Abstract
Allergen recognition by IgE antibodies is a key event in allergic inflammation.
In this study, the IgE IGHV repertoires of individuals with allergy to the major birch pollen allergen, Bet v 1, were analyzed over a four years period of allergen exposure by RT-PCR and sequencing of cDNA.
Approximately half of the IgE transcripts represented non-redundant sequences, which belonged to seventeen different IGHV genes. Most variable regions contained somatic mutations but also non-mutated sequences were identified. There was no evidence for relevant increases of somatic mutations over time of allergen exposure. Highly similar IgE variable regions were found after four years of allergen exposure in the same and in genetically non-related individuals.
Our results indicate that allergens select and shape a limited number of similar IgE variable regions in the human IgE repertoire.
Keywords: Allergy, Allergen, Antibodies, IgE repertoire
1. Introduction
Immunoglobulin E occurs in extremely low concentrations in serum and other body fluids but can induce severe inflammatory reactions via interaction with a variety of immune cells (Geha et al., 2003; Ishizaka et al., 1966). IgE-mediated cross-linking of FcɛRI receptors on mast cells and basophils induces the immediate release of inflammatory mediators, enzymes and pro-inflammatory cytokines and represents a key mechanism in acute allergic inflammation (Kraft and Kinet, 2007). Furthermore, IgE antibodies can present minute amounts of antigens to T cells and thus contribute to efficient T cell activation (van Neerven et al., 2006). Besides its key role in allergic inflammation, IgE may play a part in defense against parasitic infestations via activation of eosinophils (Gounni et al., 1994). The class switch to IgE production in the course of the primary immune response is induced by Th2 cytokines secreted by Th2 helper cells upon allergen encounter (Geha et al., 2003; Romagnani, 2001). Most of the allergens inducing IgE responses in man are protein antigens of which many have been characterized down to their three-dimensional structure and biological functions (Valenta, 2002). For some of these allergens evidence exists that they may promote the development of IgE responses through their intrinsic biological properties. Other studies suggest that non-allergenic components present in allergen sources, certain allergen carriers, Toll-receptor agonists/antagonists or environmental factors may induce or hamper the development of Th2 and IgE responses (Blumer et al., 2005; Traidl-Hoffmann et al., 2005).
Due to the extremely low concentrations of IgE in serum only few studies have investigated the molecular basis of allergen-specific IgE responses and the repertoire of IgE antibodies. In fact, respiratory allergen contact has been shown to up-regulate strongly IgE production in allergic subjects (Niederberger et al., 2007). Several studies have analyzed the IgE repertoire in allergic subjects but all the analyzed individuals mounted IgE responses against numerous and unknown allergens. Hence it has been impossible to draw conclusions regarding the repertoire of IgE antibodies specific for defined allergens (Bando et al., 2004; Coker et al., 2005; Davies and O’Hehir, 2004; Janezic et al., 1998; Lim et al., 2007; Snow et al., 1995; Tilgner et al., 1997; van der Stoep et al., 1993). Those studies reporting the molecular characterization of allergen-specific IgE antibodies have analyzed only few allergen-specific IgE antibodies (Andreasson et al., 2006; Edwards et al., 2002; Flicker et al., 2002, 2006; Niemi et al., 2007; Steinberger et al., 1996). To gain insight into the allergen-specific IgE IGHV repertoire and its development over several years of repeated allergen contact (i.e., secondary IgE response) we have established a model for allergen-specific IgE responses in man. We identified allergic subjects with IgE reactivity against one defined protein allergen, the major allergen of birch pollen, Bet v 1 (Breiteneder et al., 1989). The fact that birch pollen allergic subjects are exposed to antigen during defined periods of the year allowed us to study the IgE IGHV repertoire over several years and episodes of antigen contact. Our finding that highly similar IgE IGHV transcripts were used in different allergic subjects after four years of allergen exposure strongly suggests that allergens directly select and stimulate allergen-specific IgE production (i.e., secondary IgE responses) via activation of a limited and pre-defined set of IgE memory cells.
2. Methods
2.1. Pollen counts, characterization of allergic subjects
Five hundred allergic subjects were screened to identify six individuals with exclusive allergic sensitization to birch pollen using a multi-allergen test system (MAST CLA allergen-specific IgE assay, Hitachi Chemical Diagnostics) containing 46 allergen sources (Alder pollen, Almond, Alternaria, Apple, Aspergillus, Birch pollen, Carrot, Casein, Cat dander, Celery, Cladosporium, Cockroach, Codfish, Dermatophagoides farinae, Dermatophagoides pteronyssinus, Dog dander, Grass pollen mix, Guinea pig dander, Hamster dander, Hazel pollen, Hazelnut, Horse dander, Juniper, Latex, Milk protein, Mugwort pollen, Olive tree pollen, Parietaria pollen, Peach, Peanut, Penicillium, Pine mix, Plantain pollen, Plume mix, Potato, Rabbit, Ragweed pollen, Rye pollen, Rye flour, Sesame, Shrimp, Soy bean, Tomato, Walnut, Wheat flour, whole Egg). Blood samples from the six allergic subjects were obtained in spring and summer 2002 and 2005. At each appointment, Bet v 1-specific IgE levels were quantified in plasma by CAP-RAST measurements (Phadia, Uppsala, Sweden) and allergic symptoms and anti-allergic medication were recorded. None of the selected subjects had received any kind of allergen-specific immunotherapy. Birch pollen exposure in the individuals living area was recorded as described (Drachenberg et al., 2001).
2.2. HLA typing of the allergic subjects
HLA typing of allergic subjects was performed using a commercially available typing HLA kit based on HLA-sequence specific primers (PCR-SSP) according to the manuals provided by the manufacturer (Dynal AllSet+™ SSP DR “low resolution”; Invitrogen Corporation, Carlsbad, CA 92008, U.S.A.). Ambiguous HLA typing results where further clarified by direct DNA sequencing after HLA group-specific amplification with an automated DNA sequencer (ABI Prism 310 Genetic Analyzer; PE Applied Biosystems, Foster City, CA 94404, U.S.A.) as described elsewhere (McGinnis et al., 1997).
2.3. Allergen-specific IgE antibodies
To confirm that all IgE in the allergic subjects is directed exclusively against the major birch pollen allergen, Bet v 1, IgE inhibition experiments with recombinant Bet v 1 were performed. Recombinant Bet v 1, (Biomay, Vienna, Austria), was coupled to CNBr-activated sepharose 4B (GE Healthcare Bio-Sciences AB) in a concentration of five mg protein per ml medium according to the manufacturer's instructions. Samples of 1500 μl of plasma from each of the six allergic persons were incubated with 500 μl of allergen-coupled gel by end-over-end rotation overnight at 4 °C. Serum was recovered by centrifugation (4 °C, 5 min, 5000 × g). IgE levels against food allergen mix (egg white, milk protein, codfish, wheat flour, peanut and soy bean) and respiratory mix (mugwort, birch pollen, parietaria, timothy grass and ribwort) as well as IgE levels against birch pollen extract and rBet v 1 were determined before and after the depletion by CAP-RAST measurements (Phadia). In the IgE inhibition experiments three subjects (B, D, H) were identified who reacted exclusively with Bet v 1 in birch pollen.
2.4. PBMC isolation and RT-PCR amplification of IgE transcripts
Peripheral mononuclear cells were isolated by Ficoll density-gradient centrifugation at the time of serum collection. Total cellular RNA was isolated using the guanidine isothiocyanate method and CsCl gradient centrifugation (Eibensteiner et al., 2000). IgE transcripts were generated by the SuperScript™ One-Step RT-PCR with Platinum® Taq (Invitrogen) using VH1–VH6 family specific primers together with a primer specific for the first constant region of the IgE heavy chain (Eibensteiner et al., 2000) (Table 2). Primers used for the experiments in 2002 had accessory Eco RI restriction sites (Table 2 in parenthesis). The PCR amplification procedure consisted of an initial step reverse transcription of 30 min at 47 °C and 5 min at 94° followed by 40 cycles of 20 s 94 °C, 30 s 59 and 1 min 72 °C with final extension of 5 min at 72 °C. All PCR products were agarose gel purified using the Wizard® SV Gel and PCR Clean-Up System (Promega) according to the manufacturer's instructions. Subsequently cDNA was cloned into the AccepTor™ Vector (Novagen) and transformed into Escherichia coli XL1-blue. Plasmid DNA was purified from 3 ml overnight cultures containing 100 μg/ml ampicillin using Wizard® Plus SV Miniprep DNA Purification System (Promega) and digested with the restriction enzymes KpnI and SacI (Roche). Plasmids with inserts of the correct size were sequenced (Microsynth AG, Buchs, Switzerland). Non-redundant sequences are defined as non-identical sequences from one PCR reaction.
Table 2.
Oligonucleotides used for RT-PCR amplification of cDNAs coding for IgE variable regions. Oligonucleotides used for RT-PCR amplification of cDNAs coding for IgE variable regions. Names, specificities and sequences of the PCR primers are displayed. Eco RI restriction sites are underlined.
| Name | Specificity | Sequence 5′–3′ |
|---|---|---|
| VH1 | hu VH1 gene family | (GGA ATT) CAC TCC CAG GTG CAG CTG CTC GAG TCT GG |
| VH2 | hu VH2 gene family | (GGA ATT C)GT CCT GTC CCA GGT CAA CTT ACT CGA GTC TGG |
| VH3 | hu VH3 gene family | (GGA ATT C)GT CCA GGT GGA GGT GCA GCT GCT CGA GTC TGG |
| VH4 | hu VH4 gene family | (GGA ATT C)GT CCT GTC CCA GGT GCA GCT GCT CGA GTC GGG |
| VH5 | hu VH5 gene family | (GGA ATT C)GT CTG TGC CGA GGT GCA GCT GCT CGA GCT CGG |
| VH6 | hu VH6 gene family | (GGA ATT C)GT CCT GTC ACA GGT ACA GCT GCT CGA GTC AGG |
| IgEC1 | hu ɛ-chain first constant region | (GAG AGG AAT TC)G CTA CTA GTT TTG TTG TCG ACC CAG TCT GTG |
2.5. Sequence analysis of IgE transcripts
Analyses of immunoglobulin rearrangement, N-diversity and somatic mutations were performed using SoDA (Volpe et al., 2006) and IMGT/V-QUEST (Giudicelli et al., 2004) (Version 3.0.0). For VDJ germline fitting the consensus of IMGT/V-QUEST and SoDA analyses was used. Mutation frequencies are listed based on the IMGT/V-QUEST results, taking into account consistency with SoDA mutation rates. Identification of sequences with more than 98% identity to each other (Table 4) has been performed using Cd-hit (Li and Godzik, 2006).
Table 4.
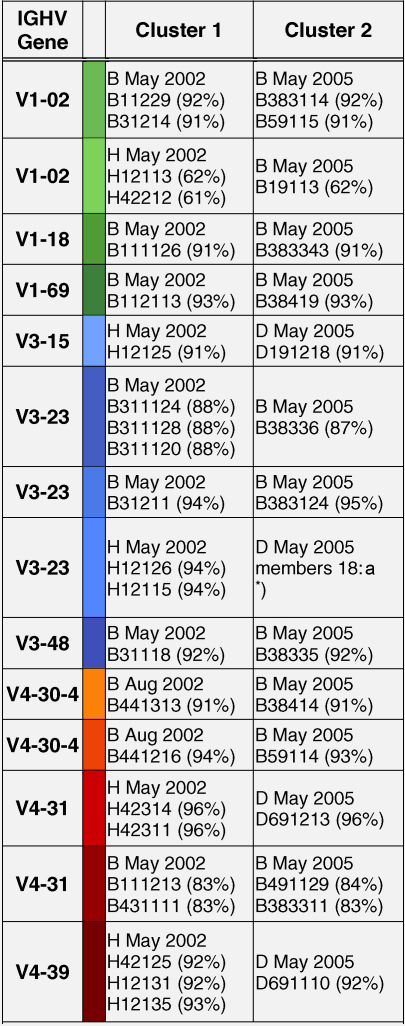
Identification of IgE sequences in different patients and different years (cluster 1 and cluster 2) with an identity of more than 98% to each other. The sequences have been grouped according to IGHV subgroups (IGHV1: green; IGHV3: blue; IGHV4: red) and the sequence identities with the corresponding germline sequences are displayed. Designations of IgE variable regions and the colour codes correspond to those in the Supplemental Figure 1. The first letter indicates the patient (B, D, H) and the time point of blood sampling is identified by the second number (1, 2, 3: May 2002; 4: August 2002; 8, 9: May 2005).
3. Results
3.1. Allergic patients with a defined IgE response against a protein allergen
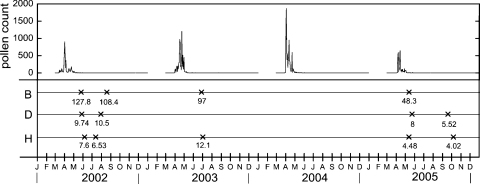
Most of the allergic subjects exhibit IgE reactivity against a plethora of different allergenic proteins. In order to identify allergic subjects with IgE reactivity against one defined allergen we screened allergic patients for IgE antibody responses against 46 allergen sources covering the allergen repertoire in Europe and identified subjects who were monosensitized to birch pollen. Using recombinant Bet v 1 we finally identified three genetically unrelated subjects B, D, H (B: HLA-A *02, *32; HLA-B *39, *51; HLA-Cw *07, *14; HLA-DR *03, *08; HLA-DQ *02, *04; D: HLA-A *01, *02; HLA-B *08, *51; HLA-Cw *07, *15; HLA-DR *03, *07; HLA-DQ *02; H: HLA-A *01, *30; HLA-B *13, *44; HLA-Cw *06, *07; HLA-DR *11, *13; HLA-DQ *03, *06) with exclusive IgE reactivity to Bet v 1. The three subjects suffered from respiratory allergy to birch pollen in spring. They had never received any form of allergen-specific immunotherapy. Their Bet v 1-specific immune response therefore must have been induced by natural allergen exposure. Pre-adsorption of their sera with rBet v 1 depleted completely IgE antibodies against Bet v 1 and birch pollen extracts and either completely depleted IgE antibodies specific for a respiratory allergen mix (subject H) or reduced IgE levels close to the cut off detection level (0.35 kUA/L) for subjects B and D (Table 1). No IgE antibodies were detected against a food allergen mix in any of the three individuals (B, D, H; Table 1). The subjects were studied over a period of four years (2002–2005) with seasonal pollen exposure. The registration of pollen counts during the full study in the living area of the subjects demonstrates that they have been exposed to birch pollen in the spring of each year with the pollen exposure being lowest in spring 2005 (Fig. 1). Bet v 1-specific IgE antibodies were detected in serum of the three individuals and symptoms of respiratory birch pollen allergy were recorded in the first two years and at the end of the study (2005).
Table 1.
IgE antibodies in the allergic subjects B, D and H are mainly directed to Bet v 1. Quantitative measurements of IgE levels (kUA/L) against inhalant allergens (sx1), food allergens (fx5), birch pollen, rBet v 1 and total IgE before (pre) and after (post) pre-adsorption of sera with rBet v 1 in the allergic subjects (B, D, H). The percentage inhibition of IgE achieved by pre-adsorption with rBet v 1 is displayed. The cut-off level for the detection of specific IgE is 0.35 kUA/L.
| B | D | H | |
|---|---|---|---|
| sx1-pre | 41.1 | 5.79 | 4.53 |
| sx1-post | 0.36 | 0.37 | 0 |
| Inhibition | 99.12% | 93.61% | 100% |
| fx5 pre | neg | neg | neg |
| fx5 post | neg | neg | neg |
| Inhibition | – | – | – |
| Birch pollen pre | 74.7 | 10.1 | 5.6 |
| Birch pollen post | 0 | 0 | 0 |
| Inhibition | 100% | 100% | 100% |
| rBet v 1 pre | 65.7 | 9.78 | 4.92 |
| rBet v 1 post | 0 | 0 | 0 |
| Inhibition | 100% | 100% | 100% |
Fig. 1.
Allergen exposure and allergen-specific IgE levels in the four years study period. Birch pollen exposure expressed as pollen counts per m3 of air (y-axis: pollen count) in the years 2002–2005 (x-axis: months are abbreviated). The time points when PBMCs were obtained from allergic subjects (B, D, H) for the characterization of IgE IGHV regions are indicated (×) and the IgE levels specific for the major birch pollen allergen, Bet v 1 are displayed in kUA/L.
3.2. Bet v 1-allergic subjects use a relatively limited and similar IgE-IGHV gene repertoire
From peripheral blood mononuclear cells of the three allergic subjects 296 IgE transcripts (2002: n = 121; 2003: n = 2; 2005: n = 173) were sequenced (B: n = 130; D: n = 136; H: n = 30). Among the 296 sequences, 152 non-redundant sequences (B: n = 99; D: n = 27; H: n = 26) were identified and compared with the germ line sequences deposited in the IMGT (Giudicelli et al., 2004) and SoDA (Volpe et al., 2006) database. Non-redundant sequences have been submitted to GenBank and accession numbers are displayed in the Supplemental list 1. Each of these sequences was derived from IgE-encoding transcripts due to the use of a 3′-primer located in the end of DNA coding for the first constant region of IgE. The presence of the IgE-encoding DNA in each of the sequences was verified by sequence comparison.
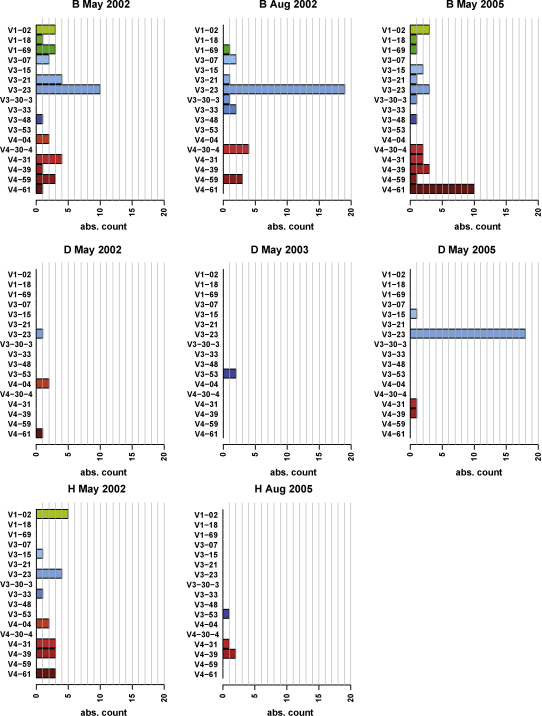
Fig. 2 displays the frequencies of the closest related V-genes for each subject and each point of time. The human germ line IGHV locus contains in principle 129 genes belonging to 7 subgroups of which 45 have been reported as functional genes (Pallares et al., 1999). We found that the three Bet v 1-allergic subjects used only 17 different IGHV genes belonging to three IGHV families (IGHV1, 3 and 4) (Fig. 2). From the 9 functional IGHV1 genes 3 were used. Eight of the 20 functional IGHV3 genes were utilized and 6 of the 10 IGHV4 genes. IGHV3-23 (n = 55) was the most frequently used IGHV gene among the sequenced clones followed by IGHV4-61 (n = 15), IGHV1-2 (n = 11) and IGHV4-31 (n = 11) (Fig. 2). Several IGHV genes were used by each of the three subjects. They included IGHV3-15, IGHV3-23, IGHV4-4, IGHV4-31, IGHV4-39 and IGHV4-61. The findings are therefore consistent with previous observations according to which IGHV3-23 is the most commonly used human V-gene (Brezinschek et al., 1997).
Fig. 2.
Frequency of the usage of V genes in the IgE repertoires of the allergic subjects over a four years period. Frequencies (x-axes: absolute numbers of non-redundant sequences) of V genes are displayed for allergic subjects (B, D, H) at different dates (top of the figures).
The human germ line IGHD locus comprises 27 (Lefranc, 2001) genes on chromosome 14 of which 19 were used by the three subjects. More than two-thirds of all D genes could be identified. IGHD 3-3 was the most frequently used D gene and was found in 20% of the Bet v 1 specific IgE sequences (data not shown). Regarding the D genes it has to be noted that the annotation of the transcripts to the closest germ line sequences was hampered by the high mutation rates in this part of the transcripts and their short length. From the six J genes described for the human IGHJ locus (Lefranc, 2001), five were used by the Bet v 1-allergic subjects (data not shown). J4 was by far the most frequently J gene used (n = 88) followed by J6 and J3. The results obtained for the usage of D and J genes were thus comparable to those obtained in a study analyzing the heavy chain repertoire of human peripheral B cells by single cell PCR (Brezinschek et al., 1995).
3.3. Usage of similar IgE-VH genes in Bet v 1-allergic subjects during four years of allergen exposure
Fig. 2 shows that most of the identified V-genes are used in 2002 and after three additional seasons of pollen exposure in the year 2005. In the case of subject B PBMC were analyzed at three points of time and all but two of the V genes were used at least twice in the observation period. IGHV3-23 was the most frequently IGHV gene used by subject D and found in 2002 and 2005. Likewise, subject H used certain IGHV genes in 2002 and 2005 (e.g., IGHV4-31, IGHV4-39). The variation of the number of clones obtained for each IGHV gene at different time points might be due to a repertoire shift or to the fact that the primers used in 2002 and 2005 differed regarding the presence of an additional Eco RI site which might have affected PCR amplification.
3.4. Varying degree of somatic mutations in IgE variable regions: some IgE variable regions are highly mutated whereas others contain no somatic mutations
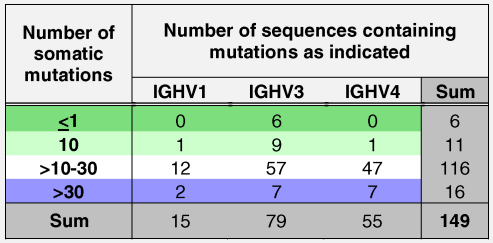
A comparison of the IgE variable DNA sequences with germ line sequences was performed in order to analyze the presence of somatic mutations in the FR1, FR2, FR3, CDR1 and CDR2. Since in the CDR3 somatic mutations cannot be unambiguously distinguished from junctional diversity, this part was not included in the analysis. The Supplemental Table 1 shows the detailed frequency and distribution of somatic mutations in each of the 152 IgE variable regions. Replacement and silent mutations are also shown there. Table 3 gives an overview how many variable regions of each IGHV family contained a certain number of somatic mutations in the FR1, FR2, FR3, CDR1 and CDR2 together. The majority of variable regions contained >10–30 somatic mutations (i.e., 116 out of 149 sequences). Variable regions with a number of somatic mutations between >10–40 were evenly distributed among the three IGHV families (IGHV1: 12/15; IGHV3: 57/77; IGHV4: 47/55). In each of the three IGHV families sequences containing more and less somatic mutations were detected. Interestingly, 6 sequences, which contained 1 or 0 mutations, were found in the IGHV3 family-derived sequences (Table 3). Silent as well as non-silent somatic mutations were evenly distributed over the CDR's as well as in the FR's in the analyzed sequences (Supplemental Table 1). Three sequences belonging to the IGHV1 family have been excluded from mutation analysis due to high differences in their length.
Table 3.
Number of sequences containing a certain number of somatic mutations displayed for the three IGHV families.
3.5. Highly similar IgE IGHV transcripts can be detected in genetically unrelated patients and in the same patient after several years
When we performed a multiple sequence alignment of all non-redundant IgE variable regions (Supplemental Figure 1) we found several variable region-encoding cDNAs with more than 98% identity in different patients and in the same patient after four years of allergen exposure. In patients D and H several highly similar IGHV sequences were retrieved, e.g. sequence D191218 IGHV3-15 that was isolated from patient D in May 2005 was highly similar to sequence H12125 (patient H) isolated in May 2002 (Table 4). Likewise, sequence H12115 (May 2002) and sequence D19111 (May 2005), D691110 isolated from patient D in May 2005 and H12131 (patient H) from May 2002 (Table 4) showed more than 98% identity.
Interestingly, variable region transcripts with more than 98% identity were also found to occur in the very same patient but at different points of time. For example, transcript B111126 (IGHV1-18) detected in patient B in May 2002 was isolated as identical sequence (B383343) in May 2005. Additionally some other sequences, e.g. B11229-IGVH1-2 May 2002; B31214-IGHV1-2 May 2002; B111216-IGHV4-31 May 2002; B441313-IGHV4-30-4 August 2002, which had been found to reach more than 98% identity (Table 4 and Supplemental Figure 1).
4. Discussion
We have analyzed the IgE variable sequences in allergic subjects who were sensitized against one defined allergen, the major allergen of birch pollen Bet v 1 (Breiteneder et al., 1989). Similar as for most other important respiratory allergens, IgE antibodies of allergic patients recognize exclusively conformational epitopes on the Bet v 1 allergens which require the intact three-dimensional structure of the protein for IgE recognition (Valenta and Kraft, 2001). The IgE response to Bet v 1 thus resembles the features of typical IgE responses to other important respiratory allergens from pollens, house dust mites, animal dander and moulds. Since allergic subjects mostly exhibit IgE antibody reactivity against several independent allergen molecules allergic patients were screened for “IgE mono-reactivity” to Bet v 1 and its cross-reactive allergens in order to establish a human system of allergic sensitization to one defined antigen. It can thus be assumed that most of the IgE variable sequences in these patients are specific for Bet v 1 but outliers cannot be excluded. The advantage of this human system over experimental animal systems is that it is based on natural sensitization to physiological doses (i.e., ng) of inhaled allergen, whereas most animal systems require injection of adsorbed artificial antigens or inhalation/contact with non-physiologically high doses of antigen in mostly inbred strains (Epstein, 2004). Although Bet v 1-monosensitized patients are rare in middle Europe, we do not think that the patients studied by us represented a unique subgroup of allergic patients. Similar as poly-sensitized patients our patients did not change their molecular reactivity profile over time. Their clinical symptoms to birch pollen were not different from those of poly-sensitized patients and each of them had a different genetic background. Interestingly, we found highly similar IgE variable regions in the three different patients. Moreover, these sequences reappeared unchanged in the same patients again after several years of pollen exposure. We interpret this finding as a molecular imprint of the antigenic signature of the Bet v 1 allergen in the human IgE repertoire. Interestingly, we found a rather limited usage of IGHV sequences in the allergic subjects which has been suggested also for grass pollen allergic patients (Andreasson et al., 2006). In eight blood samples taken over a period of four years only 152 different non-redundant VH sequences were identified among more than 296 sequenced PCR products. The non-redundant sequences belonged to three IGHV families (i.e., IGHV1, IGHV3, IGHV4) indicating that there is no bias towards the use of one certain IGHV family in the human IgE response (Bando et al., 2004; Davies and O’Hehir, 2004; Edwards et al., 2002; Eibensteiner et al., 2000; Janezic et al., 1998; Lim et al., 2007; Tilgner et al., 1997).
There is evidence that IgE antibodies bind much tighter to allergen than IgG antibodies and that they recognize epitopes which are either different from IgE epitopes or overlap only partially (Niemi et al., 2007). The latter would explain why allergen-IgE immune complexes could form even in the presence of higher amounts of allergen-specific IgG antibodies in allergic patients. It was therefore quite surprising to discover also a considerable number of IgE variable sequences containing few or almost no somatic mutations, which are thought to contribute to high affinity binding.
There is evidence that the secondary IgE response is strongly triggered by allergen contact but this response seems to be already independent of T cell help (Linhart et al., 2007; Niederberger et al., 2007).
The identification of identical and highly similar IgE variable sequences at different points of time (i.e., after several periods of allergen contact) may be interpreted in at least two ways. Either allergen contact reactivates a pool of IgE memory cells that then undergo development to IgE-secreting plasma cells. Consistent with this assumption it has been observed that allergen-contact strongly boosts allergen-specific IgE production in allergic subjects (Niederberger et al., 2007). It is also possible that an established pool of IgE-secreting plasma cells is the main source for continuous IgE production (Radbruch et al., 2006) but this hypothesis would not explain the strong boosts of IgE production which occur after allergen contact because plasma cells do not respond to antigen contact (Niederberger et al., 2007).
The fact that the allergen-specific IgE repertoire is rather limited opens interesting possibilities for therapeutic approaches targeting the allergen-specific IgE production. It is well established that the number of important disease-eliciting allergens is limited and most of the relevant allergens have been isolated and characterized down to their three-dimensional structure (Valenta, 2002; Valenta and Kraft, 2001). It should thus be possible to characterize the corresponding IgE variable sequences and to develop allergen mimicks, which target as anti-idiotypic reagents specifically IgE producing cells. Such a therapeutic approach would allow eliminating selectively those IgE producing cells that are responsible for allergic symptoms in patients and leave IgE specificities with potentially physiological roles untouched.
Conflict of interest
The authors declare that they have no conflicts of interest regarding the data presented in the paper. R.V. is a scientific consultant of Phadia, Uppsala, Sweden and Biomay, Vienna, Austria.
Acknowledgments
This study was supported by grant F1815 of the Austrian Science Fund and by the Christian Doppler Research Association, Vienna, Austria.
MN was supported by the Austrian Federal Ministry of Science and Research (GEN-AU Bioinformatics Integration Network).
The authors thank Meinrad Busslinger for critical reading of the manuscript and regarding advice in the interpretation of the results.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.molimm.2010.05.285.
Appendix A. Supplementary data
Multiple sequence alignment of all non-redundant IgE variable sequences (n = 152) grouped according to IGHV families (IGHV1, IGHV3, IGHV4). Variable sequences from each IGHV family exhibiting a more than 98% sequence identity have been grouped and coloured (IGHV1 genes: green colours; IGHV3 genes: blue colours; IGHV4: red colours).
References
- Andreasson U., Flicker S., Lindstedt M., Valenta R., Greiff L., Korsgren M., Borrebaeck C.A., Ohlin M. The human IgE-encoding transcriptome to assess antibody repertoires and repertoire evolution. J. Mol. Biol. 2006;362:212–227. doi: 10.1016/j.jmb.2006.06.062. [DOI] [PubMed] [Google Scholar]
- Bando Y., Shimizu A., Ra C. Characterization of VHepsilon gene expressed in PBL from children with atopic diseases: detection of homologous VH1-69 derived transcripts from three unrelated patients. Immunol. Lett. 2004;94:99–106. doi: 10.1016/j.imlet.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Blumer N., Herz U., Wegmann M., Renz H. Prenatal lipopolysaccharide-exposure prevents allergic sensitization and airway inflammation, but not airway responsiveness in a murine model of experimental asthma. Clin. Exp. Allergy. 2005;35:397–402. doi: 10.1111/j.1365-2222.2005.02184.x. [DOI] [PubMed] [Google Scholar]
- Breiteneder H., Pettenburger K., Bito A., Valenta R., Kraft D., Rumpold H., Scheiner O., Breitenbach M. The gene coding for the major birch pollen allergen Betv1, is highly homologous to a pea disease resistance response gene. EMBO J. 1989;8:1935–1938. doi: 10.1002/j.1460-2075.1989.tb03597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezinschek H.P., Brezinschek R.I., Lipsky P.E. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J. Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- Brezinschek H.P., Foster S.J., Brezinschek R.I., Dorner T., Domiati-Saad R., Lipsky P.E. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(−)/IgM+ B cells. J. Clin. Invest. 1997;99:2488–2501. doi: 10.1172/JCI119433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker H.A., Harries H.E., Banfield G.K., Carr V.A., Durham S.R., Chevretton E., Hobby P., Sutton B.J., Gould H.J. Biased use of VH5 IgE-positive B cells in the nasal mucosa in allergic rhinitis. J. Allergy Clin. Immunol. 2005;116:445–452. doi: 10.1016/j.jaci.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Davies J.M., O’Hehir R.E. VH gene usage in immunoglobulin E responses of seasonal rhinitis patients allergic to grass pollen is oligoclonal and antigen driven. Clin. Exp. Allergy. 2004;34:429–436. doi: 10.1111/j.1365-2222.2004.01900.x. [DOI] [PubMed] [Google Scholar]
- Drachenberg K.J., Wheeler A.W., Stuebner P., Horak F. A well-tolerated grass pollen-specific allergy vaccine containing a novel adjuvant, monophosphoryl lipid A, reduces allergic symptoms after only four preseasonal injections. Allergy. 2001;56:498–505. doi: 10.1034/j.1398-9995.2001.056006498.x. [DOI] [PubMed] [Google Scholar]
- Edwards M.R., Brouwer W., Choi C.H., Ruhno J., Ward R.L., Collins A.M. Analysis of IgE antibodies from a patient with atopic dermatitis: biased V gene usage and evidence for polyreactive IgE heavy chain complementarity-determining region 3. J. Immunol. 2002;168:6305–6313. doi: 10.4049/jimmunol.168.12.6305. [DOI] [PubMed] [Google Scholar]
- Eibensteiner P., Spitzauer S., Steinberger P., Kraft D., Valenta R. Immunoglobulin E antibodies of atopic individuals exhibit a broad usage of VH-gene families. Immunology. 2000;101:112–119. doi: 10.1046/j.1365-2567.2000.00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein M.M. Do mouse models of allergic asthma mimic clinical disease? Int. Arch. Allergy Immunol. 2004;133:84–100. doi: 10.1159/000076131. [DOI] [PubMed] [Google Scholar]
- Flicker S., Steinberger P., Ball T., Krauth M.T., Verdino P., Valent P., Almo S., Valenta R. Spatial clustering of the IgE epitopes on the major timothy grass pollen allergen Phl p 1: importance for allergenic activity. J. Allergy Clin. Immunol. 2006;117:1336–1343. doi: 10.1016/j.jaci.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Flicker S., Steinberger P., Norderhaug L., Sperr W.R., Majlesi Y., Valent P., Kraft D., Valenta R. Conversion of grass pollen allergen-specific human IgE into a protective IgG(1) antibody. Eur J. Immunol. 2002;32:2156–2162. doi: 10.1002/1521-4141(200208)32:8<2156::AID-IMMU2156>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Geha R.S., Jabara H.H., Brodeur S.R. The regulation of immunoglobulin E class-switch recombination. Nat. Rev. Immunol. 2003;3:721–732. doi: 10.1038/nri1181. [DOI] [PubMed] [Google Scholar]
- Giudicelli V., Chaume D., Lefranc M.P. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res. 2004;32:W435–W440. doi: 10.1093/nar/gkh412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounni A.S., Lamkhioued B., Ochiai K., Tanaka Y., Delaporte E., Capron A., Kinet J.P., Capron M. High-affinity IgE receptor on eosinophils is involved in defence against parasites. Nature. 1994;367:183–186. doi: 10.1038/367183a0. [DOI] [PubMed] [Google Scholar]
- Ishizaka K., Ishizaka T., Hornbrook M.M. Physicochemical properties of reaginic antibody. V. Correlation of reaginic activity with gamma-E-globulin antibody. J. Immunol. 1966;97:840–853. [PubMed] [Google Scholar]
- Janezic A., Chapman C.J., Snow R.E., Hourihane J.O., Warner J.O., Stevenson F.K. Immunogenetic analysis of the heavy chain variable regions of IgE from patients allergic to peanuts. J. Allergy Clin. Immunol. 1998;101:391–396. doi: 10.1016/S0091-6749(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Kraft S., Kinet J.P. New developments in FcepsilonRI regulation, function and inhibition. Nat. Rev. Immunol. 2007;7:365–378. doi: 10.1038/nri2072. [DOI] [PubMed] [Google Scholar]
- Lefranc M.P. Nomenclature of the human immunoglobulin heavy (IGH) genes. Exp. Clin. Immunogenet. 2001;18:100–116. doi: 10.1159/000049189. [DOI] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lim A., Luderschmidt S., Weidinger A., Schnopp C., Ring J., Hein R., Ollert M., Mempel M. The IgE repertoire in PBMCs of atopic patients is characterized by individual rearrangements without variable region of the heavy immunoglobulin chain bias. J. Allergy Clin. Immunol. 2007;120:696–706. doi: 10.1016/j.jaci.2007.05.035. [DOI] [PubMed] [Google Scholar]
- Linhart B., Bigenzahn S., Hartl A., Lupinek C., Thalhamer J., Valenta R., Wekerle T. Costimulation blockade inhibits allergic sensitization but does not affect established allergy in a murine model of grass pollen allergy. J. Immunol. 2007;178:3924–3931. doi: 10.4049/jimmunol.178.6.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis M.B.C., Chadwick R., Conrad M., Kronick M., Iovannisci D., Bellissimo D., Maurer D., Larsen N., Wu J., Williams T., Schaeffer V., Charron D., Van der Zwan A.W., Rozemuller E., Tilanus M.J.G. HLA sequencing based typing: multi-site validation of DRB1 region sequencing-based typing using the 12th International Histocompatibility Workshop panel. In: Charron D., editor. Genetic Diversity of HLA: Functional and Medical Implication. 1997. p. 238ff. [Google Scholar]
- Niederberger V., Ring J., Rakoski J., Jager S., Spitzauer S., Valent P., Horak F., Kundi M., Valenta R. Antigens drive memory IgE responses in human allergy via the nasal mucosa. Int. Arch. Allergy Immunol. 2007;142:133–144. doi: 10.1159/000096439. [DOI] [PubMed] [Google Scholar]
- Niemi M., Jylha S., Laukkanen M.L., Soderlund H., Makinen-Kiljunen S., Kallio J.M., Hakulinen N., Haahtela T., Takkinen K., Rouvinen J. Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen. Structure. 2007;15:1413–1421. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Pallares N., Lefebvre S., Contet V., Matsuda F., Lefranc M.P. The human immunoglobulin heavy variable genes. Exp. Clin. Immunogenet. 1999;16:36–60. doi: 10.1159/000019095. [DOI] [PubMed] [Google Scholar]
- Radbruch A., Muehlinghaus G., Luger E.O., Inamine A., Smith K.G., Dorner T., Hiepe F. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat. Rev. Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- Romagnani S. T-cell responses in allergy and asthma. Curr. Opin. Allergy Clin. Immunol. 2001;1:73–78. doi: 10.1097/01.all.0000010988.60715.c8. [DOI] [PubMed] [Google Scholar]
- Snow R.E., Chapman C.J., Frew A.J., Holgate S.T., Stevenson F.K. Analysis of Ig VH region genes encoding IgE antibodies in splenic B lymphocytes of a patient with asthma. J. Immunol. 1995;154:5576–5581. [PubMed] [Google Scholar]
- Steinberger P., Kraft D., Valenta R. Construction of a combinatorial IgE library from an allergic patient. Isolation and characterization of human IgE Fabs with specificity for the major timothy grass pollen allergen, Phl p 5. J. Biol. Chem. 1996;271:10967–10972. doi: 10.1074/jbc.271.18.10967. [DOI] [PubMed] [Google Scholar]
- Tilgner J., Golembowski S., Kersten B., Sterry W., Jahn S. VH genes expressed in peripheral blood IgE-producing B cells from patients with atopic dermatitis. Clin. Exp. Immunol. 1997;107:528–535. doi: 10.1046/j.1365-2249.1997.d01-960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traidl-Hoffmann C., Mariani V., Hochrein H., Karg K., Wagner H., Ring J., Mueller M.J., Jakob T., Behrendt H. Pollen-associated phytoprostanes inhibit dendritic cell interleukin-12 production and augment T helper type 2 cell polarization. J. Exp. Med. 2005;201:627–636. doi: 10.1084/jem.20041065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta R. The future of antigen-specific immunotherapy of allergy. Nat. Rev. Immunol. 2002;2:446–453. doi: 10.1038/nri824. [DOI] [PubMed] [Google Scholar]
- Valenta R., Kraft D. Recombinant allergen molecules: tools to study effector cell activation. Immunol. Rev. 2001;179:119–127. doi: 10.1034/j.1600-065x.2001.790112.x. [DOI] [PubMed] [Google Scholar]
- van der Stoep N., van der Linden J., Logtenberg T. Molecular evolution of the human immunoglobulin E response: high incidence of shared mutations and clonal relatedness among epsilon VH5 transcripts from three unrelated patients with atopic dermatitis. J. Exp. Med. 1993;177:99–107. doi: 10.1084/jem.177.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Neerven R.J., Knol E.F., Ejrnaes A., Wurtzen P.A. IgE-mediated allergen presentation and blocking antibodies: regulation of T-cell activation in allergy. Int. Arch. Allergy Immunol. 2006;141:119–129. doi: 10.1159/000094714. [DOI] [PubMed] [Google Scholar]
- Volpe J.M., Cowell L.G., Kepler T.B. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics. 2006;22:438–444. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of all non-redundant IgE variable sequences (n = 152) grouped according to IGHV families (IGHV1, IGHV3, IGHV4). Variable sequences from each IGHV family exhibiting a more than 98% sequence identity have been grouped and coloured (IGHV1 genes: green colours; IGHV3 genes: blue colours; IGHV4: red colours).