Abstract
The Stanford Microarray Database (SMD) stores raw and normalized data from microarray experiments, and provides web interfaces for researchers to retrieve, analyze and visualize their data. The two immediate goals for SMD are to serve as a storage site for microarray data from ongoing research at Stanford University, and to facilitate the public dissemination of that data once published, or released by the researcher. Of paramount importance is the connection of microarray data with the biological data that pertains to the DNA deposited on the microarray (genes, clones etc.). SMD makes use of many public resources to connect expression information to the relevant biology, including SGD [Ball,C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Issel-Tarver,L., Kasarskis,A., Scafe,C.R., Sherlock,G., Binkley,G., Jin,H. et al. (2000) Nucleic Acids Res., 28, 77–80], YPD and WormPD [Costanzo,M.C., Hogan,J.D., Cusick,M.E., Davis,B.P., Fancher,A.M., Hodges,P.E., Kondu,P., Lengieza,C., Lew-Smith,J.E., Lingner,C. et al. (2000) Nucleic Acids Res., 28, 73–76], Unigene [Wheeler,D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Nucleic Acids Res., 28, 10–14], dbEST [Boguski,M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) Nature Genet., 4, 332–333] and SWISS-PROT [Bairoch,A. and Apweiler,R. (2000) Nucleic Acids Res., 28, 45–48] and can be accessed at http://genome-www.stanford.edu/microarray.
INTRODUCTION
Microarray experiments are routinely performed to examine gene expression (1) or DNA copy number (2) on a genomic scale. Typically many thousands of DNA samples are arrayed on a glass slide, and labeled cDNA or genomic DNA from control and experimental samples are competitively hybridized to the array. Images of the slide are then acquired and processed to produce a data file that contains dozens of values per spot for several thousand spots. Although the salient information for each spot is the ratio between the experimental and control samples, the other values may be used as filtering criteria for determining which data are reliable. Thus access to all the data for each spot is required for its comprehensive analysis. A single microarray of 20 000 spots may generate in the order of a million pieces of information and an experimental series (e.g. 3) may therefore generate more than 50 million data points. A major goal of the Stanford Microarray Database (SMD) is to organize this vast amount of data, such that a researcher can filter their data to retrieve only that which he or she wishes to work with, and then perform analyses on that data.
IMPLEMENTATION
SMD is accessed over the Internet using a web browser, which allows for significant flexibility and remote access, without the need for special software installation on client computers. Updates made to the software that runs on the server machine are therefore automatically reflected for all users. Although a few features do require a recent, JavaScript enabled browser, multiple platforms (MacOS, UNIX and Windows 95/98/2000) can access SMD without difficulty. SMD runs on a Sun server running the Solaris operating system, and uses Oracle 8 as the database management system. Scripts are implemented using the Perl language (www.perl.com), in conjunction with the DBI module (http://search.cpan.org/search?module=DBI), that allows the scripts to connect to the database, and the CGI (http://stein.cshl.org/WWW/software/CGI/) and GD (http://stein.cshl.org/WWW/software/GD/) modules. For improved performance some of the more processor intensive tasks (e.g. clustering, image generation) are implemented in the C programming language. All of the source code that is used by SMD will be made freely available to academic researchers who wish to set up their own database using SMD’s model. Since SMD uses Oracle as its database management system, it is implemented as a relational database, table specifications for which can be found at: http://genome-www4.stanford.edu/MicroArray/SMD/doc/db_specifications.html.
DATA LOADING
SMD’s loading program allows Stanford users to load their experiments into the database via a web form, specifying the location of their data and image files. The user may load experiments either individually or via a batch procedure, to minimize user time spent loading. The database accepts data produced by both GenePix (http://axon.com/GN_GenePixSoftware.html) and Scanalyze (http://rana.Stanford.EDU/software/), and loading is implemented via a queuing system, which allows users to monitor the progress of their experiment loading, and provides advanced and robust recovery procedures should a problem occur during loading. Original 16 bit TIFF images are archived. In addition, during loading, a proxy GIF image is generated from the two TIFF images. This GIF image is stored on the file system, and allows the user to visualize and assess their original data (see below). Data normalization is performed during loading and both the normalized and original data are stored.
DATA PROCESSING AND EDITING
The data and associated experimental information loaded into the database are not static, but instead may be later modified by the owner of the experiment. The owner may opt to modify or add to any of the associated experimental information (experiment name, channel descriptions, category, subcategory and experiment description) that describe the types of questions being asked. In addition the owner may visually inspect their data, by means of the proxy GIF image, and then flag or unflag their data based on whether a spot appears to contain reliable information, either visually, or by some filtering criteria. Users may also renormalize their data using one of two automated methods or can enter their own normalization factor. Normalized data will be recalculated, and the new results stored in the database.
If there is a systematic problem with a microarray print run (e.g. a PCR failure occurred for some clones, or some clones are found to be contaminated) database curators can modify the details of that print run. All experiments that used microarrays from that print run are automatically updated to reflect the current information.
SPOT TO GENE ASSOCIATIONS
One of the larger yet more important challenges facing SMD is the association of known biological information with individual spots on an array. Since microarray data cannot be analyzed meaningfully in the absence of biological context, a large part of our effort is expended to associate the microarray data with the most current biological annotation available. SMD currently contains results for arrays of DNA from eight organisms, and it is therefore necessary to use several resources to obtain the biological information about each DNA sample spotted. For the systematically sequenced ORFs of Saccharomyces cerevisiae, SMD stores gene names, biological process and molecular function from the Saccharomyces Genome Database (SGD) (4) (http://genome-www.stanford.edu/Saccharomyces/), and these are automatically updated when SGD itself is updated. For Caenorhabditis elegans ORFs, SMD uses the title lines that are provided by Proteome as part of their WormPD database (5), which are well-curated information (http://www.proteome.com/databases/index.html). For human and mouse clones, the association between an EST and a gene is often still being determined and likely to change. SMD therefore stores human and mouse Unigene (http://www.ncbi.nlm.nih.gov/UniGene/) in a relational format within the database, and updates this data as new Unigene builds are posted. By storing the accession numbers of each clone that has been arrayed, SMD is able to connect a clone to its latest gene assignment through Unigene. Thus when users retrieve or analyze data, they can always see it in its most current biological context. SMD also provides hyperlinks to external databases, where users can view additional information about their genes or clones of interest.
SEARCHING SMD
The large number of experiments in SMD (9022, as of November 13, 2000, of which 313 are public) necessitates simple, intuitive interfaces that enable the user to narrow down the experiments with which they are dealing. For each experiment, SMD records the name of the researcher, a category and subcategory that describe the biological nature of the experiment, and the organism that served as the source of the DNA spotted on the microarray. Each of these criteria may be used simultaneously to create a query that will narrow down the number of experiments for subsequent examination or analysis. A researcher may also limit the experiments of interest based on the print run during which a set of microarrays was fabricated. After selecting the search criteria a researcher may select whether to look at arrays individually, or whether to combine the results of the selected arrays for retrieval and analysis.
SMD offers researchers several options when examining arrays individually. A researcher may download all the data for an individual array to a local machine, or examine the data online, sorted and filtered according to various criteria. Additionally a user can view the proxy GIF image to evaluate the placement of the grid used during data acquisition, and elect to change the flag status of any of the spots. Since the GIF is a clickable image map, the user can browse the individual spots, viewing both the data acquired during scanning, and the associated biological information (Fig. 1).
Figure 1.
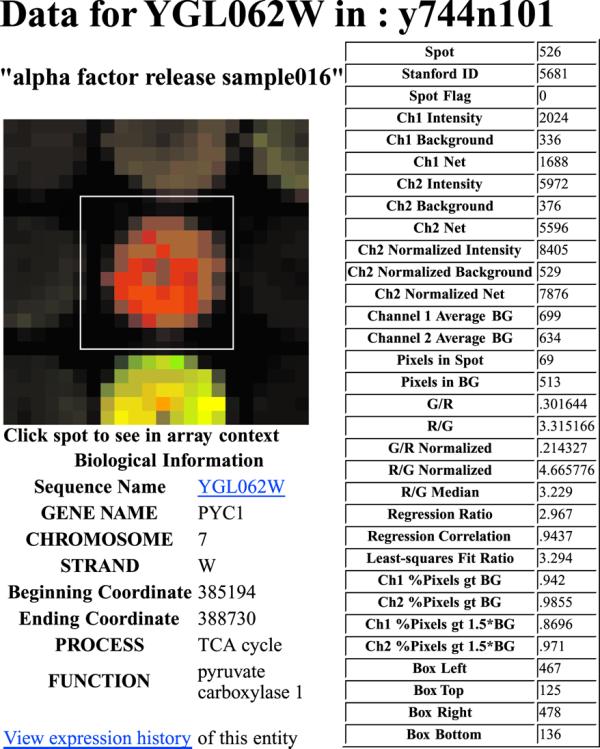
Zoomed spot image. A close-up of an individual spot, generated from the proxy GIF image, is shown, with all of the spot parameters, and the associated biological annotation.
To analyze microarray data comprehensively, the results of many microarrays are typically combined. When retrieving data from multiple arrays simultaneously, a user may apply several filters to the data as it is processed for retrieval, and select what biological annotation to have associated with the data. Filters may be applied on a spot by spot basis (e.g. minimum signal intensity, or flag status) or on a global basis (e.g. only retrieve data for genes whose expression varies by a certain amount). The spot by spot filters may also be combined using Boolean AND and OR operators, such that a researcher could request data, for example, from spots that have normalized ratio > 2 AND (channel 1 intensity greater > 150 OR channel 2 intensity > 150). After retrieval, data may be preprocessed, e.g. the data may be log transformed, or a mathematical operation such as centering the data for each gene (or experiment) by the median (or mean) expression value for that gene may be carried out. A file is then generated, which the user may download for analysis on their local machine, or the user may continue with online analysis.
Currently SMD provides online hierarchical clustering (6) and Self-Organizing Maps (7,8), which use XCluster underneath (9) (http://genome-www.stanford.edu/~sherlock/cluster.html). SMD will in future support k-means clustering (10), and the use of Singular Value Decomposition to find patterns in the microarray data (11). In addition methods for comparison of these analysis tools are being developed to allow the researcher to better understand the similarities and differences in analyses using different methods.
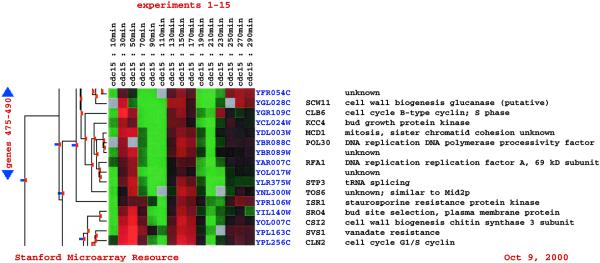
After the data have been analyzed, files that are compatible with TreeView (http://rana.Stanford.EDU/software/) may be downloaded, or the results may be viewed online. Online browsing of the clustered data is facilitated by clickable maps, ORF name searches, and display of the joining correlation between genes or experiments, and links to external biological databases (Fig. 2).
Figure 2.
Hierarchical cluster. A portion of a hierarchical cluster, which can be easily navigated, is shown. Red indicates up-regulation in the experimental sample, and green indicates down-regulation in the experimental sample, with respect to the control. The intensity of the color indicates the magnitude of up- or down-regulation (see 6).
Thus, during a typical analysis run, a user would first select the experiments in which they were interested, then select the filtering criteria they wanted to use to retrieve data for those experiments, and what preprocessing they wanted for the resulting dataset. Finally they could choose to cluster the resulting file, and visualize the clustered data in their web browser, with the biological data in the cluster to help them interpret the expression patterns.
SMD AS A RESOURCE FOR THE BIOLOGY COMMUNITY
The enormous quantity of data produced by microarray experiments also poses a challenge for the public dissemination of the results. Many current publications of microarray results require supplemental web pages in order to fully release the data to interested researchers. Furthermore it is in the best interest of the scientific community at large that the data and tools are available to all. Therefore SMD is endeavoring to provide a public interface for data release to the biological community. The aim is that upon publication, or at the experiment owner’s discretion, data will be made world-viewable. Published data will be organized into curated datasets that can be either analyzed online or downloaded. The scientific community will be able to search and analyze experiments by their criterion of interest, whether it is by organism, by publication, or by category of experiment. In addition, in collaboration with the Arabidopsis Functional Genomics Consortium (http://afgc.stanford.edu/), results from plant microarrays are provided. SMD will not however act as a pubic repository for data, and instead will make all of its source code available to enable other institutions to set up their own microarray databases using SMD’s model. SMD will be supported as long as the microarray community at Stanford University supports and uses it.
THE FUTURE OF SMD
Once SMD has met its immediate goals of providing a database that can be used by both local and public researchers to analyze and retrieve microarray data, the project will embark on some more long-term goals. Of key importance is the annotation of the experiments themselves. Currently only a limited amount of information is stored for each experiment. In the future we hope to allow researchers to store as much information as would be needed to reproduce their experiments. Although the storage of these data will be invaluable for people other than the experimenter themselves, its entry should not be too onerous on the researcher. A second goal is to implement a flexible way such that results of analyses can also be stored in the database. To be useful, this system would need to capture all the various criteria and parameters that were used in the analyses—in essence the database would store the results of computer-based experiments, in addition to the microarray ones, with full information on how to repeat the experiment. Finally, and closely related to the first two future goals, is the support of a data exchange format that will allow researchers to easily exchange microarray data. SMD intends to support and help define the format being discussed by the Array XML working group (http://beamish.lbl.gov/).
References
- 1.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 2.Pollack J.R., Perou,C.M., Alizadeh,A.A., Eisen,M.B., Pergamenschikov,A., Williams,C.F., Jeffrey,S.S., Botstein,D. and Brown,P.O. (1999) Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genet., 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 3.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Issel-Tarver,L., Kasarskis,A., Scafe,C.R., Sherlock,G., Binkley,G., Jin,H., Kaloper,M., Orr,S.D., Schroeder,M., Weng,S., Zhu,Y., Botstein,D. and Cherry,J.M. (2000) Integrating functional genomic information into the Saccharomyces genome database. Nucleic Acids Res., 28, 77–80. Updated article in this issue: Nucleic Acids Res. (2001), 29, 80–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costanzo M.C., Hogan,J.D., Cusick,M.E., Davis,B.P., Fancher,A.M., Hodges,P.E., Kondu,P., Lengieza,C., Lew-Smith,J.E., Lingner,C., Roberg-Perez,K.J., Tillberg,M., Brooks,J.E. and Garrels,J.I. (2000) The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res., 28, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohonen T. (1995) Self Organizing Maps. Springer, Berlin.
- 8.Tamayo P., Slonim,D., Mesirov,J., Zhu,Q., Kitareewan,S., Dmitrovsky,E., Lander,E.S. and Golub,T.R. (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl Acad. Sci. USA, 96, 2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherlock G. (2000) Analysis of large-scale gene expression data. Curr. Opin. Immunol., 12, 201–205. [DOI] [PubMed] [Google Scholar]
- 10.Everitt B. (1974) Cluster Analysis 122. Heinemann, London.
- 11.Alter O., Brown,P.O. and Botstein,D. (2000) Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl Acad. Sci. USA, 97, 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]