SUMMARY
Asf1 is a highly conserved chaperone of histones H3/H4 that assembles or disassembles chromatin during transcription, replication, and repair. The structure of the globular domain of Asf1 bound to H3/H4 determined by X-ray crystallography to a resolution of 1.7 Å shows how Asf1 binds the H3/H4 heterodimer, enveloping the C-terminus of histone H3 and physically blocking formation of the H3/H4 heterotetramer. Unexpectedly, the C-terminus of histone H4 that forms a mini-beta sheet with histone H2A in the nucleosome, undergoes a major conformational change upon binding to Asf1 and adds a beta strand to the Asf1 beta-sheet sandwich. Interactions with both H3 and H4 were required for Asf1 histone chaperone function in vivo and in vitro. The Asf1-H3/H4 structure suggests a “strand-capture” mechanism whereby the H4 tail acts as a lever to facilitate chromatin disassembly / assembly that may be used ubiquitously by histone chaperones.
INTRODUCTION
The packaging of the eukaryotic genome into chromatin is essential for normal growth, development, and differentiation. The repeating unit of chromatin is the nucleosome core particle, which comprises 147 bp of DNA wound around a histone octamer (Luger et al., 1997). Chromatin is a dynamic structure that tightly regulates transcription, replication, repair, and recombination. In the context of these processes, the most severe alteration of chromatin structure is the removal of histone proteins from DNA (chromatin disassembly) or the deposition of histone proteins onto naked DNA (chromatin assembly). The ordered packaging of DNA into chromatin is thought to involve the initial deposition of a heterotetramer of histones H3/H4 followed by two heterodimers of histones H2A/H2B to form the nucleosome. This process is mediated by histone chaperone proteins that regulate the association of the basic histone proteins with the DNA, which permits the nucleosome to form in an ordered and controlled manner (Akey and Luger, 2003; Loyola and Almouzni, 2004).
The histone chaperone Anti-silencing function 1 (Asf1) is the only histone chaperone that is implicated in both replication-dependent and replication-independent chromatin assembly (Nakatani et al., 2004). Asf1 is also a critical factor in multiple other cellular processes. For example, Asf1 is a histone H3/H4 chaperone that assists Chromatin Assembly Factor 1 (CAF-1) during the assembly of newly-synthesized DNA into chromatin in vitro (Mello et al., 2002; Smith and Stillman, 1991; Tyler et al., 1999) and is required for replication-independent chromatin assembly together with the Hir histone chaperone (Green et al., 2005; Tagami et al., 2004). Asf1 also mediates chromatin disassembly from promoters in budding yeast during transcriptional activation (Adkins et al., 2004) and chromatin disassembly and reassembly during transcriptional elongation (Schwabish and Struhl, 2006). In fact, all non DNA-bound histones are bound to Asf1 (Groth et al., 2005; Tagami et al., 2004), underscoring its fundamental role as a central histone chaperone in eukaryotes.
The function and structure of Asf1 are highly conserved among eukaryotes. The N-terminal 155 residues of Asf1 form a globular core that consists of an immunoglobulin-like fold (Daganzo et al., 2003; Mousson et al., 2005) with highly conserved acidic patches that are predicted to mediate interactions with histone H3. The structures of histones H3 and H4 have also been determined as components of the nucleosome core particle and histone octamer (Arents et al., 1991; Luger et al., 1997). The yeast and Xenopus laevis nucleosome structures underscore the structural and functional conservation of this complex throughout evolution (White et al., 2001). Thus, it is well known how the individual histones interact with each other and with DNA, but there is little information about the interaction of the histones with each other or with chaperones in the absence of DNA. As a result, the mechanisms by which the nucleosomes are assembled and disassembled are not well understood.
Recently, we biophysically characterized the Asf1-H3/H4 complex and demonstrated that Asf1 binds to a heterodimer of histones H3 and H4 (English et al., 2005). In addition, yeast two-hybrid analysis suggests that the C-terminus of histone H3 interacts with Asf1 (Munakata et al., 2000), and NMR chemical-shift mapping revealed interactions of an H3 peptide (residues 122-135) with a highly conserved and acidic patch on the concave surface of Asf1 (Mousson et al., 2005). This histone H3 peptide corresponds to helix 3, which is a crucial part of a four-helix bundle that forms the H3:H3 dimerization interface in the nucleosome (Luger et al., 1997). Furthermore, a disruptive mutation in the middle of this H3-interacting region of Asf1, V94R, abolished the interaction with histone H3 (Mousson et al., 2005). These data raised the possibility that the region that mediates H3:H3 dimerization in the H3/H4 heterotetramer may also be the region of H3 that binds to Asf1.
In this study, we have determined the crystal structure of Asf1 bound to histones H3/H4, revealing how a histone chaperone binds histones and how the structures of H3 and H4 differ outside of the histone octamer. We have confirmed that Asf1 binds to a heterodimer of histones H3/H4 and that Asf1 not only interacts with histone H3 but also with the C-terminal tail of histone H4 in vivo and in vitro. Of interest, the C-terminal tail of H4 undergoes a major conformational change upon binding to Asf1, suggesting a mechanism by which the H4 tail may act as a handle for the assembly and disassembly of the H3/H4 heterotetramers by Asf1.
RESULTS
Asf1, H3, and H4 form a heterotrimeric complex
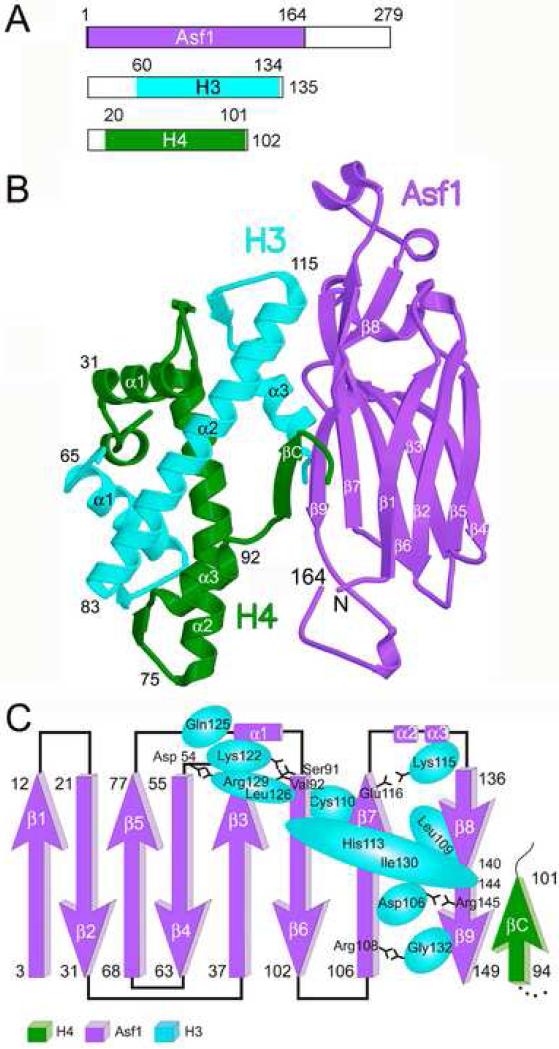
In order to investigate how Asf1 interacts with histones H3 and H4, we determined the structure of the globular core of Asf1 bound to a histone H3/H4 heterodimer at a resolution of 1.7 Å (Fig. 1). N-terminal truncation of the histones and co-expression of the three proteins were necessary to obtain the crystals, which contained the conserved region of Saccharomyces cerevisiae Asf1 (yAsf1, 1-169) together with Xenopus laevis histones H3 (60-135) and H4 (20-102) (Fig. 1A). A single Asf1-H3/H4 complex formed the asymmetric unit in the crystals, which belong to space group P3121 (Suppl. Table 1). The structure was solved by molecular replacement with the structures of Asf1 and the histone H3/H4 dimer from the nucleosome (Fig. 1B) (Daganzo et al., 2003; Luger et al., 1997). The final refined model (Suppl. Table 1) contains yAsf1 (1-164), Xenopus H3 (61-134), Xenopus H4 (21-101), solvent molecules, and ions.
Figure 1. Asf1-H3/H4 structure.
A. Regions of the Asf1, H3 and H4 proteins that appear in this structure are shown as colored regions of the boxes representing the full-length proteins. B. The ribbon diagram shows the overall structure of the Asf1-H3/H4 complex, with Asf1 in violet, H3 in cyan, and H4 in green. The major secondary structure elements are labeled. C. Topology diagram of Asf1 with contacts between Asf1 and H3 mapped to indicate the extent of H3-Asf1 interactions. The amino acids of histone H3 that significantly contribute to the Asf1 interface are represented by the cyan ovals, whose sizes are proportional to the extent of the buried surface, and are labeled accordingly.
The overall structure of the Asf1-H3/H4 complex reveals an extensive interaction interface of Asf1 with both histones H3 and H4 (Fig. 1B; Suppl. Table 2). As previously described for the structures of free Asf1 (Daganzo et al., 2003; Mousson et al., 2005), the core of Asf1 is an elongated β-sandwich domain with three α helices in the loops between the beta strands. This structure has a concave face comprising beta strands β3, β4, and β6-9, with high sequence-conservation across species and a distinctly negative charge. In fact, this is the site of histone H3 binding (Figs 1C and 2). The contacts between H3 and Asf1 are extensive and result in a buried surface area of 909 Å2. The histones retain the configuration of the conserved histone fold (Luger, 2003) with a few important exceptions. Much of the alpha helical region (aa 60-85) of H4 is near, but not in direct contact with Asf1 (Fig. 1C). However, the C-terminus of H4 forms a new beta strand (H4 βC) with the core of Asf1, which creates a hydrogen-bonding network connecting all three proteins, mediated by T147 of Asf1 (Fig. 2). In addition, the H4 C-terminus interacts with the core of Asf1 between β1 and β8-9, which greatly contributes to the total Asf1-H4 buried surface area of 506 Å2.
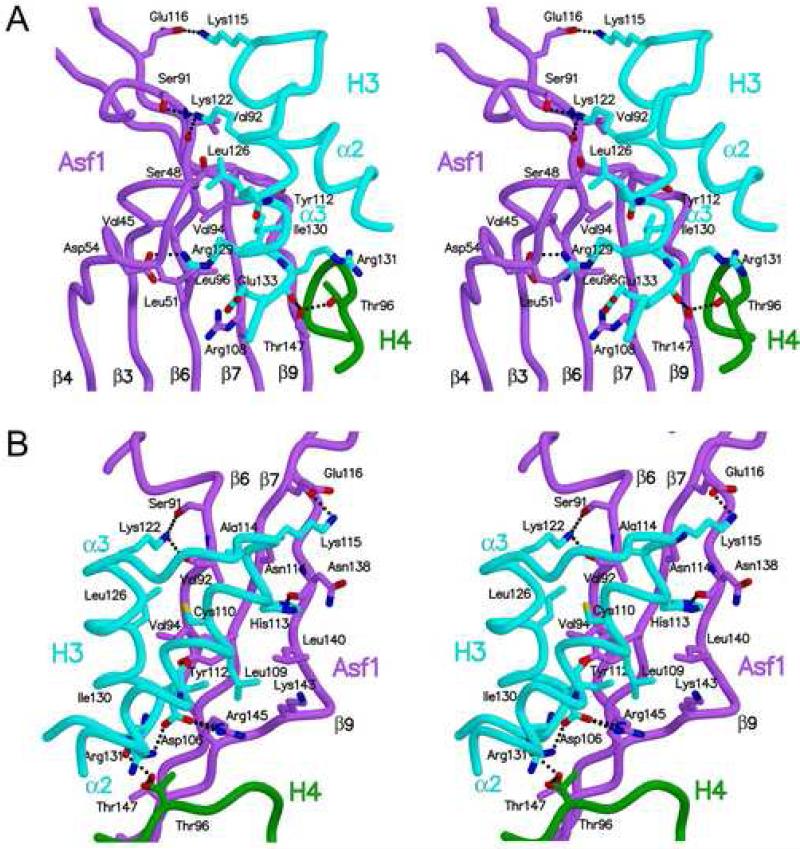
Figure 2. Interactions of Asf1 with histone H3.
A. The stereodiagram shows the details of the interface between Asf1 and H3 focusing on the interactions mediated by alpha helix (α3) of histone H3. Asf1 is colored in violet, H3 in cyan, and H4 in green, with several residues labeled, and dotted lines to indicate inferred hydrogen bonds. B. The view in this stereodiagram is rotated approximately 120° clockwise about the vertical axis and 25° toward the viewer about the horizontal axis compared to panel A, to show the details of the interface between Asf1 and alpha helix (α2) of histone H3, colored as in A.
Asf1 directly interacts with H3
Analysis of the Asf1-H3 interface reveals that the extensive interaction of histone H3 with Asf1 includes H3 α2 in addition to α3 (Fig. 1B,C). The region of the Asf1-H3 interface that is mediated by the C-terminal helix of H3 (α3, residues 122-134) makes extensive contacts with each of the Asf1 beta strands on the concave face of the protein (Fig. 2A). Asf1 V94, which is an important residue for the interaction with histone H3, lies within this region, and it is clear why substituting this residue with an arginine would disrupt H3 binding (Mousson et al., 2005). However, the extent of the contacts between Asf1 and H3 is not limited to α3 of histone H3, as was previously thought, because the C-terminal part of H3 α2 (106-115) and the loop connecting α2-α3 (aa 114-122) interact with residues in β7-9 of Asf1 (Fig. 2B). Specifically, there are a number of inferred electrostatic interactions and hydrogen bonds that we would expect to play important roles in the stability of the Asf1-H3 complex, such as D54Asf1-R129H3, R145Asf1-D106H3, and T147Asf1-R131H3 (Fig. 2A and B; Suppl. Fig 1; Suppl. Table 2). In addition, many van der Waals interactions also contribute to the stabilization of this complex (Suppl. Table 2). We conclude that Asf1 and H3 form an extensive interface with a high degree of shape complementarity and specificity due to a large number of specific hydrogen bonds, ion pairs, and van der Waals interactions.
Asf1 directly interacts with H4
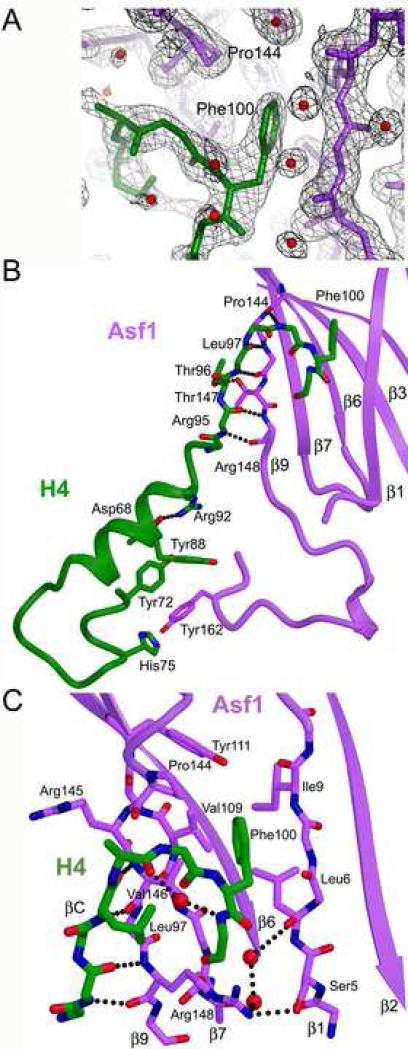
It has been widely assumed that the interaction of Asf1 with histones H3 and H4 is mediated solely by the C-terminal helix of histone H3 (Mousson et al., 2005; Munakata et al., 2000), but the Asf1-H3/H4 structure clearly shows that Asf1 also directly binds to histone H4. Figure 3A shows the electron density map of the C-terminal tail of H4, with F100 binding in the hydrophobic core of Asf1, to illustrate the quality of the data in this region. H4 interacts with two separate parts of Asf1. Residues in the histone-fold core of H4, specifically in α2 (Y72 and H75) and α3 (Y88), interact with Y162 in the C-terminal part of Asf1 (Fig. 3B). However, these residues are also involved in significant crystal-packing contacts with symmetry-related H3 and H4 molecules, which may influence the details of the structure in this region.
Figure 3. Interactions of H4 with Asf1.
A. The electron density map (σA weighted 2Fo-Fc map contoured at 1σ) is shown with Asf1 in violet, solvent molecules in red, and H4 in green. B. The diagram illustrates the two regions of H4 that bind to Asf1. The side chains of H4 that interact directly with Asf1 are shown with other portions of the proteins shown in a ribbon representation. The figure is colored as in A, with oxygen atoms in red, nitrogen atoms in blue, and inferred hydrogen bonds are shown as dotted lines. C. Close-up view of the H4 C-terminal interaction with the core of Asf1. The view is zoomed in and rotated slightly clockwise about the vertical axis compared to panel B.
The more extensive and intriguing interaction between Asf1 and histone H4 occurs within the last 10 residues of the H4 protein. H4 residues 95-98 form an additional anti-parallel beta strand (βC) with β9 of Asf1 through several main-chain hydrogen bonds (Fig. 3B,C, Suppl. Table 2). Within this beta strand, T96H4 Oγ1 forms a hydrogen bond with T147Asf1 Oγ1 that in turn hydrogen bonds to the main chain of H3, linking all three proteins. In addition, the five final amino acids of H4 interact with Asf1 behind the newly formed beta strand, inserting into a hydrophobic pocket created between beta strands β1 and β9 (Fig. 3C). An important contributor to this interaction is the F100 side chain of H4 (Fig. 3A and C), which itself buries 172 Å2 of surface area, nearly 35% of the total surface area of the Asf1-H4 interface, through van der Waals interactions with Asf1 L6, V146, Y111, V109, and P144 (Fig. 3C). Therefore, H4 significantly contributes to the binding of the H3/H4 dimer to Asf1 through several main-chain and side-chain hydrogen bonds as well as burial of a key hydrophobic residue.
Structural changes occur in the formation of the Asf1-H3/H4 complex
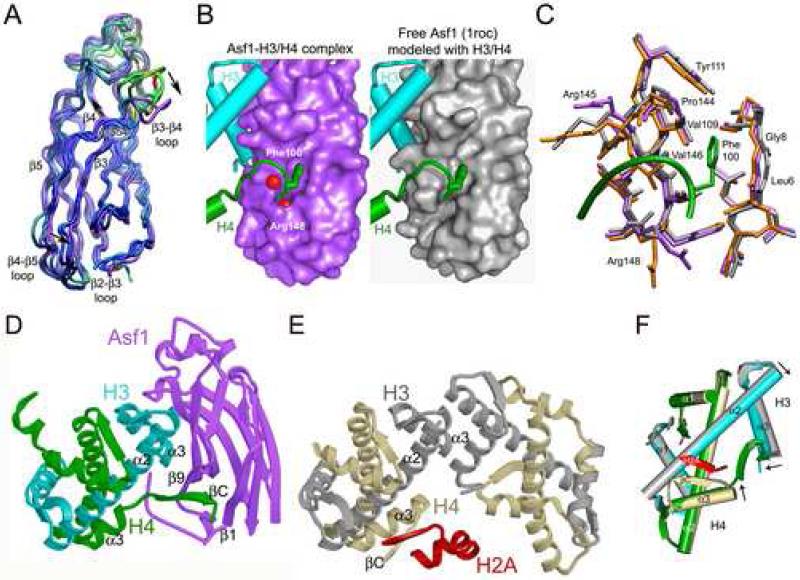
Comparison of the Asf1-H3/H4 structure with the structures of free Asf1 reveals subtle conformational changes that occur in Asf1 upon binding to the H3/H4 dimer (Fig. 4A). The overall root mean squared deviation (r.m.s.d) of main-chain atoms of Asf1 in the Asf1-H3/H4 complex compared to the free proteins is 2.42 Å, 1.79 Å, and 1.42 Å for the yAsf1 structure at 2.95 Å resolution (PDB ID 1wg3 (Padmanabhan et al., 2005)), yAsf1 structure at a resolution of 1.5 Å (PDB ID 1roc, (Daganzo et al., 2003)), and hAsf1a structure determined by NMR (PDB ID 1tey (Mousson et al., 2005)), respectively. The largest differences between the structures occur in the loop regions, as expected for structures determined by different methods or crystallized in different space groups. However, the β3-β4 loop moves approximately 6 Å closer to H3 from its position in free Asf1 (PDB ID 1roc), and the strands β3 and β4 move nearly 2 Å away from H3 around residue D54 in β4, which together appear to facilitate the formation of the D54Asf1-R129H3 ion pair and surrounding contacts. The movements about this pivot point (D54) in β3 and β4 result in an overall less twisted beta sheet and a shift of up to 3.5 Å between β3 and β4 of Asf1, and in the associated loops (Fig. 4A). However, this region of Asf1 (aa 35-77) is also involved in crystal packing contacts with H3 (aa 60-72) and H4 (aa 20-31) of symmetry related molecules, so that the significance of these structural differences is difficult to evaluate in the context of this crystal.
Figure 4. Asf1 and histones undergo structural changes to accommodate binding.
A. The overlay diagram of Asf1 from this study (in violet) has the three other Asf1 structures superimposed. The free Asf1 models are colored according to r.m.s.d. value from red (most different) to blue (most similar). Arrows indicate the directions and positions of notable structural changes, and Asp54 is indicated. B. Surface representations of Asf1 in the current structure on the left and free Asf1 modeled with histones (from PDB 1roc (Daganzo et al., 2003)). H3 is in cyan, H4 is in green, and free Asf1 is in grey. Water molecules are shown as red spheres. C. The superimposed Asf1 proteins are from two Asf1 structures (1roc is in grey and 1wg3 is in orange) overlaid onto the Asf1-H3/H4 complex. D. Ribbon diagram of the Asf1-H3/H4 complex, with Asf1 colored in violet, H3 in cyan, and H4 in green. E. Ribbon diagram showing the histone H3/H4 heterotetramer from the nucleosome core particle (PDB ID 1k5x (Davey et al., 2002)) oriented so that the H3/H4 dimer on the left is superimposed with the H3/H4 dimer in panel A, with coloring of H3 in silver, H4 in tan, and H2A in red. F. Cartoon diagram showing the superposition of the H3/H4 dimer from the Asf1-H3/H4 complex onto one H3/H4 dimer with the C-terminus of H2A from the nucleosome, as in panels A and B. Black arrows indicate structural differences between the helices of the histones that occur in the different environments.
The structural changes that take place in Asf1 to accommodate the binding of the H4 C-terminus are minor. As seen in Figure 4B, which shows surface representations of Asf1 in the complex compared to free Asf1, very small structural changes occur for insertion of H4 F100 into the core of Asf1 (< 0.9 Å), but these result in noticeably better shape complementarity in the complex. Although no significant changes in the backbone conformation in this region of Asf1 occur in the formation of the βCH4-β9Asf1 anti-parallel beta-sheet extension, there are slight side chain movements that facilitate this interaction. For example, R145 rotates 180° about χ1 to create space for the H4 tail to enter the core of Asf1 (Fig. 4C), and the side chain of R148 rotates about χ3 to further enclose the binding pocket around F100. Without these subtle changes, H4 F100 would clash with hydrophobic residues that line the cleft of the Asf1 core (Fig. 4B). Interestingly, the minor adjustments that would be necessary in Asf1 for H4 binding are suggestive of a lock and key binding mechanism, which would be an energetically favorable interaction.
In contrast, there is a profound structural change for histone H4 in the Asf1-H3/H4 complex when compared to the structure of H4 in the nucleosome core particle or histone octamer (Fig. 4D-F). Panel E in Figure 4 shows that the C-terminal tail of histone H4 in the nucleosome folds back over α3 of H4 and that residues 96-99 of H4 form a two-stranded parallel beta sheet with residues 100-103 of histone H2A. However, in complex with Asf1, α3 in the H4 C-terminus terminates one turn earlier than in the nucleosome and extends 180° in the opposite direction so that residues G94 to L97 can form the anti-parallel beta sheet and insert into the core of Asf1 (Panel D, Figure 4). In fact, the r.m.s.d. value for the backbone atoms of the H4 protein in the nucleosome (PDB ID 1k5x chain B) compared to the H4 protein in the Asf1-H3/H4 structure is 1.80 Å (Fig 4D-F). This value is high for two identical proteins and is due solely to the drastic conformational change of the C-terminus, with a small portion attributed to deviations at the N-terminus. Specifically, the r.m.s.d. for the H4 C-terminal tail (aa 92-102) compared to nucleosomal H4 is 3.15 Å and without the C-terminal residues the r.m.s.d. is 0.72 Å. Importantly, when superimposed onto the H4 protein in the nucleosome (1kx5), the octamer (1tzy), the nucleosome with H2AZ (1f66), or the nucleosome with macro H2A (1u35), all H4 proteins have the same C-terminal tail structure except for H4 bound to Asf1.
The C-terminus of histone H3 in the Asf1-H3/H4 complex has a slightly different structure in comparison to its structure in the nucleosome (Fig. 4D-F). In the nucleosome, α3 of H3 extends to the very end of the protein. However, when bound to Asf1, the last few residues of α3 of H3 unfold so that they do not clash with Asf1. In fact, the r.m.s.d. for the overall H3 protein is 0.94 Å, but for the residues in H3 α3 (121-134), the r.m.s.d. is higher (1.31 Å). There is also a change in the interhelical angle between α2 and α3, which can be described as a scissoring motion with an approximate decrease in the scissoring angle of 8° for H3 in the Asf1-H3/H4 complex compared with the nucleosome. A smaller scissoring motion is observed for histone H4 (Fig. 4F). Thus, the histone fold exhibits some flexibility in α3 of both histones, regions that are important in the structure of the nucleosome.
Disruption of Asf1 function by mutations in the histone-binding interface of Asf1
To assess the validity and contribution of the Asf1-H3/H4 interactions revealed by the structure, amino acid substitutions were made in the budding yeast Asf1 protein (Table 1). In order to weaken the interaction between Asf1 and H3, we made substitutions T147E, R145E, Y112A, and S48R in Asf1. The interaction between Asf1 and histone H4 was tested with mutants T147E, V109M, V146L, P144Y, L6M, and Y162A in the core of Asf1 and C-terminal tail (Fig. 3). None of these Asf1 mutations significantly affected the expression level of Asf1 (Suppl. Fig. 2; data not shown).
Table 1.
Summary of phenotypes of Asf1, H3, and H4 mutants in the interaction interface.
| Growth | Silencing | MMS resistance | HU resistance | Zeocin resistance | PHO5 induction | |
|---|---|---|---|---|---|---|
| ASF1 WT | +++ | + | ++ | ++ | +++ | ++++ |
| asf1 Δ | + | - | - | - | + | - |
| Asf1 mutants: | ||||||
| T147E | +++ | ++ | nd | nd | +++ | nd |
| R145E | +++ | +/- | nd | nd | +++ | nd |
| L6M | +++ | ++ | nd | nd | +++ | nd |
| V109M | +++ | ++ | ++ | ++ | +++ | +++ |
| V146L | +++ | ++ | ++ | ++ | +++ | ++ |
| P144Y | +++ | + | nd | nd | +++ | nd |
| Y112A | +++ | ++ | nd | nd | +++ | nd |
| S48R | +++ | ++ | ++ | ++ | +++ | ++++ |
| V94R | + | - | - | - | +++ | - |
| Y162A | +++ | + | nd | nd | +++ | nd |
| T147E/R145E | + | +/- | - | - | +++ | - |
| Y112A/R145E | + | - | - | - | +++ | - |
| Y112A/P144Y | +++ | + | ++ | ++ | +++ | nd |
| Y112A/T147E | + | - | - | - | + | - |
| Y112A/V146L | +++ | + | ++ | ++ | +++ | nd |
| S48R/T147E | +++ | + | ++ | ++ | +++ | nd |
| S48R/R145E | ++ | - | + | + | +++ | + |
| S48R/P144Y | +++ | ++ | nd | nd | nd | nd |
| H4 mutants: | ||||||
| F100A | +++ | - | ++ | + | +++ | +++ |
| R92A1 | +++ | +* | ++ | ++ | - | ++ |
| H75Y2 | +++ | ++* | ++ | ++ | + | +++ |
| Δ92-102 | - | n/a | n/a | n/a | n/a | n/a |
| Y72G3 | + | nd | ++ | + | + | +++ |
| Y88G3 | ++ | nd | ++ | + | - | ++++ |
| H3 mutants: | ||||||
| K122Q2 | +++ | -* | ++ | ++ | - | ++++ |
| K115A2 | +++ | +* | ++ | + | - | ++ |
| K122A2 | +++ | -* | ++ | ++ | - | ++ |
The indicated mutants were previously described in the following papers:
The phenotypes designated with * indicate phenotypes determined from these previous publications. n/a= not applicable, nd= not determined.
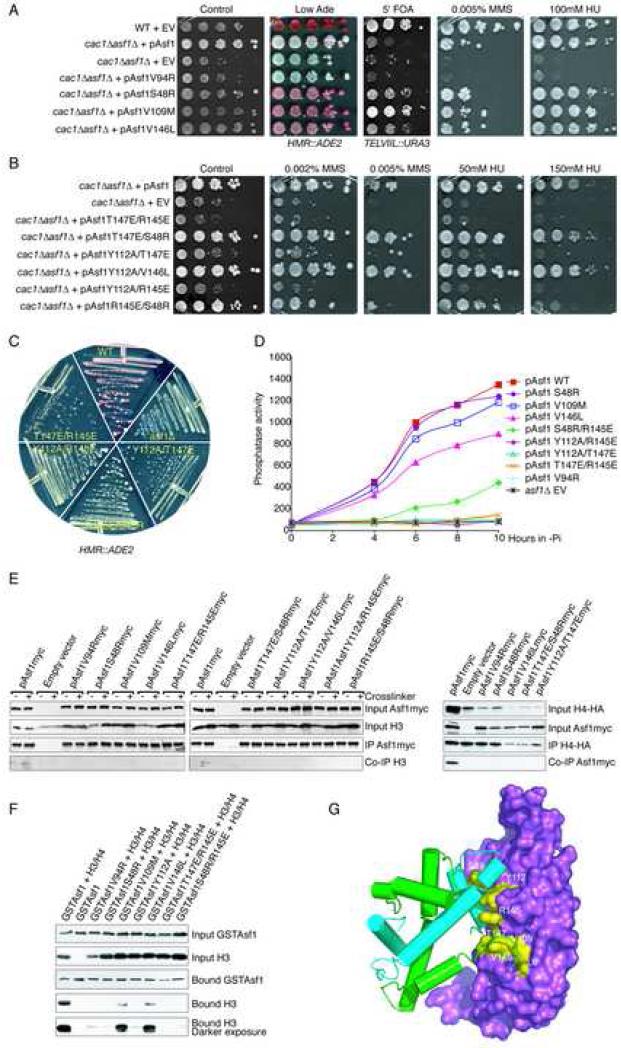
We examined the effects of the single Asf1 mutations on the function of Asf1 in transcription, cell cycle, DNA repair, and replication. The deletion of ASF1 leads to a slow-growth phenotype (Tyler et al., 1999), which is linked to the sensitivity of Asf1 mutants to DNA damaging agents (methyl methane sulfonate, MMS) and replication stress (hydroxyurea, HU) (Ramey et al., 2004). None of the single mutations in Asf1 led to pronounced defects in growth (Table 1). However, L6M, S48R, V109M, Y112A, V146L, and T147E led to enhanced transcriptional silencing of an HMR::ADE2 reporter (apparent from the red colonies in Fig. 5A; Suppl. Fig. 3), as compared to the normal Asf1 protein. Additionally, these mutants had enhanced telomeric silencing of an TELVIIL::URA3 reporter, as indicated by their increased ability to grow on 5’ fluoroorotic acid (5’FOA) (Fig. 5A; data not shown).
Figure 5. Disruption of Asf1 functions by mutations in the histone binding interfaces.
A. Asf1 mutations that lead to enhanced transcriptional silencing. A wild type (“WT”) strain containing pRS314 (“EV”) and cac1Δasf1Δ containing pRS314 (“EV”) or the wild type or mutagenized pAsf1 plasmid (as indicated) were analyzed by 10 fold serial-dilution analysis onto the indicated plates. B. Asf1 mutations that inactivate Asf1. cac1Δasf1Δ containing pRS314 (“EV”), pAsf1 plasmid, or mutagenized pAsf1 (as indicated) were analyzed as in A. C. Analyses of growth and silencing in Asf1 mutants. D. Ability of asf1 mutants to activate the PHO5 gene. Phosphate was depleted from the media of strains cac1Δasf1Δ carrying the indicated plasmids at time 0, and samples were taken and assayed for acid phosphatase activity. E. Disruption of the Asf1-histone interaction by Asf1 mutations in yeast. Co-immunoprecipitation analysis (left panel) was performed using an anti-myc antisera from strain ROY1169 carrying the indicated plasmids. The inclusion of the DSP crosslinker is indicated by the “+” sign. The input and immunoprecipitation (“IP”) samples were western blotted as indicated. Co-immunoprecipitation analysis (right panel) was performed using anti-HA antisera from strain ROY1169 carrying the indicated plasmids and overexpressing HA-H4. F. Disruption of the Asf1-histone interaction by Asf1 mutations in vitro. The inputs of the indicated co-expressed proteins and the material bound to the Glutathione affinity column were western blotted, as indicated. G. Locations of the Asf1 substitutions that alter Asf1 function.
We also conducted phenotypic analysis with pairs of Asf1 mutations. T147E/R145E, Y112A/T147E, and Y112A/R145E had a growth defect similar to that of yeast lacking Asf1 (asf1Δ), as revealed by the small colony size on rich media (Fig. 5B, C). Consistent with their growth defect, we found that the pairs of Asf1 mutations T147E/R145E, Y112A/T147E, and Y112A/R145E led to MMS and HU sensitivity that was similar to asf1Δ (Fig. 5B). The ability of the Asf1 T147E/R145E, Y112A/T147E, and Y112A/R145E proteins to mediate transcriptional silencing of the HMR::ADE2 reporter was also indistinguishable from asf1Δ, as these strains yielded white colonies (Fig. 5C). The R145E/S48R mutant of Asf1 had intermediate activity, producing yeast colonies that were smaller than wild type, being sensitive to elevated concentrations of MMS and HU and having a silencing defect (Fig. 5B,C).
To determine the role of the interactions between Asf1 and histones H3/H4 in chromatin disassembly, we measured the ability of asf1 mutants to activate the PHO5 gene. Asf1-mediated chromatin disassembly of the PHO5 gene promoter is required for transcriptional activation (Adkins et al., 2004). Asf1 mutations that inactivate Asf1 function in all the assays shown above, i.e. V94R, Y112A/R145E, Y112A/T147E, and T147E/R145E, completely failed to activate the PHO5 gene, indicating that these mutations also disrupt the chromatin disassembly activity of Asf1 (Fig. 5D). S48R resembled wild type Asf1 for PHO5 activation, V109M was slightly defective at activating PHO5, and V146L had a more significant defect in PHO5 activation (Fig. 5D).
To ascertain whether the mutations alter the binding of Asf1 to histones H3/H4, we examined their effects on the Asf1-H3/H4 complex in vivo and in vitro. All of the Asf1 mutations prevented our ability to detect co-immunoprecipitating histones from yeast (Fig. 5E). Similarly, in the reciprocal immunoprecipitation analysis all the mutations prevented our ability to detect Asf1 co-immunoprecipitating with histone H4 (Fig. 5E). We further tested the effect of the Asf1 mutations on the ability to purify the recombinant Asf1-H3/H4 complex from E. coli. At similar loadings of GST-Asf1 onto the glutathione-affinity resin, all the Asf1 mutants that were examined had greatly reduced amounts of co-purifying H3/H4 when compared to wild type Asf1 (Fig. 5F). In particular, the binding of Asf1 mutants S48R, Y112A and S48R/R145E to H3/H4 was drastically reduced to levels equivalent to the V94R disruptive mutation of Asf1, as was the binding of T147E, R145E, and Y112A/T147E (data not shown). Asf1 mutations V109M and V146L also showed significantly reduced binding to H3/H4. Asf1 T147E/R145E had the greatest reduction in H3/H4 binding seen for any Asf1 mutant to date. As such, the mutations in Asf1 that were designed to decrease histone H3/H4 binding (Fig. 5G) do indeed weaken the Asf1-histone H3/H4 interaction.
Disruption of Asf1 function by mutations in the Asf1-binding interfaces of histones H3/H4
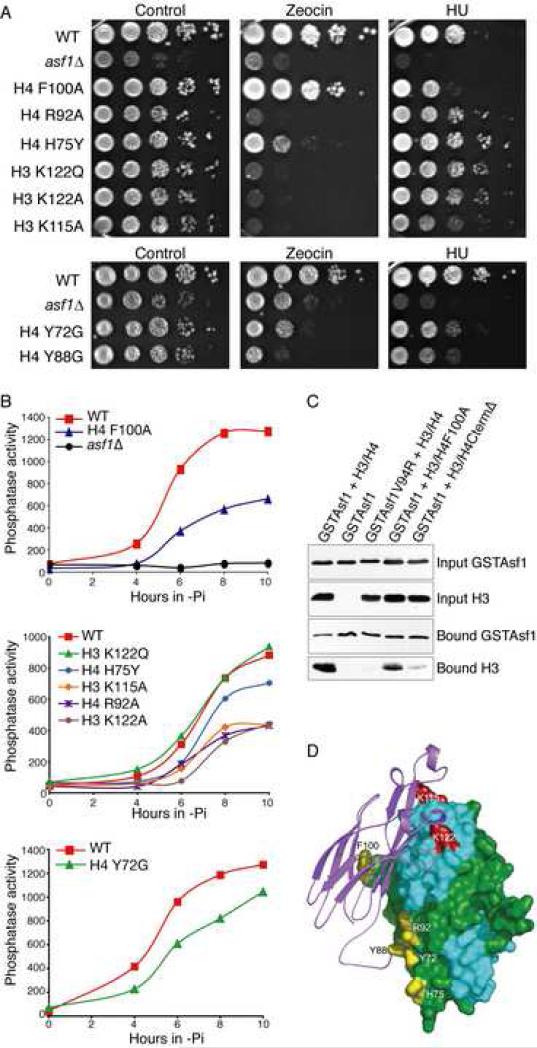
In order to further examine the physiological relevance of the interaction between Asf1 and histone H4, we created mutations in the region of H4 predicted to alter the Asf1-H4 interaction. Amino acids 92-102 were deleted from H4 in order to eliminate binding of H4 to Asf1. H4 Δ92-102 was found to be inviable as the sole source of histone H4 in the cell (Table 1), indicating that the C-terminus of histone H4 may perform an important role in the structure of the nucleosome per se. Additionally, F100A was created to alter the contacts between H4 and the pocket of Asf1 (Fig. 4B,C). The F100A mutation caused no significant growth defect (Table 1), indicating that H4 F100 does not mediate critical contacts within the nucleosome. However, H4 F100A caused slight sensitivity to replication stress (Fig. 6A) and led to a defect in transcriptional silencing comparable to deletion of the ASF1 gene (Suppl. Fig. 4). The H4 F100A mutation also led to a reduced ability to disassemble chromatin from the PHO5 promoter (Fig. 6B), indicating a role for the Asf1-H4 F100 interaction in chromatin disassembly. When we examined the effect of these H4 mutations on the ability of H3/H4 to co-purify with GSTAsf1 from E. coli, we observed a defect resulting from the F100A mutation (Fig. 6C). H4 Δ94-102 also greatly reduced the ability of H3/H4 to co-purify with Asf1. As such, the C-terminus of H4 plays an important role in the interaction of Asf1 with H3/H4 and the function of Asf1 in vivo.
Figure 6. Disruption of Asf1 functions by mutations of H3/H4.
A. Sensitivity of histone mutants to DNA damaging agents and replication stress, as described in Fig. 5A. B. Phosphatase activity was measured as described in Fig. 5D. C. The stability of the Asf1-H3/H4 complex is reduced by the F100A mutation or deletion of residues 92-102 of histone H4. The analysis was performed as described in Fig. 5F. D. Location of the H3 (in red) and H4 residues (in yellow) whose mutation alters Asf1 function.
We also analyzed other mutations in histone H4, H75Y, R92A, Y88G, and Y72G, which were expected to weaken the binding of H4 to Asf1 (Hyland et al., 2005; Santisteban et al., 1997; Xu et al., 2005) (Fig. 3B). We found that the growth defects caused by H4 Y72G and Y88G (Santisteban et al., 1997) were similar to that of yeast deleted for ASF1 (Suppl. Fig. 5). The most pronounced effect of these H4 mutations was their sensitivity to the radiomimetic Zeocin that results in double-strand DNA breaks (Fig. 6A). In order to examine the ability of these H4 mutants to disassemble chromatin, they were compared to their isogenic wild type strains for their ability to activate PHO5. We found that H4 R92A had a strong defect in PHO5 activation, while H4 H75Y and H4 Y72G had minor defects in PHO5 activation (Fig. 6B, Table 1). Taken together, these results demonstrate that four mutations in histone H4 in the region that binds to Asf1 (Fig. 6D) reduce the ability of Asf1 to mediate chromatin disassembly, indicating the importance of the Asf1-H4 interaction in chromatin disassembly.
To further characterize the physiological relevance of the interactions between Asf1 and histone H3, we examined available mutations in regions of histone H3 (Hyland et al., 2005; Santisteban et al., 1997; Xu et al., 2005) that we predicted would weaken the interaction with Asf1 (Fig. 2B). We found that H3 K115A led to sensitivity to HU to the same extent as deletion of ASF1, whereas K115A, K122A, and K122Q led to sensitivity to Zeocin that was even more pronounced than that resulting from deletion of ASF1 (Fig. 6A). Furthermore, we found that K115A and K122A, but not K122Q, led to a defect in Asf1-mediated chromatin disassembly (Fig. 6B). Importantly, the altered activity of the histone mutants was not due to potential differences in protein stability, as the mutants were all expressed to the same level as wild type histones (Suppl. Fig. 6). These results indicate that the interaction between Asf1 and histone H3 contributes to the function of Asf1 during DNA replication and DNA repair (presumably in chromatin assembly) and to chromatin disassembly during transcriptional activation.
DISCUSSION
Here we present the only structure to date of a histone chaperone bound to histones. The structure of Asf1 bound to a heterodimer of H3/H4 was confirmed to be the physiologically relevant form of the Asf1-H3/H4 complex by extensive mutagenesis analyses in yeast and in vitro. This complex reveals the altered conformations of the histones outside of the nucleosome. Although the Asf1 structure was not drastically altered by binding to the histones, larger conformational changes occurred in both histones H3 and H4 in the Asf1 complex, as compared to their structures within the nucleosome. Remarkably, there was a dramatic change in the conformation of the C-terminal tail of H4 upon binding to Asf1 through beta-strand addition (Fig. 4D-F).
Functional investigation of the Asf1-H3/H4 interactions
The ubiquitous function of Asf1 in Eukaryotes is highlighted by the sequence conservation of the residues involved in the interactions between Asf1 and histones H3/H4 (Suppl. Figs. 1 and 7). Budding yeast Asf1 is 56% identical to Xenopus Asf1 in the conserved core, and the Xenopus histones are 88% and 92% identical to H3 and H4 from yeast, respectively (Suppl. Fig. 1). Only three residues of Xenopus H3 that contact Asf1 differ in other species, C110 (Ala in yH3), Q125 (Lys in yH3), and I130 (Leu in yH3), and these substitutions would appear to cause only minor and possibly compensatory differences in inter-protein packing. Furthermore, none of these interspecies differences occur in residues of H4 that contact Asf1. Therefore, the interactions observed in this structure will likely be applicable to Asf1-histone H3/H4 complexes from different species.
The Asf1 histone chaperone forms extensive contacts with both histones H3 and H4. The Asf1-H3/H4 structure reveals the details of the interface between Asf1 and α3 of H3 and has identified a new interaction between Asf1 and α2 of H3. The implications of the mutagenesis study, with regard to Asf1 and H3, are that disruption of this intricate interface has severe consequences in the context of the cellular activity. For example, mutations in the regions of Asf1 that bind to only H3 (R145E/S48R, Y112A/R145E, V94R, and S48R) or the region of H3 that binds to Asf1 (K115, K122) weakened the interaction between Asf1 and H3 and disrupted Asf1 function in vivo and in vitro (Fig. 5,6). As such, the interaction between histone H3 and Asf1 is clearly critical for its cellular functions.
The Asf1-H3/H4 structure shows extensive contacts between Asf1 and histone H4 (Fig. 3). This interface has two parts: the globular core of Asf1 interacts with the C-terminal tail of H4 to form a strand-swapped dimer and the C-terminal tail of Asf1 binds to the histone-fold region (α3) of histone H4. These interactions are also important because mutations in residues of Asf1 that contact H4 (T147, L6, V109, V146) weaken histone binding (Fig. 5E,F) and alter the functions of Asf1 in yeast (Fig. 5, Table 1) (Recht et al., 2006). Similarly, mutation of histone H4 residues R92, H75, Y72, Y88, and F100 that contact Asf1 in the Asf1-H3/H4 structure reduces the chromatin assembly and / or disassembly functions of Asf1 in vivo (Fig. 6). Clearly, interactions of Asf1 with both histones H3 and H4 are required for Asf1 function and neither interaction is sufficient.
The mutations that affect the interaction between Asf1 and H3/H4 fall into two distinct functional classes, those that reduce the function of Asf1 and those that cause a gain-of-function phenotype. The former was expected, but the latter uncovered specific mutations that overcome the requirement for CAF-1 in transcriptional silencing. These include Asf1 S48R, V109M, Y112E, and V146L that weaken the interaction with histones H3/H4 in vivo and in vitro. Interestingly, the histone H4 H75Y mutation that had reduced Asf1-mediated chromatin disassembly activity and Zeocin sensitivity (Fig. 6) has also been previously shown to bypass the requirement for CAF-1 in silencing (Xu et al., 2005). We recently observed the same ability to bypass the requirement for CAF-1 in silencing by truncations or insertion mutations in the C-terminus of Asf1 (Tamburini et al., 2006). Specifically, we found that inactivation of CAF-1 led to reduced histone deposition onto DNA, while additional mutations in the C-terminus of Asf1 restored the histone deposition onto DNA (Tamburini et al., 2006). Although the C-terminus of Asf1 is not present in our structure, it may extend toward histone H4 from its current location in the structure and may contribute further to histone-binding affinity. It is possible that the Asf1 L6M, S48R, V109M, Y112E, V146L, and T147E mutations enhance transcriptional silencing by the same mechanism as the C-terminal mutations in Asf1.
Implications for function of other histone chaperones
Histone chaperones exhibit common structural features that suggest a similar mode of histone binding. For example, a variant of the anti-parallel beta-sheet region of Asf1, which interacts with histones H3 and H4 in the Asf1-H3/H4 structure, is also found in Nap1 and all histone chaperone structures (Park and Luger, 2006), including nucleoplasmin (Np and nucleoplasmin-like proteins) (Namboodiri et al., 2004; Namboodiri et al., 2003). Interestingly, the RbAp48 and p60 subunits of CAF-1 (Kaufman et al., 1995; Verreault et al., 1996) and the histone chaperone HIRA (Desilva et al., 1998) are composed of WD-40 repeats that are predicted to adopt β-propeller structures with exposed beta sheets. In the Asf1-H3/H4 structure, an acidic face of this conserved beta sheet in Asf1 forms the platform for interaction of H3 and may also contribute to orientation of the complex through electrostatic steering. Thus, for the other histone chaperones, it is possible that α3 of H2B or H3 (or another region of the histone dimer) also binds to the acidic and exposed face of these beta sheets.
A prominent general feature of the histone chaperone structures is the exposure of the edges of beta-sheets in these proteins (Park and Luger, 2006). One exposed beta-sheet edge of Asf1 forms the site for beta-strand addition by the C-terminus βC of H4 (Fig. 3B). Furthermore, the exposure of the edges of beta sheets is common among the histone chaperones; it occurs either as part of a beta-sheet sandwich (Asf1), a double-pentamer of beta-sheet sandwiches (Np, Nlp, NO38) (Namboodiri et al., 2004; Namboodiri et al., 2003), or as a single sheet with at least one edge exposed (Nap1 and WD-40 repeat proteins) (Park and Luger, 2006). Interestingly, both histones H4 and H2A also have the propensity for beta-strand formation in their C-termini (Luger et al., 1997). In the structure of Np there is acidic character to the exposed beta sheet, which is a favorable site for beta-strand addition by either H4 or H2A, particularly with the βH domain or the β1 strand of Np (Namboodiri et al., 2003). There is also a hydrophobic pocket in the neighboring Np monomer that contains several well-conserved residues, F30, L47, L45, I94, W19, and F102, and which resembles the hydrophobic pocket where H4 inserts into Asf1. As such, our structure provides a framework for understanding a possible mode in which other histones chaperones may bind to histones.
Implications for the assembly of chromatin
The Asf1-H3/H4 structure clearly demonstrates that Asf1 escorts H3/H4 histones as a heterodimer, rather than the heterotetramer that is seen in the nucleosome. It has been a widely held belief that soluble histones H3 and H4 exist as H3/H4 heterotetramers in the cell because extraction of histones from cells yielded H3/H4 in a heterotetrameric form (Moss et al., 1976). The Asf1-H3-H4 structure shows that Asf1 binds to an H3/H4 heterodimer, physically blocking formation of the H3/H4 heterotetramer, by enveloping precisely the same residues of H3 that are involved in the formation of the four-helix bundle formed by the H3:H3 dimer in the nucleosome (Luger et al., 1997). This discovery raises the question of whether the H3/H4 heterotetramer is assembled from two H3/H4 heterodimers prior to deposition onto the DNA or whether two H3/H4 heterodimers are deposited sequentially or concertedly onto DNA, and the possibility that other H3/H4 chaperones may assist Asf1 in the transfer of the H3/H4 dimer or tetramer to DNA. Clearly, the mechanism of formation of the H3/H4 heterotetramer on DNA must involve removal of Asf1 from histone H3 to enable H3:H3 dimerization. The trigger for removal of Asf1 from histone H3 is unknown, but it may be the presence of DNA, as the addition of DNA to histone-bound Asf1 results in chromatin assembly in vitro (English et al., 2005; Krawitz et al., 2002). However, the specific role of Asf1-DNA interactions during this process is unclear (Padmanabhan et al., 2005). As such, the mechanism of how chromatin is assembled and disassembled needs to be further investigated.
A proposed mechanism for the disassembly of nucleosomes
A key feature of the novel interaction between Asf1 and histone H4 is the formation of the strand-swapped anti-parallel beta sheet, which is a unique conformation, not seen for H4 in any nucleosome or octamer structures. The C-terminus of H4 normally forms a parallel-beta sheet with histone H2A in the nucleosome (Luger et al., 1997) that stabilizes the interaction between the H3/H4 heterotetramer and H2A/H2B heterodimers within the nucleosome. However, in the Asf1-H3/H4 complex, the H4 C-terminus rotates almost 180° to form an anti-parallel beta sheet with strand β9 of Asf1 (Fig. 4). This raises the intriguing possibility that the C-terminus of H4 can act as a handle for chromatin disassembly.
In this “strand-capture” model for disassembly of histone H3/H4 tetramers, Asf1 captures the C-terminus of H4 and uses it to aid in formation of the Asf1-H3/H4 complex. This interaction could potentially occur even when H4 is sequestered in the H3/H4 heterotetramer. Therefore, Asf1 binding to the C-terminus of H4 may be the first step and the trigger for splitting the H3/H4 heterotetramer into Asf1-bound H3/H4 heterodimers during chromatin disassembly. The molecular mechanism of the “strand-capture” model could be either that Asf1 alone or together with another remodeling activity uses the H4 C-terminus as a lever to unzip the H3/H4 heterotetramer, or that the association of Asf1 with the C-terminus of H4 could tether Asf1 so that binding to the H3 dimerization interface could occur more readily. The model also suggests that the formation of a new beta-strand in the H4 C-terminus and the lock-and key binding observed for F100 of H4, that fits snugly into the pocket on the surface of Asf1, would provide sufficient binding energy to sequester the H3/H4 tetramer, but this binding affinity is currently unknown (Fig. 4B).
This strand capture model is supported by the in vivo and in vitro data. Disruption of the interaction of the H4 C-terminal tail with both the binding pocket of the Asf1 core and with the C-terminal region of Asf1 led to defects in chromatin disassembly, transcriptional silencing and deficiencies in Asf1-H3/H4 complex formation in vivo and in vitro (Fig. 5,6). Indeed, the involvement of H4 F100 and the Asf1 pocket in chromatin disassembly is seen by the defects in PHO5 gene activation resulting from substitutions H4 F100A, Asf1 V109M, and Asf1 V146L that disrupt this interaction (Fig. 3C,5,6). A feature of this chromatin disassembly model is that Asf1 can only access the C-terminal tail of H4 once the H2A/H2B dimers are removed, potentially by other histone chaperones. This model is in accord with recent studies that Asf1 alone cannot mediate histone disassembly from nucleosomes in vitro (Lorch et al., 2006), suggesting that an H2A/H2B chaperone is necessary to remove H2A/H2B first. Furthermore, the C-terminal tail of H4 in the nucleosome would not be accessible to Asf1 without unwinding the outermost gyre of the DNA, indicating that an ATP-dependent chromatin remodeling activity may be required prior to or concomitant with Asf1-mediated removal of H3/H4 from the DNA.
Conclusions
Characterization of the Asf1-H3/H4 complex provides the basis for interpretation of a wealth of functional and structural studies. The structure is consistent with all of the biological data and mutagenesis analyses on Asf1 and histones to date. In addition to providing novel insights into the structure of histone chaperones bound to histones, this work gives insights into new potential mechanisms of nucleosome assembly and disassembly. Furthermore, it poses hypotheses about histone-carrying mechanisms that may be generally applicable to other histone chaperone proteins.
EXPERIMENTAL PROCEDURES
Crystallization, Structure Determination, and Analysis
The mutant and wild type Asf1-H3/H4 complexes were coexpressed and purified as described in (English et al., 2005). Crystallization conditions were identified using the Fluidigm system (Hansen and Quake, 2003). Data from crystals (space group P3121; cell dimensions, a=b=95.75 Å, c=110.68 Å) were collected on beamline 4.4.2 at the Advanced Light Source (Berkeley, CA) and were processed using D*Trek (Pflugrath, 1999). The structure was solved by molecular replacement using Phaser (McCoy et al., 2005) and refined using Refmac (CCP4) (Bailey, 1994) and O (Kleywegt and Jones, 1996) (Table 1). The model contains residues 1-164 of Asf1 (chain A), 60-134 of H3 (chain B), and 20-101 of H4 (chain C), and the side chains of H3 Y98, K133 and Asf1 N156 were built as alanine. Structural analyses, used CCP4, Superpose (Maiti et al., 2004), and LSQMAN (Kleywegt, 1996) programs, and structure diagrams were made using MOLSCRIPT (Kraulis, 1991), VMD (Humphrey et al., 1996), and PyMOL (DeLano, 2002). Coordinates have been deposited with the PDB ID 2hue.
Yeast Analyses
Yeast strains and plasmids are described in the Supplemental data. Resistance to methyl methane sulfonate (MMS), Zeocin, and hydroxyurea (HU) was determined by 10 fold dilution analysis of 1 OD 600 nm logarithmically growing cultures of yeast strains onto plates containing the indicated amount of the agents. Transcriptional silencing of the HMR::ADE2 and the TELVIIL::URA3 reporters was measured by plating onto 1 mg/ml 5’fluoroorotic acid and onto low adenine plates respectively. The ability to activate the PHO5 gene was measured as previously described, as an indicator of Asf1-mediated promoter chromatin disassembly (Adkins et al., 2004). Co-immunoprecipitation analysis of Asf1 and histones was performed as described previously (Tamburini, 2005).
Supplementary Material
Acknowledgements
We thank members of the Tyler and Churchill labs, Leslie Krushel, Rui Zhao, and Jeff Kieft for critical reading of the manuscript. We are very grateful to Karolin Luger for histone plasmid constructs and advice. We thank Jeff Boeke, Jim Broach, and Mitch Smith for yeast strains, and Rui Zhao, Jeff Kieft, Jill Waters, and Sarah Roemer for technical support and advice. We thank Jay Nix and the staff at beamline 4.2.2 at the Advanced Light Source, Lawrence Berkeley National Lab. The UCDHSC Biomolecular X-ray Crystallography Center was supported in part by funding from the HHMI, the UC Cancer Center and NIH. This work was supported by NIH grant GM064475 to J.K.T., and by AHA grant 0355468Z to M.E.A.C.. J.K.T. is a Leukemia and Lymphoma Society Scholar.
References
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol. 2003;13:6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Daganzo SM, Erzberger JP, Lam WM, Skordalakes E, Zhang R, Franco AA, Brill SJ, Adams PD, Berger JM, Kaufman PD. Structure and function of the conserved core of histone deposition protein Asf1. Curr Biol. 2003;13:2148–2158. doi: 10.1016/j.cub.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Desilva H, Lee K, Osley MA. Functional dissection of yeast Hir1p, a WD repeat-containing transcriptional corepressor. Genetics. 1998;148:657–667. doi: 10.1093/genetics/148.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Maluf NK, Tripet B, Churchill ME, Tyler JK. ASF1 Binds to a Heterodimer of Histones H3 and H4: A Two-Step Mechanism for the Assembly of the H3-H4 Heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Hansen C, Quake SR. Microfluidics in structural biology: smaller, faster... better. Curr Opin Struc Biol. 2003;13:538–544. doi: 10.1016/j.sbi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–&. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Cryst. 1996;D52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Efficient rebuilding of protein structures. Acta Crystallogr. 1996;D52:829–832. doi: 10.1107/S0907444996001783. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript - a Program to Produce Both Detailed and Schematic Plots of Protein Structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- Krawitz DC, Kama T, Kaufman PD. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol Cell Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Luger K. Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maiti R, Van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nuc Acids Res. 2004;32:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T, Cary PD, Crane-Robinson C, Bradbury EM. Physical studies on the H3/H4 histone tetramer. Biochem. 1976;15:2261–2267. doi: 10.1021/bi00656a003. [DOI] [PubMed] [Google Scholar]
- Mousson F, Lautrette A, Thuret JY, Agez M, Courbeyrette R, Amigues B, Becker E, Neumann JM, Guerois R, Mann C, Ochsenbein F. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc Natl Acad Sci U S A. 2005;102:5975–5980. doi: 10.1073/pnas.0500149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata T, Adachi N, Yokoyama N, Kuzuhara T, Horikoshi M. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells. 2000;5:221–233. doi: 10.1046/j.1365-2443.2000.00319.x. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Ray-Gallet D, Quivy JP, Tagami H, Almouzni G. Two distinct nucleosome assembly pathways: dependent or independent of DNA synthesis promoted by histone H3.1 and H3.3 complexes. Cold Spring Harb Symp Quant Biol. 2004;69:273–280. doi: 10.1101/sqb.2004.69.273. [DOI] [PubMed] [Google Scholar]
- Namboodiri VMH, Akey IV, Schmidt-Zachmann MS, Head JF, Akey CW. The structure and function of Xenopus NO38-core, a histone chaperone in the nucleolus. Structure. 2004;12:2149–2160. doi: 10.1016/j.str.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Namboodiri VMH, Dutta S, Akey IV, Head JF, Akey CW. The crystal structure of drosophila NLP-core provides insight into pentamer formation and histone binding. Structure. 2003;11:175–186. doi: 10.1016/s0969-2126(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Kataoka K, Umehara T, Adachi N, Yokoyama S, Horikoshi M. Structural Similarity between Histone Chaperone Cia1p/Asf1p and DNA-Binding Protein NF-{kappa}B. J Biochem (Tokyo) 2005;138:821–829. doi: 10.1093/jb/mvi182. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci U S A. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol Cell Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban MS, Arents G, Moudrianakis EN, Smith MM. Histone octamer function in vivo: mutations in the dimer-tetramer interfaces disrupt both gene activation and repression. Embo J. 1997;16:2493–2506. doi: 10.1093/emboj/16.9.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 Mediates Histone Eviction and Deposition during Elongation by RNA Polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. Embo J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tamburini B, Carson J, Linger JG, Tyler JK. Dominant mutants of the Saccharomyces cereivisiae ASF1 histone chaperone bypass CAF-1 mediated nucleosome assembly and Sir protein recruitment during transcriptional silencing. Genetics. 2006 doi: 10.1534/genetics.105.054783. Accepted, pending revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Carson JJ, Adkins MW, Tyler JK. Functional conservation and specialization among Eukaryotic Anti-silencing function 1 histone chaperones. Eukaryotic Cell. 2005;4:1583–1590. doi: 10.1128/EC.4.9.1583-1590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. Embo J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Bi X, Holland MJ, Gottschling DE, Broach JR. Mutations in the nucleosome core enhance transcriptional silencing. Mol Cell Biol. 2005;25:1846–1859. doi: 10.1128/MCB.25.5.1846-1859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.