Abstract
Here we show a novel mechanism by which FLICE-like inhibitory protein (c-FLIP) regulates apoptosis induced by tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) and one of its receptors, DR5. c-FLIP is a critical regulator of the TNF family of cytokine receptor signaling. c-FLIP has been postulated to prevent formation of the competent death-inducing signaling complex (DISC) in a ligand-dependent manner, through its interaction with FADD and/or caspase-8. In order to identify regulators of TRAIL function, we used the intracellular death domain (DD) of DR5 as a target to screen a phage-displayed combinatorial peptide library. The DD of DR5 selected from the library a peptide that showed sequence similarity to a stretch of amino acids in the C terminus of c-FLIPL. The phage-displayed peptide selectively interacted with the DD of DR5 in in vitro binding assays. Similarly, full-length c-FLIP (c-FLIPL) and the C-terminal p12 domain of c-FLIP interacted with DR5 both in in vitro pull-down assays and in mammalian cells. This interaction was independent of TRAIL. To the contrary, TRAIL treatment released c-FLIPL from DR5, permitting the recruitment of FADD to the active DR5 signaling complex. By employing FADD-deficient Jurkat cells, we demonstrate that DR5 and c-FLIPL interact in a FADD-independent manner. Moreover, we show that a cellular membrane permeable version of the peptide corresponding to the DR5 binding domain of c-FLIP induces apoptosis in mammalian cells. Taken together, these findings indicate that c-FLIPL interacts with the DD of DR5, thus preventing death signaling by DR5 prior to the formation of an active DISC. Because TRAIL and DR5 are ubiquitously expressed, the interaction of c-FLIPL and DR5 indicates a mechanism by which tumor selective apoptosis can be achieved through protecting normal cells from undergoing death receptor-induced apoptosis.
Tumor necrosis factor (TNF)1-related apoptosis-inducing ligand (TRAIL), also known as Apo-2 ligand, is a type II membrane protein belonging to the TNF family (1–5). TRAIL has been shown to preferentially induce apoptosis in a variety of tumor and transformed cells, but not in normal cells (1, 3, 5–9). Unlike Fas ligand (FasL) and TNF, which have a restricted tissue distribution, TRAIL mRNA is expressed in a wide variety of tissues and cell types. Recently, the functional expression of TRAIL has been observed on the surface of various immune cells that were previously known to induce apoptosis of target cells by an unidentified mechanism. These cells include natural killer cells (NK), dendritic cells (DC), monocytes, and T cells that have been stimulated by cellular transformation (10–15), suggesting a role for TRAIL in the selective induction of apoptosis in tumor cells.
TRAIL function is mediated through its interaction with TRAIL receptors. TRAIL can bind to five receptors, two of which are signaling receptors that carry a functional death domain (DD), whereas the remaining three are decoy receptors. The signaling receptors are named death receptor 4 (DR4), or TRAIL-R1/APO2A (16–18), and death receptor 5 (DR5), or TRAIL-R2 (also known as KILLER and TRICK 2) (17–24). Shortly after treatment with TRAIL, the DD of DR4 or DR5 associates with a similar domain found in the adaptor protein Fas-associated protein with death domain (FADD), resulting in the recruitment of pro-caspase-8 or -10 and subsequent formation of the death-inducing signaling complex (DISC) (25, 26). Once formed, DISC promotes the autoproteolytic activation and release of active caspase-8, which in turn can cleave and activate downstream executioner caspases, thus leading to apoptosis (14, 25, 26). The TRAIL-induced activation of caspase-8 can also culminate in the cleavage of Bid and activation of the mitochondrial apoptotic pathway (27–30).
Because of the tumor selectivity of TRAIL-induced apoptosis and its ubiquitous expression, it has been postulated that the apoptosis pathway induced by TRAIL is tightly regulated by several mechanisms to prevent spontaneous cell death. One such mechanism involves the cellular inhibitor of apoptosis, cellular FLICE inhibitory protein (c-FLIP), which is also known as I-FLICE, FLAME, CASPER, or CASH (31–38). c-FLIP exists as a long (c-FLIPL) or as a short (c-FLIPS) splice variant (1, 8, 39–42). c-FLIPL contains two death effector domains (DED) and an inactive caspase domain composed of p17 and p12 subunits (39, 40). On the other hand, c-FLIPS lacks the entire caspase domain (39, 40), but retains the two DEDs. DED is a critical protein interaction domain that recruits caspases into complexes with members of the TNF receptor family (39, 40). Furthermore, there is a unique caspase cleavage site at position 341 (LEVD) of c-FLIPL that is thought to be involved in the regulation of c-FLIP function by caspases. Indeed, another c-FLIP variant, c-FLIPp43 is the product of caspase cleavage of full-length c-FLIPL (39, 40).
c-FLIP has been shown to interfere with TNF, Fas, and TRAIL-induced death receptor signaling pathways upstream of mitochondrial events by binding to FADD and/or FLICE/caspase-8 at the level of the DISC, thereby inhibiting subsequent activation of the caspase cascade (17,31,33–38,42). However, it is now thought that the c-FLIP variants may have distinct roles in regulating apoptosis (39, 40, 43, 44). For instance, whereas c-FLIPS has been shown to inhibit TRAIL-induced DISC formation and apoptosis (45, 46), c-FLIPL appears to have dual opposing functions wherein it inhibits Fas-induced caspase-8 recruitment and activation when it is present at a high level, but promotes activation of caspase-8 at low concentrations (39, 40, 43, 44). Thus, c-FLIP proteins are critical regulators of death ligand-induced apoptosis, and the levels and activities of different c-FLIP variants can modulate death ligand-induced caspase activation at the DISC.
Given that TRAIL and TRAIL receptors are expressed in a variety of tissues and that their functions must be tightly regulated in order to inhibit unwanted cell death, it is imperative to characterize the mechanisms that are involved in the regulation of the physiological function and tumor selectivity of TRAIL-induced apoptosis. This need is further highlighted by the implication of TRAIL in various immunological processes and its ability to induce tumor selective cell death. To identify regulators of TRAIL function, we screened a phage display combinatorial library with the death domain of DR5. Here, we report the identification of a peptide ligand that corresponds to a region within the caspase domain of c-FLIPL and specifically interacts with the death domain of DR5. We confirmed that c-FLIPL interacts with the death domain of DR5 both in in vitro pull-down assays and in co-immunoprecipitation studies of endogenous proteins in mammalian cells. This interaction occurs in the absence of TRAIL and in a FADD-independent manner. Besides, TRAIL treatment results in the release of c-FLIPL from DR5, permitting the recruitment of FADD and caspase-8 complex to the receptor. Finally, we demonstrate that a cellular membrane permeable from the c-FLIP peptide corresponding to the DR5 binding domain of c-FLIPL induces spontaneous, ligand-independent apoptosis. Taken together, these observations suggest that c-FLIPL directly interacts with DR5 receptor and regulates TRAIL-induced apoptosis prior to the formation of the DISC.
MATERIALS AND METHODS
Biological Reagents
Anti-FLAG M2 antibody was from Sigma-Aldrich, DR5 antibodies were HS201, and ALX-210-743-C200 from Alexis Corp. (Santa Cruz, CA). The following antibodies were used in immunocomplex and DISC analysis: caspase-8 1C12 (Cell Signaling Technology, Beverly, MA), caspase-10 N-19 (Santa Cruz Biotechnology, Santa Cruz, CA), FADD H181 (Santa Cruz Biotechnology), FLIP AF821 (R&D Systems, Minneapolis, MN), FLIP G11 (Santa Cruz Biotechnology), and FLIP Dave 2 (Alexis Corporation). Monoclonal anti-biotin FITC-conjugated antibody was from Sigma. Recombinant TRAIL was from R&D Systems. Synthetic peptides were purchased from Anaspec (San Jose, CA). The sequences of the peptides are as follows: TAT (biotin-LCYGRKKRRQRRR-NH2) and TAT-FLIP (biotin-LCYGRKKRRQRRRREADFFWSLCTADMS-NH2).
Cell Culture
Human embryonic kidney (HEK) 293 cells were maintained in Dulbecco’s modified Eagle’s medium containing 2 mm glutamine, 50 units/ml penicillin, 50 µg/ml streptomycin, 1mm pyruvate, and 10% fetal calf serum. HEK 293 cells were transfected by the calcium phosphate method as previously described (47, 48). Cells of the human leukemic T-cell line Jurkat A3 and FADD-deficient Jurkat cells (generous gifts from Dr. John Blenis, Harvard Medical School) were maintained in RPMI 1640 medium containing 50 units/ml penicillin, 50 µg/ml streptomycin, 1 mm pyruvate, and 10% fetal calf serum.
Expression Vectors
The cDNA encoding the death domain of DR4, DR5 (DR4 and DR5 were generous gifts from Dr. Ashkenazi, Genentech, South San Francisco, CA and Dr. El-Deiry, University of Pennsylvania, Philadelphia, PA), Fas (generous gift from Dr. Yang, University of Pennsylvania, Philadelphia, PA), the SH3 domains of Src and Abl, the PDZ domain of hNOS, p53N, and E6 were subcloned downstream and in-frame with the GST open reading frame in the pGEX-2TK vector. Full-length c-FLIP cDNA (generous gift from Dr. Tschopp, University of Lausanne, Switzerland) was subcloned in pAX-FLAG plasmid (generous gift from Dr. Robert Kay, Terry Fox Laboratory, Vancouver, British Columbia, Canada) (49).
Isolation and Characterization of Death Domain-binding Phage
GST fusion proteins of the death domain of human DR5 were prepared in the following manner. Oligonucleotides were designed to flank the death domain, and this region was selectively amplified from cDNA clones by polymerase chain reaction. The amplified segments were subcloned in-frame between the Bam-HI and EcoRI sites of pGEX4T-2 (Amersham Biosciences), and recombinants were verified by DNA sequencing. GST fusion proteins were expressed and purified according to the manufacturer’s instructions (Amersham Biosciences).
To screen the phage-displayed combinatorial 12-mer peptide library (50–53) (54), microtiter plate wells were coated with 1 µg of a GST fusion protein of DR5 death domain, blocked with SuperBlock® (Pierce), and then 1011 phage particles were added. After incubation for 2 h at room temperature, the wells were washed of excess phage, and the bound phage was eluted by the addition of 50 µl of 0.2 m glycine, pH 2.0. After three rounds of affinity selection, isolates were tested for binding to GST-DR5 (DD) fusion protein by ELISA (Amersham Biosciences). The DNA sequences of bound phages were determined, and the peptide sequences deduced with MacVector® software (IBI-Kodak, New Haven, CT).
The binding properties of the isolated phage were studied by phage ELISA assay. Microtiter plate wells were coated with equal amounts of various GST fusion proteins, washed, blocked with bovine serum albumin, and then incubated with 1010 plaque-forming units. The amount of bound phage was quantitated with anti-M13 phage antibodies conjugated to horseradish peroxidase (Amersham Biosciences).
Alanine Scan of the DR5 Death Domain Ligand
Each amino acid of the peptide ligand selected in the screen was sequentially substituted by alanine and displayed at the N terminus of M13 phage. Each phage mutant was tested for its ability to bind GST fusion proteins of the DR5 death domain immobilized on microtiter plates. The wells were coated with 0.5 µg of purified DR5 death domain fusion protein and blocked with 1% bovine serum albumin in 0.1 m NaHCO3, pH 8.3, for 1 h at room temperature. The wells were then incubated for 2 h at room temperature with 1010 recombinant phage particles. After extensive washing with phosphate-buffered saline, pH 7.4, and 0.1% Tween 20, the amount of retained phage peptide was quantitated in a phage ELISA using an anti-M13 antibody (53).
GST Pull-down Assay
Cell lysates were prepared from either HEK 293 cells that were transfected with FLAG-tagged constructs of c-FLIPL, or from Jurkat cells (parental or FADD−/−) in lysis buffer (48). Lysates (250–400 µg) were then incubated with 10 µg of GST fusion protein of DR5 death domain immobilized on glutathione beads for 1 h at 4 C. The beads were washed three times in cold lysis buffer and processed for SDS-PAGE followed by Western blot analysis. c-FLIPL was detected by immunoblotting with anti-FLAG antibody or c-FLIP antibody.
Immunoprecipitation and Immunoblotting
Jurkat cells or transfected HEK 293 cells were incubated for 30 min at 4 °C in lysis buffer (48). Lysates were cleared at 10,000 × g for 10 min at 4 °C, and supernatants were incubated for 16 h with antisera against DR5 antibody that were preabsorbed on protein A-Sepharose and protein G-Sepharose. The immunocomplexes were washed three times in cold lysis buffer. Proteins were resolved by SDS-PAGE and transferred to Immobilon membranes (Millipore, Bedford, MA) using standard procedures. Immunoblot analysis was performed with the indicated antibodies. Bound antibodies were revealed with horseradish peroxidase-coupled immunoglobulins using the Enhanced Chemiluminescence (ECL) Western blotting detection system (Amersham Biosciences) according to the manufacturer’s instructions.
DISC Analysis
The DISC analysis was performed as previously described (55). Briefly, a total of 5 × 107 Jurkat T lymphocyte cells were starved for 12 h using 0.5% fetal bovine serum RPMI medium, 10 ng/ml TRAIL was added for 1 h (or indicated times) at 37 °C, and then the cells were lysed in lysis buffer (30 mm Tris-HCl, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100, 10% glycerol, and a protease inhibitor mixture) (55) (stimulated condition), or the cells were first lysed and then TRAIL was added (unstimulated condition). TRAIL antibody (Alexis Corp.) immobilized on protein A-Sepharose (Sigma-Aldrich) was prepared. The TRAIL-R complex was immunoprecipitated for 2 h at 4 °C using protein A-Sepharose-coupled beads. The immune complex was spun down, washed, and resuspended in SDS-gel sample buffer, and boiled at 100 °C for 5 min. Immunoprecipitates were separated by SDS-PAGE and immunoblotted with the following antibodies: caspase-8 1C12, caspase-10 N-19, FADD H181, TRADD N-19, FLIP G11, and FLIP Dave 2.
Immunofluorescence
Jurkat cells were incubated without peptide (Mock), with 15 µm TAT, or with 15 µm TAT-FLIP in growth medium for 2 h. Cells were then fixed, permeabilized with ethanol/acetic acid (9:1) for 5 min at −20 °C, blocked for 30 min with 10% fetal calf serum in phosphate-buffered saline, and incubated for 1 h with FITC-conjugated biotin antibody. Cells were then washed with phosphate-buffered saline and visualized by epifluorescence microscopy.
Apoptosis Assay
4 × 105 Jurkat cells were grown in RPMI in the media in the absence (Mock) or presence of cellular permeable TAT peptide or TAT-FLIP peptide for 2 or 16 h. The cells were then washed once with phosphate-buffered saline and re-suspended in DNA-staining buffer containing 50 µg/ml propidium iodide (PI) (Roche Diagnostics, Basel, Switzerland), 0.1% Triton X-100, and 0.1% sodium citrate and incubated for 1 h at 4 °C. The cell population was assayed with FACS (Becton Dickinson Immunocytometry System, FACS Calibur, San Jose, CA), and Cell Quest software was used for measuring the sub-G1 apoptotic cell population. Additionally, annexin V-PE was used to confirm the specificity of apoptosis. In these studies, cells were washed with phosphate-buffered saline followed by re-suspension in annexin V binding buffer. Annexin V-PE was then added to the cells, and apoptotic cells were detected by flow cytometry using the FL-2 channel.
RESULTS
Isolation and Characterization of a DR5-interacting Peptide from a Combinatorial Phage-displayed Library
The death domains of death receptors are protein-protein interaction domains that are involved in recruitment and activation of intracellular apoptotic pathways. In order to identify proteins that directly interact with and regulate the function of DR5, we screened a phage-displayed combinatorial peptide library with a GST fusion protein of the death domain of DR5. The library of ~109 primary recombinants displayed combinatorial 12-mer peptides at the N terminus of bacteriophage M13 protein III (50–53). After three rounds of affinity selection, a single peptide sequence was isolated that bound to the death domain of DR5 in an ELISA-based assay. The DNA from the phage isolate with specific binding to DR5-DD was purified, and DNA sequence analysis demonstrated that the binding peptide sequence was RGSFWWLETAPP.
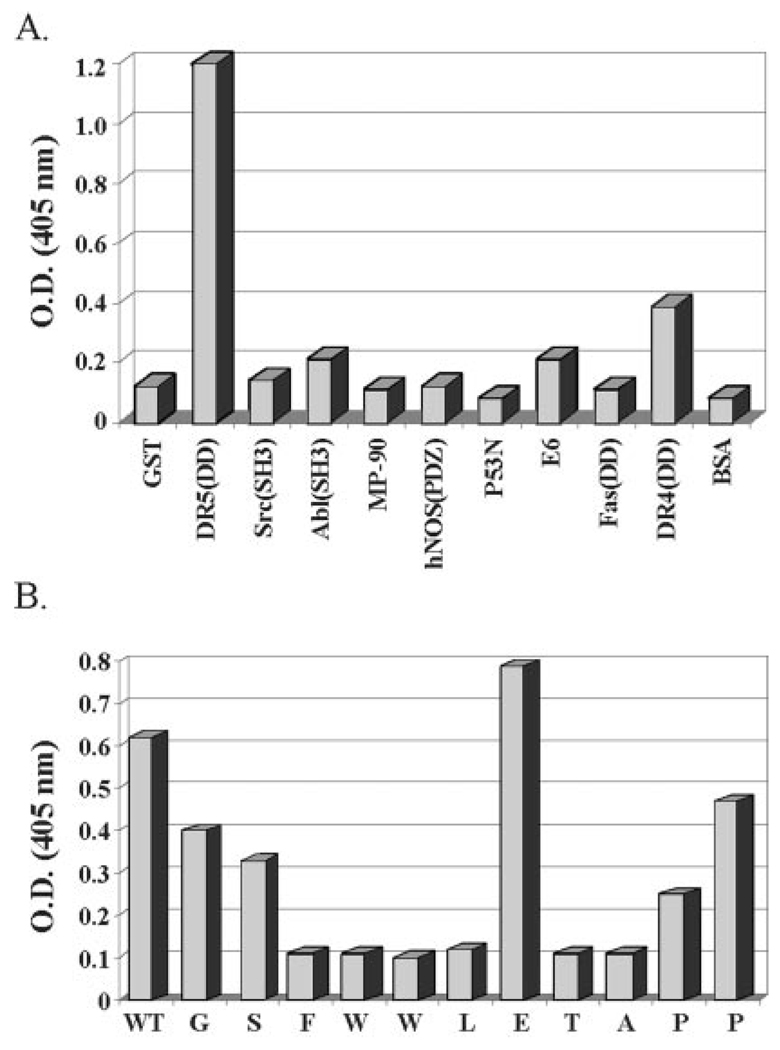
The isolated peptide ligand was further tested in an in vitro phage binding assay for its ability to specifically interact with the death domain of DR5. This was achieved by coating micro-titer wells with equivalent amounts of GST, GST-DR5, or a variety of other unrelated GST fusion proteins (Src-SH3, Abl-SH3, p53N, E6, MP90, hNOS(PDZ), Fas-DD, DR4-DD). These proteins were incubated with equivalent amounts of phage isolate (1010 plaque-forming units). The binding of phage particles was then assayed by anti-M13 phage ELISA. The isolated peptide was found to interact specifically with the death domain of DR5 (Fig. 1A). Interestingly, this peptide does not interact with the death domain of Fas and interacts only weakly with the death domain of DR4 (the receptor with 64% homology to DR5) (Fig. 1A).
FIG. 1.
A, isolated peptide interacts specifically with DR5. Using a phage-displayed combinatorial peptide library, we have identified a peptide that interacts with DR5. Equivalent numbers of phage particles were added to microtiter wells coated with 1 µg each of GST fusion proteins of DR5(DD), DR4(DD), Fas(DD), Src(SH3), Abl (SH3), MP-90, hNOS(PDZ), p53N, E6, or negative controls GST and bovine serum albumin as indicated. The strength of interaction was determined in an ELISA with anti-M13 phage antibody. The isolated peptide specifically interacts with DR5(DD) and only very weakly with the highly homologous relative, DR5(DD) protein. The y-axis is a measure of optical density at 405 nm. B, alanine scanning of the phage peptide reveals the DR5 binding sequence. Equivalent amounts (1 µg/well) of DR5 fusion proteins were immobilized in microtiter plates and incubated with 1010 phage particles from variants of the isolated phage peptide sequence RGSFWWLETAPP. These variants have one mutation each, sequentially substituting each amino acid in the peptide to alanine. Except for the Glu → Ala substitution, which enhances binding, sequential alanine substitutions of the FWWLET peptide sequence causes loss of binding.
Alanine Scanning of Peptide Sequence Displayed by DD-binding Phage Isolate
In order to characterize the critical residues in the isolated peptide that constitute the DR5-DD binding site, we carried out alanine scanning of the isolated peptide by sequential substitution of each amino acid to alanine. These substitutions were generated by constructing M13 phages with sequential point mutations corresponding to alanine. As seen in Fig. 1B, with the exception of the Glu → Ala substitution, which increases the affinity of the peptide for DR5-DD, alanine substitution at most of the amino acid residues of the peptide abolished its interaction with DR5-DD. From these studies, we deduced that the FWWLXTA sequence constitutes the specific binding domain of a DR5-DD-interacting protein (Fig. 1B).
Identification of the Protein That Corresponds to the Phage-displayed Peptide
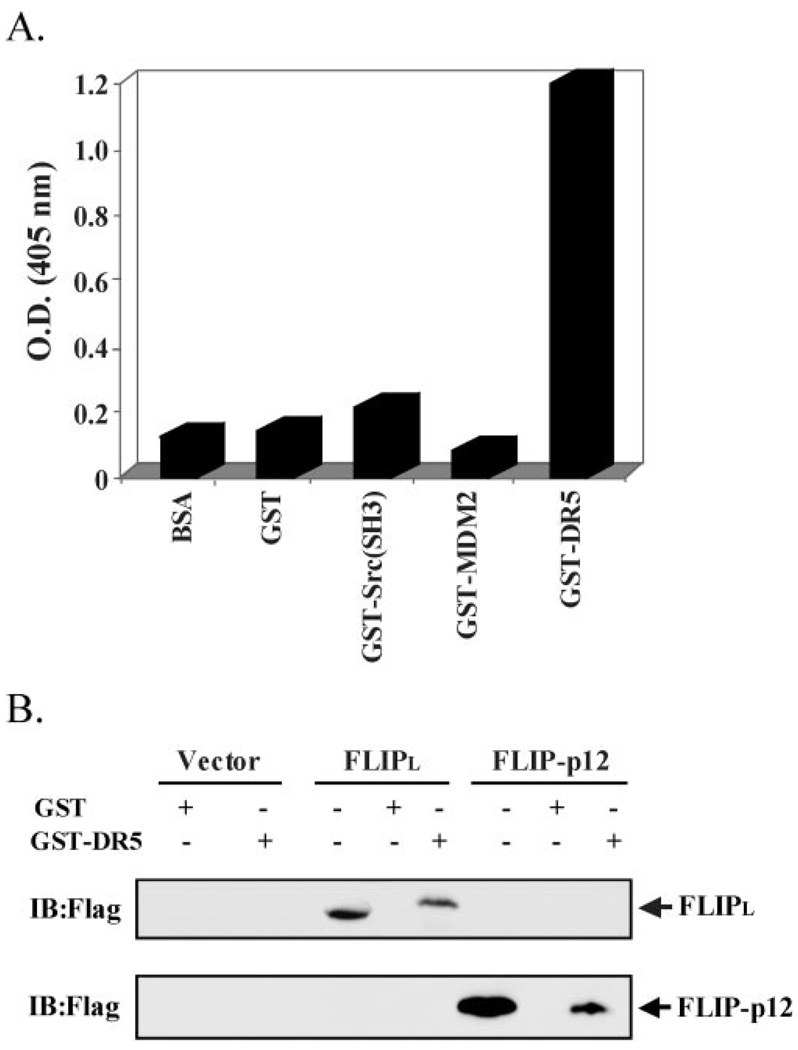
Following a genomic data base search for ESTs with similarity to the isolated phage DNA that interacted with the death domain of DR5, we identified c-FLIPL as a candidate protein. This contention was based on a region of c-FLIPL that is analogous to the isolated peptide. We, therefore, genetically engineered and displayed on the surface of M13 phage the c-FLIP peptide (READFFWSLSTADMS) corresponding to the c-FLIP/DR5 binding domain and tested its ability for binding to DR5 in an ELISA assay. As seen in Fig. 2, DR5-DD has a high affinity interaction with the synthesized c-FLIP peptide. The isolated peptide can be mapped to the p12 region in the caspase domain of c-FLIPL.
FIG. 2. DR5 and c-FLIPL interact in vitro.
A, interaction of a c-FLIP peptide with DR5 protein. We displayed on the phage a c-FLIP peptide segment resembling the sequence of the peptide ligand selected by the DD of DR5 from a phage-displayed combinatorial peptide library. In this format, the c-FLIP peptide specifically interacts with GST-DR5 protein, but not to unrelated GST fusion proteins, in an ELISA. B, interaction of full-length c-FLIPL with DR5 in a GST pull-down assay. Cell lysates were prepared from HEK 293 cells ectopically expressing FLAG-tagged c-FLIPL or FLAG-tagged c-FLIPp12. These cell lysates were used in a GST pull-down assay with GST fusion proteins of DR5 and immunoblotted with an anti-FLAG antibody. Total cell lysate is used as a control to monitor expression of c-FLIPL and c-FLIPp12.
c-FLIPL Interacts with DR5 Both in Vitro and in Vivo
While c-FLIP has previously been detected at the DISC, a direct interaction between c-FLIPL and DR5 that is independent of TRAIL-mediated DISC formation has not yet been demonstrated. In order to confirm a direct association between c-FLIPL and DR5, we tested the binding of full-length c-FLIP with DR5 in vitro and in mammalian cells. In vitro binding assays were carried out using GST fused to either the death domain of DR5 or of Fas protein in pull-down experiments with cell lysates from c-FLIPL-transfected HEK 293 cells. This binding is specific as c-FLIPL was pulled down with the death domain of DR5 (Fig. 2B), but not with the death domain of Fas (data not shown). Most likely the binding element maps to the C-terminal p12 domain of c-FLIP, as it interacts with DR5 in a pull-down assay (Fig. 2B).
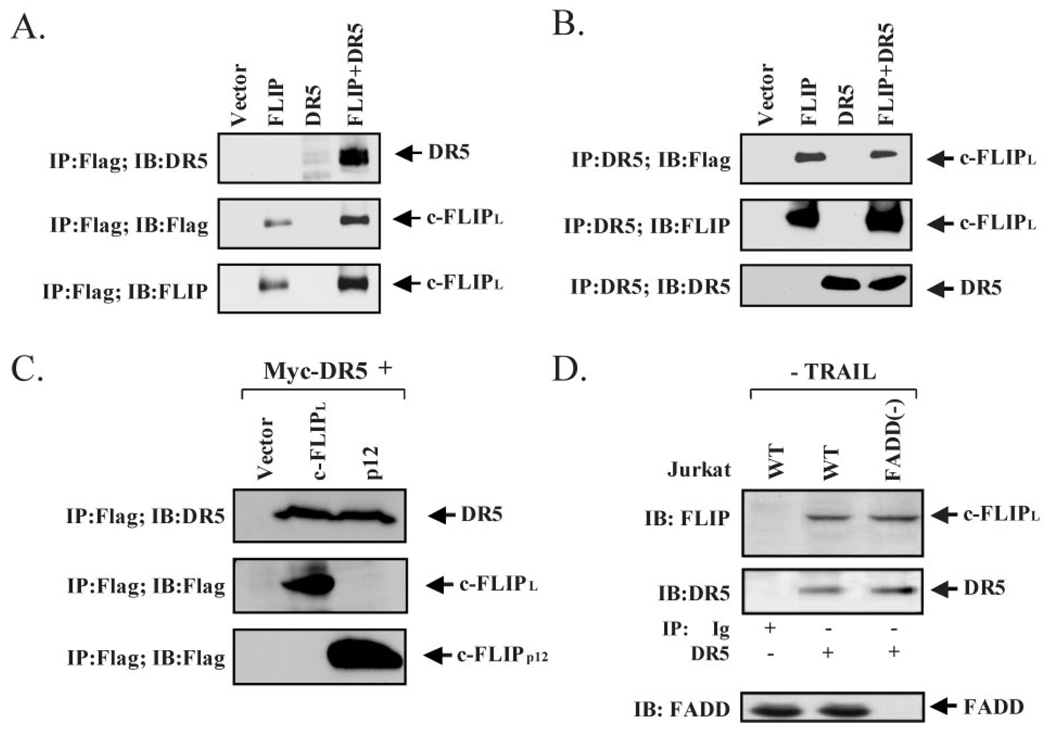
Next, we confirmed the interaction between c-FLIPL and c-FLIPp12 with DR5 in co-immunoprecipitation studies in HEK 293 cells. In these studies, HEK 293 cells were transiently transfected with vector alone, FLAG-tagged construct of c-FLIPL, c-FLIPp12, Myc-tagged construct of DR5, or both c-FLIP and DR5 constructs As shown in Fig. 3, DR5 co-precipitated with c-FLIPL (Fig. 3A), conversely, c-FLIPL co-precipitated with DR5 (Fig. 3B). Additionally, we observed that DR5 also co-precipitated with the p12 fragment of c-FLIP, demonstrating that the interaction domain is indeed in the carboxyl region of c-FLIP (Fig. 3C). Finally, since overexpression of proteins can lead to aberrant protein complex formation, we validated the observed interaction by studying the endogenous proteins under physiological conditions. As shown in Fig. 3D, endogenous c-FLIPL co-precipitated with endogenous DR5 in Jurkat T cells, lacking expression of DR4 (56, 57). Collectively, these data show that the C terminus of c-FLIPL mediates an interaction with the death domain of DR5, and that this interaction is likely to occur directly and in vivo.
FIG. 3. DR5 and c-FLIPL interact in cell culture.
A and B, interaction of c-FLIPL and DR5 proteins in co-immunoprecipitation studies. HEK 293 cells were co-transfected with expression vectors for vector alone, FLAG-tagged c-FLIPL, DR5, or c-FLIPL and DR5 as indicated. FLAG immunoprecipitates were analyzed for the presence of DR5 and DR5 immunoprecipitates were analyzed for the presence of c-FLIPL. The presence of DR5 and c-FLIP in cell extracts were verified by immunoblotting for DR5 or c-FLIP. C, interaction between c-FLIPp12 and DR5 in co-immunoprecipitation studies. HEK 293 cells were co-transfected with expression vectors for vector alone, FLAG-tagged c-FLIPp12, DR5, or c-FLIP12 and DR5 as indicated. FLAG immunoprecipitates were analyzed for the presence of DR5 and DR5 immunoprecipitates were analyzed for the presence of c-FLIPp12. The presence of DR5 in total cell lysate is shown by Western blot analysis with an anti-Myc antibody. D, interaction between endogenous c-FLIPL and endogenous DR5 is independent of FADD. Association of endogenous DR5 with endogenous c-FLIPL in parental Jurkat A3 and FADD-deficient Jurkat A3 cells is shown by immunoprecipitation with DR5 antibody followed by immunoblotting with c-FLIP antibody. The level of DR5 protein in the immunocomplex is determined by immunoblotting with DR5 antibody. An unrelated immune serum is used as a negative control. In the bottom panel, the level of FADD expression is shown for each cell line.
DR5 and c-FLIP Interaction Occurs in the Absence of TRAIL
The recruitment of FADD to the TRAIL receptors and the subsequent activation of the caspase cascade which results in the induction of apoptosis, is dependent on stimulation by TRAIL (25, 26, 58). However, we have observed that the interaction between c-FLIPL and DR5 occurs in the absence of TRAIL treatment (Figs. 3 and 4). Furthermore, we found that the interaction between c-FLIPL and DR5 is diminished by ~50% in response to TRAIL stimulation (15 min) of Jurkat cells (Fig. 4). On the other hand, there was no significant interaction observed between DR5 and FADD, caspase-8, c-FLIPp43, or c-FLIPS in the absence of TRAIL (Fig. 4).
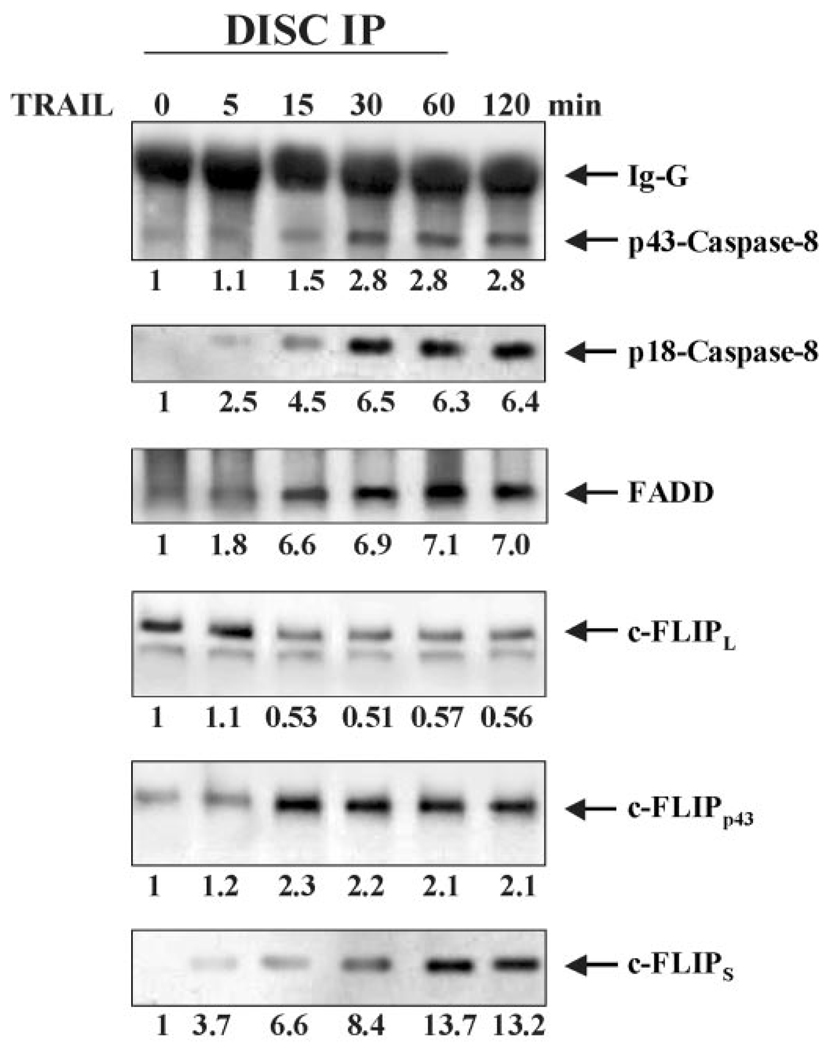
FIG. 4. c-FLIPL interacts with DR5 in the absence of TRAIL stimulation and upon formation of the FADD/caspase-8 DISC, DR5, and c-FLIPL interaction is diminished.
Time course of c-FLIPL and DR5 interaction and FADD/caspase-8 DISC formation was carried out in Jurkat cells that were treated with 10 ng/ml of TRAIL for 0, 5, 15, 30, 60, and 120 min. In stimulated cells, upon TRAIL treatment (10 ng/ml) for 1 h, cells were lysed and the assembled DISCs were immunoprecipitated with protein A and analyzed by Western blotting using antibodies to FADD, caspase-8, caspase-10, TRADD, c-FLIPL, c-FLIPp43, and c-FLIPs In unstimulated condition, cells were first lysed, and then TRAIL was added followed by analysis of the immunocomplex. In the absence of TRAIL treatment, c-FLIPL is associated with TRAIL receptor. Approximately, 5 min after TRAIL treatment FADD and caspase-8 are recruited to the DISC. At the same time, the level of c-FLIPL is diminished and the levels of c-FLIPp43 and c-FLIPS are increased in the complex.
TRAIL Stimulation Releases c-FLIPL from DR5
TRAIL stimulation leads to the formation of an apoptosis-inducing protein complex known as the TRAIL DISC (25, 26, 58). FADD is an adaptor molecule that is recruited to the apoptosis-inducing TRAIL receptors, DR4 and DR5, resulting in the activation of caspase-8 (25, 26, 58). Our observation that the interaction between c-FLIPL and DR5 is diminished in response to TRAIL ligation suggests that TRAIL treatment releases c-FLIPL from DR5, thereby allowing the recruitment of FADD to the receptor. In order to test this hypothesis, we monitored TRAIL DISC formation in Jurkat cells over time. As expected, c-FLIPL, but not FADD, interacted with DR5 in unstimulated Jurkat cells (Fig. 4). Following 15 min of stimulation with TRAIL, the interaction between c-FLIPL and DR5 was diminished by ~50% (Fig. 4). At the same time point, a significant level of FADD (increased by 6.6-fold) and activated caspase-8 (p43 and p18) (increased by 1.5- and 4.5-fold respectively) were detected within the DISC (Fig. 4). Interestingly, concomitant with the reduced level of c-FLIPL at the DISC, increased levels of c-FLIPp43 (2.3-fold) and c-FLIPS (6.6-fold) were detected in the complex (Fig. 4). This is in agreement with previous reports that c-FLIPp43 (also referred to as p43-c-FLIPL) (43) and c-FLIPS also regulate apoptosis at the Fas DISC (43).
The Interaction between DR5 and c-FLIP Is Independent of FADD
Our observation that c-FLIPL interacts with DR5 prior to stimulation with TRAIL, whereas FADD interacts with DR5 only after TRAIL treatment, suggests that the association between c-FLIPL and DR5 is independent of FADD and DR5 interaction. In order to test this hypothesis, we utilized FADD-deficient Jurkat cells (FADD−/−) (56, 57) in which the expected FADD deficiency was confirmed (Fig. 3D) (56, 57). The TRAIL receptor surface expression pattern of the deficient clones was identical to that reported for parental Jurkat A3 control cells, wherein only DR5 but not DR4 is expressed (56, 57). We immunoprecipitated endogenous DR5 from parental Jurkat A3 cells and from FADD−/− Jurkat cells. The precipitates were then separated by SDS/PAGE and immunoblotted with c-FLIP antibody. Endogenous DR5 readily co-precipitated with c-FLIPL (Fig. 3D) in parental as well as in FADD−/− Jurkat cells. Cellular levels of DR5 and c-FLIPL were visualized by direct Western blot analysis of total cell lysates with antibodies to DR5 or c-FLIPL, respectively (Fig. 3D). Taken together, these observations confirm that the interaction between DR5 and c-FLIPL is independent of FADD.
A Cellular Membrane Permeable c-FLIP Peptide Triggers Apoptosis
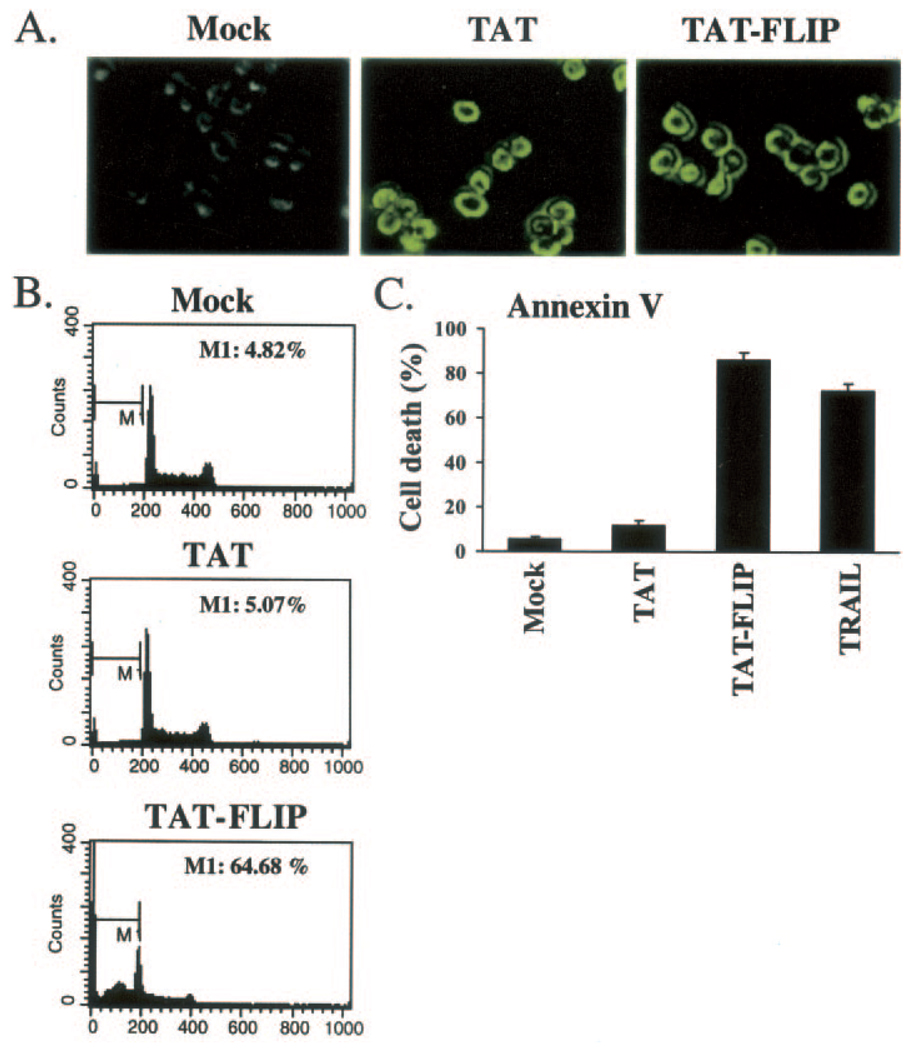
To determine the functional significance of c-FLIP and DR5 interaction in regulating cellular fate, we synthesized a cellular membrane permeable peptide corresponding to the DR5 binding domain of c-FLIP (Figs. 1 and 2). This peptide was generated by fusing the TAT peptide (YGRKKRRQRRR) sequence to the N-terminal region of the c-FLIP peptide (READFFWSLSTADMS). The TAT peptide has previously been shown to penetrate very efficiently into mammalian cells (59–61). Moreover, it has been demonstrated that the TAT peptide can translocate a variety of molecular cargoes, including proteins and nucleotides into mammalian cells (59–61). We used Jurkat cells to test the effect of internalized c-FLIP peptide on cellular fate by measuring the percentage of the sub-G1 apoptotic population and annexin V staining. While Jurkat cells treated with TAT alone did not show any significant increase in the apoptotic population (5–10%), treatment of Jurkat cells with TAT-FLIP peptide resulted in a significant increase in the apoptotic population (65–85%) (Fig. 5), indicating that there is a significant increase in spontaneous/ligand-independent apoptosis. We have also observed induction of apoptosis in HeLa cells that have been treated with TAT-FLIP peptide (data not shown). These observations indicate that inhibiting the interaction between DR5 and c-FLIPL will promote spontaneous apoptosis of Jurkat cells.
FIG. 5. A cellular-membrane permeable version of the DR5 binding domain of c-FLIP triggers apoptosis.
A–C, Jurkat cells were incubated in medium containing vehicle (Mock), 15 µm TAT, or 15 µm TAT-FLIP cellular membrane permeable peptides for 2 h (A and C) or 16 h (B). A, immunofluorescence analysis demonstrated that TAT and TAT-FLIP are internalized. B, cells were analyzed for the percentage of apoptotic sub-G1 population by flow cytometry. M1 is the G0/G1 apoptotic population. C, percent apoptosis was also determined by annexin V-PE staining and flow cytometry. Apoptotic rates from three independent experiments are shown in B and C.
DISCUSSION
Since TRAIL and TRAIL receptors are widely expressed in many tissues (1, 8, 11, 14, 41), ligand-independent spontaneous signaling from TRAIL receptors could result in inappropriate apoptosis. Thus, cellular mechanisms must exist to keep signaling from TRAIL receptors in check. c-FLIP has been generally regarded as a critical regulator of death-receptor mediated apoptosis through its association with and inhibition of caspase-8/10 at the DISC. However, it has recently been reported that c-FLIPL, when present at the DISC, functions as an activator, rather than as an inhibitor, of caspase-8 (40, 62). Therefore, the mechanism by which c-FLIP inhibits death receptor-induced apoptosis has yet to be completely elucidated (39).
In this study, we identified a phage-displayed peptide that interacts with the death domain of DR5. Interestingly, this peptide corresponds to a region in the caspase domain of c-FLIPL, although a matching sequence is not present in c-FLIPS or c-FLIPp43. Following up this observation we discovered that endogenous c-FLIPL and endogenous DR5 were pre-associated in non-stimulated Jurkat T cells (Figs. 3 and 4). Peptide specificity assays and in vitro GST fusion protein pull-down experiments, revealed that c-FLIPL binds specifically with the DD of DR5, but not with the DD of DR4 or of Fas (Figs. 1A and 2B and data not shown), suggesting that c-FLIPL may have a unique role in regulating DR5 function. Furthermore, we demonstrated that the interaction between DR5 and c-FLIPL was independent of the association between FADD and DR5 (Fig. 3D). The pre-association of c-FLIPL and DR5 in the absence of TRAIL implies a novel regulatory function of c-FLIPL in regulating death signaling upstream of the DISC at the level of the DR5 death receptor.
The role of c-FLIPL as a regulator of cell death upstream of the DISC, is analogous to the function of a recently identified protein regulator of TNF signaling known as the silencer of death domains (SODD) (63–65). SODD inhibits spontaneous signaling from TNF-R1 by pre-associating with the receptor and preventing the formation of active signaling complexes (63, 64). Upon stimulation, TNF-induced receptor trimerization causes rapid dissociation of SODD from TNFR-1, thus allowing the recruitment of TRADD and subsequent assembly of active signaling complexes (63, 64). Similarly, we have observed that in response to TRAIL stimulation, c-FLIPL dissociates from the DISC (Fig. 4). This coincides with the recruitment of FADD and caspase-8 to the DR5 DISC and the activation of caspase-8 (Fig. 4). Interestingly, the dissociation of c-FLIPL from DR5 is also accompanied by the appearance of c-FLIPp43 (which lacks the DR5 binding domain) at the DISC, suggesting that release of c-FLIPL results in its proteolytic processing and termination of its SODD-like activity (Fig. 4).
As shown in Fig. 5, the cellular-membrane permeable form of the c-FLIP peptide induces apoptosis of Jurkat cells in the absence of TRAIL. This observation indicates that the interaction between DR5 and c-FLIP plays a critical role in regulating cellular fate. In this manner, this c-FLIP peptide can inhibit the interaction between DR5 and c-FLIPL, thus promoting the ligand-independent recruitment of the DISC to DR5 and leading to spontaneous cell death. We are currently carrying out competition binding studies with this peptide. Additionally, we are investigating the utility of this peptide in suppression of cellular transformation.
The current view of the mechanism of death receptor-induced regulation of cell fate involves the formation of protein complexes (2, 41, 66, 67). The majority of protein-protein interactions that thus far identified in death receptor signaling have been through the use of isolated domains involving homotypic interaction between death domains or death effector domains (68). For example, the DD of FADD has been shown to interact with the DD of Fas or of TRAIL receptors. On the other hand, the DED domain of FADD interacts with the DED domain of caspase-8. However, in a screen for FADD mutants that are deficient in interaction with the DD of Fas, the majority of the identified mutations were within the DED domain of FADD which indicates that interactions other than homotypic associations are also involved in mediating signals from death receptors (41, 69). Interestingly, the interaction between DR5 and c-FLIPL occurs between the DD of DR5 and the inactive caspase domain of c-FLIPL, which represents yet another type of protein-protein interaction that participates in regulating death receptor-induced pathways.
Taken together, our studies demonstrate a direct interaction between DR5 and c-FLIPL, indicating an inhibition of apoptosis at the level of the death receptor. Notably, the inhibition of death signaling at the death receptor level might be one mechanism by which unwanted apoptosis is prevented in normal c-FLIPL-expressing cells. Thus, depending on the ratio of FADD/caspase-8 to c-FLIPL, DR5 might recruit either c-FLIPL or FADD/caspase-8. This idea is also supported by the observation that over-expression of c-FLIPL in some human tumor cells is thought to constitute a mechanism for resistance to TRAIL-induced apoptosis (1, 8, 39, 40, 66, 70, 71). The resistance of these tumors to TRAIL signaling could be the result of inhibition of DISC formation because of pre-association of DR5 and c-FLIPL. The recruitment of c-FLIPL to DR5 might then result in formation of anti-apoptotic signaling complexes distinct from the DISC. For example, NF-κB has previously been reported to be activated very efficiently by c-FLIPL through association of c-FLIPL, with TRAFs and IKK2 (72). Thus, disruption of DR5 and c-FLIPL interaction with the cellular-membrane permeable c-FLIP peptide (corresponding to the DR5 binding domain of c-FLIP) will result in enhanced spontaneous apoptosis. The contribution and modes of regulation of the DR5/c-FLIPL anti-apoptotic signaling complex versus the FADD/caspase-8 DISC could determine the life versus death decisions of cells.
Acknowledgments
We thank Drs. David Baltimore, Avi Ashkenazi, Shirley Stiver, and Donald Senger for helpful comments; Drs. Avi Ashkenazi, John Blenis, Wafik El-Deiry, Rob Kay, Jurg Tschopp, and Xiaolu Yang for valuable reagents; Zhen Xi Wang and Paula Livernois for valuable technical assistance; Shalini Rana for editorial assistance; and Jane Hayward for figure preparation.
Footnotes
This study was supported by Public Health Service Grants 1 R55 CA87675 and HL080192 from the National Institutes of Health, DAMD 17-02-0299 from the Department of Defense, and RSG 03-012-01-CCG from the American Cancer Society (to R. K.-F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Supported by Grant T32 H07893 from National Institutes of Health.
The abbreviations used are: TNF, tumor necrosis factor; TRAIL, TNF-related apoptosis-inducing ligand; FADD, Fas-associated protein with death domain; DED, death effector domain; HEK, human embryonic kidney cell; GST, glutathione S-transferase; DISC, death-inducing signaling complex; SODD, silencer of death domains; ELISA, enzymelinked immunosorbent assay; DD, death domain; c-FLIP, cellular FLICE-like inhibitory protein; c-FLIPL, long form of c-FLIP; c-FLIPS, short variant of c-FLIP; DR, death receptor.
REFERENCES
- 1.Abe K, Kurakin A, Mohseni-Maybodi M, Kay B, Khosravi-Far R. Ann. N. Y. Acad. Sci. 2000;926:52–63. doi: 10.1111/j.1749-6632.2000.tb05598.x. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Dixit VM. Curr. Opin. Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 3.Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. J. Biol. Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 4.Schulze-Osthoff K, Ferrari D, Los M, Wesselborg S, Peter ME. Eur. J. Biochem. 1998;254:439–459. doi: 10.1046/j.1432-1327.1998.2540439.x. [DOI] [PubMed] [Google Scholar]
- 5.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Goodwin RG. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 6.Esposti MD. J. Leukoc. Biol. 1999;65:535–542. doi: 10.1002/jlb.65.5.535. [DOI] [PubMed] [Google Scholar]
- 7.Golstein P. Curr. Biol. 1997;7:R750–R753. doi: 10.1016/s0960-9822(06)90000-1. [DOI] [PubMed] [Google Scholar]
- 8.Ozoren N, El-Deiry WS. Semin. Cancer Biol. 2003;13:135–147. doi: 10.1016/s1044-579x(02)00131-1. [DOI] [PubMed] [Google Scholar]
- 9.Almasan A, Ashkenazi A. Cytokine Growth Factor Rev. 2003;14:337–348. doi: 10.1016/s1359-6101(03)00029-7. [DOI] [PubMed] [Google Scholar]
- 10.Kayagaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, Yagita H. J. Exp. Med. 1999;189:1451–1460. doi: 10.1084/jem.189.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackay F, Kalled SL. Curr. Opin. Immunol. 2002;14:783–790. doi: 10.1016/s0952-7915(02)00407-7. [DOI] [PubMed] [Google Scholar]
- 12.Mariani SM, Krammer PH. Eur. J. Immunol. 1998;28:1492–1498. doi: 10.1002/(SICI)1521-4141(199805)28:05<1492::AID-IMMU1492>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.Martinez LM, Alava MA, Gamen S, Kim KJ, Chuntharapai A, Pineiro A, Naval J, Anel A. Eur. J. Immunol. 1998;28:2714–2725. doi: 10.1002/(SICI)1521-4141(199809)28:09<2714::AID-IMMU2714>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, El-Deiry WS. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- 15.Zamai L, Ahmad M, Bennett IM, Azzoni L, Alnemri ES, Perussia B. J. Exp. Med. 1998;188:2375–2380. doi: 10.1084/jem.188.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 17.MacFarlane M, Ahmad M, Srinivasula SM, Fernandes-Alnemri T, Cohen GM, Alnemri ES. J. Biol. Chem. 1997;272:25417–25420. doi: 10.1074/jbc.272.41.25417. [DOI] [PubMed] [Google Scholar]
- 18.Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray CL, Baker K, Wood WI, Goddard AD, Godowski P, Ashkenazi A. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 19.Walczak H, Degli EM, Johnson RS, Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA, Smith CA, Goodwin RG, Rauch CT. EMBO. 1997;16:5386–5397. doi: 10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Screaton GR, Mongkolsapaya J, Xu XN, Cowper AE, McMichael AJ, Bell JI. Curr. Biol. 1997;7:693–696. doi: 10.1016/s0960-9822(06)00297-1. [DOI] [PubMed] [Google Scholar]
- 21.Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Immunity. 1997;7:821–830. doi: 10.1016/s1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- 23.Wu GS, Burns TF, Zhan Y, Alnemri ES, El-Deiry WS. Cancer Res. 1999;59:2770–2775. [PubMed] [Google Scholar]
- 24.Sheikh MS, Huang Y. Curr. Cancer Drug Targets. 2004;4:97–104. doi: 10.2174/1568009043481597. [DOI] [PubMed] [Google Scholar]
- 25.Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J, Krammer PH, Walczak H. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 26.Bodmer JL, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P, Blenis J, Tschopp J. Nat. Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 27.Yamada H, Tada-Oikawa S, Uchida A, Kawanishi S. Biochem. Biophys. Res. Commun. 1999;265:130–133. doi: 10.1006/bbrc.1999.1641. [DOI] [PubMed] [Google Scholar]
- 28.Esposti MD. Apoptosis. 2002;7:433–440. doi: 10.1023/a:1020035124855. [DOI] [PubMed] [Google Scholar]
- 29.Zimmermann KC, Bonzon C, Green DR. Pharmacol. Ther. 2001;92:57–70. doi: 10.1016/s0163-7258(01)00159-0. [DOI] [PubMed] [Google Scholar]
- 30.Korsmeyer SJ, Gross A, Harada H, Zha J, Wang K, Yin XM, Wei M, Zinkel S. Cold Spring Harb. Symp. Quant. Biol. 1999;64:343–350. doi: 10.1101/sqb.1999.64.343. [DOI] [PubMed] [Google Scholar]
- 31.Bertin J, Armstrong RC, Ottilie S, Martin DA, Wang Y, Banks S, Wang GH, Senkevich TG, Alnemri ES, Moss B, Lenardo MJ, Tomaselli KJ, Cohen JI. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goltsev YV, Kovalenko AV, Arnold E, Varfolomeev EE, Brodianskii VM, Wallach D. J. Biol. Chem. 1997;272:19641–19644. doi: 10.1074/jbc.272.32.19641. [DOI] [PubMed] [Google Scholar]
- 33.Hu S, Vincenz C, Ni J, Gentz R, Dixit VM. J. Biol. Chem. 1997;272:17255–17257. doi: 10.1074/jbc.272.28.17255. [DOI] [PubMed] [Google Scholar]
- 34.Hu S, Vincenz C, Buller M, Dixit VM. J. Biol. Chem. 1997;272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 35.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C, Rimoldi D, French LE, Tschopp J. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 36.Shu HB, Halpin DR, Goeddel DV. Immunity. 1997;6:751–763. doi: 10.1016/s1074-7613(00)80450-1. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasula SM, Ahmad M, Ottilie S, Bullrich F, Banks S, Wang Y, Fernandes-Alnemri T, Croce CM, Litwack G, Tomaselli KJ, Armstrong RC, Alnemri ES. J. Biol. Chem. 1997;272:18542–18545. doi: 10.1074/jbc.272.30.18542. [DOI] [PubMed] [Google Scholar]
- 38.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schroter M, Scaffidi C, Krammer PH, Peter ME, Tschopp J. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 39.Krueger A, Baumann S, Krammer PH, Kirchhoff S. Mol. Cell. Biol. 2001;21:8247–8254. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Micheau O. Expert Opin. Ther. Targets. 2003;7:559–573. doi: 10.1517/14728222.7.4.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorburn A. Cell Signal. 2004;16:139–144. doi: 10.1016/j.cellsig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Guo F, Bhalla K. Cancer Biol. Ther. 2002;1:528–529. doi: 10.4161/cbt.1.5.170. [DOI] [PubMed] [Google Scholar]
- 43.Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang X. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh WC, Itie A, Elia AJ, Ng M, Shu HB, Wakeham A, Mirtsos C, Suzuki N, Bonnard M, Goeddel DV, Mak TW. Immunity. 2000;12:633–642. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 45.Burns TF, El-Deiry WS. J. Biol. Chem. 2001;276:37879–37886. doi: 10.1074/jbc.M103516200. [DOI] [PubMed] [Google Scholar]
- 46.Bin L, Li X, Xu LG, Shu HB. FEBS Lett. 2002;510:37–40. doi: 10.1016/s0014-5793(01)03222-7. [DOI] [PubMed] [Google Scholar]
- 47.Khosravi-Far R, White MA, Westwick JK, Solski PA, Chrzanowska-Wodnicka M, van Aelst L, Wigler MH, Der CJ. Mol. Cell. Biol. 1996;16:3923–3933. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Khosravi-Far R, Chang HY, Baltimore D. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitehead IP, Lambert QT, Glaven JA, Abe K, Rossman KL, Mahon GM, Trzaskos JM, Kay R, Campbell SL, Der CJ. Mol. Cell. Biol. 1999;19:7759–7770. doi: 10.1128/mcb.19.11.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurakin A, Hoffman NG, Kay BK. J. Pept. Res. 1998;52:331–337. doi: 10.1111/j.1399-3011.1998.tb00657.x. [DOI] [PubMed] [Google Scholar]
- 51.Kay BK, Kasanov J, Knight S, Kurakin A. FEBS Lett. 2000;480:55–62. doi: 10.1016/s0014-5793(00)01778-6. [DOI] [PubMed] [Google Scholar]
- 52.Sparks AB, Adey NB, Quilliam LA, Thorn JM, Kay BK. Methods Enzymol. 1995;255:498–509. doi: 10.1016/s0076-6879(95)55052-6. [DOI] [PubMed] [Google Scholar]
- 53.Gee SH, Sekely SA, Lombardo C, Kurakin A, Froehner SC, Kay BK. J. Biol. Chem. 1998;273:21980–21987. doi: 10.1074/jbc.273.34.21980. [DOI] [PubMed] [Google Scholar]
- 54.Smith GP, Schultz DA, Ladbury JE. Gene. 1993;128:37–42. doi: 10.1016/0378-1119(93)90150-2. [DOI] [PubMed] [Google Scholar]
- 55.Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Juo P, Kuo CJ, Yuan J, Blenis J. Curr. Biol. 1998;8:1001–1008. doi: 10.1016/s0960-9822(07)00420-4. [DOI] [PubMed] [Google Scholar]
- 57.Juo P, Woo MS, Kuo CJ, Signorelli P, Biemann HP, Hannun YA, Blenis J. Cell Growth Differ. 1999;10:797–804. [PubMed] [Google Scholar]
- 58.Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 59.Futaki S, Goto S, Suzuki T, Nakase I, Sugiura Y. Curr. Protein Pept. Sci. 2003;4:87–96. doi: 10.2174/1389203033487261. [DOI] [PubMed] [Google Scholar]
- 60.Takenobu T, Tomizawa K, Matsushita M, Li ST, Moriwaki A, Lu YF, Matsui H. Mol. Cancer Ther. 2002;1:1043–1049. [PubMed] [Google Scholar]
- 61.Adams JM, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 62.Meier P, Silke J. Nat. Cell Biol. 2003;5:1035–1038. doi: 10.1038/ncb1203-1035. [DOI] [PubMed] [Google Scholar]
- 63.Jiang Y, Woronicz JD, Liu W, Goeddel DV. Science. 1999;283:543–546. doi: 10.1126/science.283.5401.543. [DOI] [PubMed] [Google Scholar]
- 64.Tschopp J, Martinon F, Hofmann K. Curr. Biol. 1999;9:R381–R384. doi: 10.1016/s0960-9822(99)80233-4. [DOI] [PubMed] [Google Scholar]
- 65.Takada H, Chen NJ, Mirtsos C, Suzuki S, Suzuki N, Wakeham A, Mak TW, Yeh WC. Mol. Cell. Biol. 2003;23:4026–4033. doi: 10.1128/MCB.23.11.4026-4033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacFarlane M. Toxicol. Lett. 2003;139:89–97. doi: 10.1016/s0378-4274(02)00422-8. [DOI] [PubMed] [Google Scholar]
- 67.Scaffidi C, Schmitz I, Zha J, Korsmeyer SJ, Krammer PH, Peter ME. J. Biol. Chem. 1999;274:22532–22538. doi: 10.1074/jbc.274.32.22532. [DOI] [PubMed] [Google Scholar]
- 68.Fesik SW. Cell. 2000;103:273–282. doi: 10.1016/s0092-8674(00)00119-7. [DOI] [PubMed] [Google Scholar]
- 69.Thomas LR, Stillman DJ, Thorburn A. J. Biol. Chem. 2002;277:34343–34348. doi: 10.1074/jbc.M204169200. [DOI] [PubMed] [Google Scholar]
- 70.Leverkus M, Neumann M, Mengling T, Rauch CT, Brocker EB, Krammer PH, Walczak H. Cancer Res. 2000;60:553–559. [PubMed] [Google Scholar]
- 71.Yang X. Cancer Biol. Ther. 2002;1:407–408. doi: 10.4161/cbt.1.4.16. [DOI] [PubMed] [Google Scholar]
- 72.Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M, Burns K, Hahne M, Kennedy N, Kovacsovics M, Tschopp J. Curr. Biol. 2000;10:640–648. doi: 10.1016/s0960-9822(00)00512-1. [DOI] [PubMed] [Google Scholar]