMed] [Google Scholar]
- 9.Khosravi-Far R, Esposti MD. Death receptor signals to mitochondria. Cancer Biol Ther. 2004;3:1051–7. doi: 10.4161/cbt.3.11.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scaffidi C, Fulda S, Srinivasan A, et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–87. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dempsey PW, Doyle SE, He JQ, Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/s1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 12.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–2. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 13.Zou W, Liu X, Yue P, et al. c-Jun NH2-terminal kinase-mediated up-regulation of death receptor 5 contributes to induction of apoptosis by the novel synthetic triterpenoid methyl-2-cyano-3,12-dioxooleana-1,9-dien-28-oate in human lung cancer cells. Cancer Res. 2004;64:7570–8. doi: 10.1158/0008-5472.CAN-04-1238. [DOI] [PubMed] [Google Scholar]
- 14.Dent P, Yacoub A, Contessa J, et al. Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat Res. 2003;159:283–300. doi: 10.1667/0033-7587(2003)159[0283:sariao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Drosopoulos K, Pintzas A. Multifaceted targeting in cancer: the recent cell death players meet the usual oncogene suspects. Expert Opin Ther Targets. 2007;11:641–59. doi: 10.1517/14728222.11.5.641. [DOI] [PubMed] [Google Scholar]
- 16.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–85. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 17.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25:4798–811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer U, Voloshanenko O, Willen D, Walczak H. TRAIL: a multifunctional cytokine. Front Biosci. 2007;12:3813–24. doi: 10.2741/2354. [DOI] [PubMed] [Google Scholar]
- 19.Zamai L, Ponti C, Mirandola P, et al. NK cells and cancer. J Immunol. 2007;178:4011–6. doi: 10.4049/jimmunol.178.7.4011. [DOI] [PubMed] [Google Scholar]
- 20.Hall MA, Cleveland JL. Clearing the TRAIL for cancer therapy. Cancer Cell. 2007;12:4–6. doi: 10.1016/j.ccr.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Carlo-Stella C, Lavazza C, Locatelli A, Vigano L, Gianni AM, Gianni L. Targeting TRAIL agonistic receptors for cancer therapy. Clin Cancer Res. 2007;13:2313–7. doi: 10.1158/1078-0432.CCR-06-2774. [DOI] [PubMed] [Google Scholar]
- 22.Ricci MS, Kim SH, Ogi K, et al. Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell. 2007;12:66–80. doi: 10.1016/j.ccr.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Ren B, Yee KO, Lawler J, Khosravi-Far R. Regulation of tumor angiogenesis by thrombospondin 1. Biochim Biophys Acta. 2006;1765:178–88. doi: 10.1016/j.bbcan.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Lawler J, Hynes RO. The structure of human thrombospondin, an adhesive glycoprotein with multiple calcium-binding sites and homologies with several different proteins. J Cell Biol. 1986;103:1635–48. doi: 10.1083/jcb.103.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bornstein P, Agah A, Kyriakides TR. The role of thrombospondins 1 and 2 in the regulation of cell-matrix interactions, collagen fibril formation, and the response to injury. Int J Biochem Cell Biol. 2004;36:1115–25. doi: 10.1016/j.biocel.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Guo N, Krutzsch HC, Inman JK, Roberts DD. Thrombospondin 1 and type I repeat peptides of thrombospondin 1 specifically induce apoptosis of endothelial cells. Cancer Res. 1997;57:1735–42. [PubMed] [Google Scholar]
- 27.Volpert OV, Zaichuk T, Zhou W, et al. Inducer-stimulated Fas targets activated endothelium for destruction by anti-angiogenic thrombospondin-1 and pigment epithelium-derived factor. Nat Med. 2002;8:349–57. doi: 10.1038/nm0402-349. [DOI] [PubMed] [Google Scholar]
- 28.Reiher FK, Volpert OV, Jimenez B, et al. Inhibition of tumor growth by systemic treatment with thrombospondin-1 peptide mimetics. Int J Cancer. 2002;98:682–9. doi: 10.1002/ijc.10247. [DOI] [PubMed] [Google Scholar]
- 29.Quesada AJ, Nelius T, Yap R, et al. In vivo upregulation of CD95 and CD95L causes synergistic inhibition of angiogenesis by TSP1 peptide and metronomic doxorubicin treatment. Cell Death Differ. 2005;12:649–58. doi: 10.1038/sj.cdd.4401615. [DOI] [PubMed] [Google Scholar]
- 30.Tolsma SS, Volpert OV, Good DJ, Frazier WA, Polverini PJ, Bouck N. Peptides derived from two separate domains of the matrix protein thrombospondin-1 have anti-angiogenic activity. J Cell Biol. 1993;122:497–511. doi: 10.1083/jcb.122.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao WM, Seng WL, Duquette M, Lawler P, Laus C, Lawler J. Thrombospondin-1 type 1 repeat recombinant proteins inhibit tumor growth through transforming growth factor-β-dependent and -independent mechanisms. Cancer Res. 2001;61:7830–9. [PubMed] [Google Scholar]
- 32.Richard L, Velasco P, Detmar M. A simple immunomagnetic protocol for the selective isolation and long-term culture of human dermal microvascular endothelial cells. Exp Cell Res. 1998;240:1–6. doi: 10.1006/excr.1998.3936. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Galardi E, Duquette M, Delic M, Lawler J, Parangi S. Antiangiogenic treatment with the three thrombospondin-1 type 1 repeats recombinant protein in an orthotopic human pancreatic cancer model. Clin Cancer Res. 2005;11:2337–44. doi: 10.1158/1078-0432.CCR-04-1900. [DOI] [PubMed] [Google Scholar]
- 34.Ghaffari S, Jagani Z, Kitidis C, Lodish HF, Khosravi-Far R. Cytokines and BCR-ABL mediate suppression of TRAIL-induced apoptosis through inhibition of forkhead FOXO3a transcription factor. Proc Natl Acad Sci U S A. 2003;100:6523–8. doi: 10.1073/pnas.0731871100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song K, Benhaga N, Anderson RL, Khosravi-Far R. Transduction of tumor necrosis factor-related apoptosis-inducing ligand into hematopoietic cells leads to inhibition of syngeneic tumor growth in vivo. Cancer Res. 2006;66:6304–11. doi: 10.1158/0008-5472.CAN-05-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song K, Mariappan R, Khosravi-Far R. Analysis of TNF-related apoptosis-inducing ligand in vivo through bone marrow transduction and transplantation. Methods Enzymol. 2008;446:315–31. doi: 10.1016/S0076-6879(08)01619-4. [DOI] [PubMed] [Google Scholar]
- 37.Dawson DW, Pearce SF, Zhong R, Silverstein RL, Frazier WA, Bouck NP. CD36 mediates the in vitro inhibitory effects of thrombospondin-1 on endothelial cells. J Cell Biol. 1997;138:707–17. doi: 10.1083/jcb.138.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchida T, Kijima H, Tokunaga T, et al. Expression of the thrombospondin 1 receptor CD36 is correlated with decreased stromal vascularisation in colon cancer. Int J Oncol. 1999;14:47–51. [PubMed] [Google Scholar]
- 39.Bar-Sagi D, Rotin D, Batzer A, Mandiyan V, Schlessinger J. SH3 domains direct cellular localization of signaling molecules. Cell. 1993;74:83–91. doi: 10.1016/0092-8674(93)90296-3. [DOI] [PubMed] [Google Scholar]
- 40.Dvorak HF. Vascular permeability factor/vascular endothelial growth factor: a critical cytokine in tumor angiogenesis and a potential target for diagnosis and therapy. J Clin Oncol. 2002;20:4368–80. doi: 10.1200/JCO.2002.10.088. [DOI] [PubMed] [Google Scholar]
- 41.Nor JE, Christensen J, Mooney DJ, Polverini PJ. Vascular endothelial growth factor (VEGF)-mediated angiogenesis is associated with enhanced endothelial cell survival and induction of Bcl-2 expression. Am J Pathol. 1999;154:375–84. doi: 10.1016/S0002-9440(10)65284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhyay D, Datta K. Multiple regulatory pathways of vascular permeability factor/vascular endothelial growth factor (VPF/VEGF) expression in tumors. Semin Cancer Biol. 2004;14:123–30. doi: 10.1016/j.semcancer.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 43.Alon T, Hemo I, Itin A, Pe’er J, Stone J, Keshet E. Vascular endothelial growth factor acts as a survival factor for newly formed retinal vessels and has implications for retinopathy of prematurity. Nat Med. 1995;1:1024–8. doi: 10.1038/nm1095-1024. [DOI] [PubMed] [Google Scholar]
- 44.Gerber HP, McMurtrey A, Kowalski J, et al. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3′-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J Biol Chem. 1998;273:30336–43. doi: 10.1074/jbc.273.46.30336. [DOI] [PubMed] [Google Scholar]
- 45.Gerber HP, Dixit V, Ferrara N. Vascular endothelial growth factor induces expression of the antiapoptotic proteins Bcl-2 and A1 in vascular endothelial cells. J Biol Chem. 1998;273:13313–6. doi: 10.1074/jbc.273.21.13313. [DOI] [PubMed] [Google Scholar]
- 46.Fujio Y, Walsh K. Akt mediates cytoprotection of endothelial cells by vascular endothelial growth factor in an anchorage-dependent manner. J Biol Chem. 1999;274:16349–54. doi: 10.1074/jbc.274.23.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayes PA, Campbell L, Ricci MS, Plastaras JP, Dicker DT, El-Deiry WS. Modulation of TRAIL-induced tumor cell apoptosis in a hypoxic environment. Cancer Biol Ther. 2005;4:1068–74. doi: 10.4161/cbt.4.10.2255. [DOI] [PubMed] [Google Scholar]
- 48.Brenner C, Le Bras M, Kroemer G. Insights into the mitochondrial signaling pathway: what lessons for chemotherapy? J Clin Immunol. 2003;23:73–80. doi: 10.1023/a:1022541009662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.