Abstract
Cross-cultural anthropologists have increasingly used phylogenetic methods to study cultural variation. Because cultural behaviours can be transmitted horizontally among socially defined groups, however, it is important to assess whether phylogeny-based methods—which were developed to study vertically transmitted traits among biological taxa—are appropriate for studying group-level cultural variation. Here, we describe a spatially explicit simulation model that can be used to generate data with known degrees of horizontal donation. We review previous results from this model showing that horizontal transmission increases the type I error rate of phylogenetically independent contrasts in studies of correlated evolution. These conclusions apply to cases in which two traits are transmitted as a pair, but horizontal transmission may be less problematic when traits are unlinked. We also use the simulation model to investigate whether measures of homology (the consistency index and the retention index) can detect horizontal transmission of cultural traits. Higher rates of evolutionary change have a stronger depressive impact on measures of homology than higher rates of horizontal transmission; thus, low consistency or retention indices are not necessarily indicative of ‘ethnogenesis’. Collectively, these studies demonstrate the importance of using simulations to assess the validity of methods in cross-cultural research.
Keywords: cultural traits, cross-cultural comparison, phylogeny, consistency index, correlated evolution, simulation study
1. Introduction
Human cultural traits exhibit a rich kaleidoscope of forms. Across societies, we exhibit variation in marriage systems, the types of shelters that protect us from the elements, the foods that we cook and how we cook them and decorations to our skin, bodies and clothing. Globally, linguists have identified over 6000 languages (Gordon 2005)—which is more than the 4500 or so extant mammalian species (Bininda-Emonds et al. 2007)—and humans are thought to practise more than 4300 religions (faith groups). Many human cultural traits are likely to be adaptive, such as those related to resource allocation and health practices, and are thus subject to natural selection (Mesoudi et al. 2004). Other cultural traits, such as decorations on pottery, are probably driven less by natural selection, but they may provide social or sexual benefits that indirectly translate to higher reproduction. As with many biological species, cultural diversity is disappearing at a rapid clip (Sutherland 2003), and many of the factors that influence biological diversity also influence cultural diversity (Pagel et al. 1991; Mace & Pagel 1995; Moore et al. 2002).
Anthropologists are interested in documenting differences in the configurations of human cultural traits, and in understanding how and why particular sets of traits arise. Comparison has long played a central role in this endeavour, with the first formalized approach to cross-cultural comparison developed by Tylor (1889). Tylor was interested in developing systematic approaches to investigate cross-cultural variation, including correlations—or what Tylor called ‘adhesions’—among different traits. He was criticized for this approach on statistical grounds by Francis Galton (Naroll 1961), yet his work spawned a number of followers who developed an empirically and theoretically rich approach that flourishes to this day (Murdock & White 1969; Burton & White 1984; Borgerhoff Mulder et al. 2001; Pagel & Mace 2004; Mace & Holden 2005; Mace et al. 2005; Lipo et al. 2006).
Systematic comparisons are used, for example, to test hypotheses for why some cultures cook with more spices than others (Billing & Sherman 1998), or why some pass wealth to sons and others to daughters (Hartung 1982). As in biology (Harvey & Pagel 1991; Nunn & Barton 2001), however, a critical issue in all of these studies involves the non-independence of data points when conducting comparative tests (Mace & Pagel 1994; Mace & Holden 2005). Indeed, this is precisely the criticism levelled by Galton, and thus known as ‘Galton's problem’ among anthropologists. The essence of the problem can be summarized as follows. Because populations tend to share traits through descent from a common ancestor, the data points that make up a cross-cultural analysis will lack statistical independence. Phylogeny-based methods provide a means to deal with non-independence that arises through ‘vertical’ transmission of traits from ancestral to descendent populations (Mace & Pagel 1994; Mace & Holden 2005; Nunn et al. 2006).
Phylogenetic methods assume that cultural traits behave like genetic traits, particularly with regard to the prominence of vertical trait transmission from parent generations to later generations. In contrast to the genetic transmission of most elements in biology, however, cultural traits can spread ‘horizontally’ among unrelated individuals. While ‘vertical’ and ‘horizontal’ most aptly refer to transmission of traits between individuals (whether individuals learn from their parents or other community members), a similar distinction can be made at the population level, namely whether cultures develop by a tree-like splitting process (phylogenesis) or by admixture (ethnogenesis). Thus, under phylogenesis, cultural change results from the transmission of ideas and practices from parental to daughter cultural taxa (White et al. 1981; Moore & Romney 1994; Holden 2002; Tehrani & Collard 2002). Under ethnogenesis, cultural evolution occurs through the borrowing and blending of ideas and practices and the trade of objects among contemporary societies (e.g. Terrell et al. 1997), resulting in a weak phylogenetic signal (e.g. Hurles et al. 2003; Jordan & Shennan 2003; Moylan et al. 2006).
Ethnogenesis creates its own form of non-independence, and a form that requires different approaches from those used for investigating phylogenesis (Borgerhoff Mulder et al. 2006; Nunn et al. 2006). This is not because borrowed traits fail to shed light on adaptation; indeed if pastoralists in arid environments ‘borrow’ camel-keeping from a neighbouring ethnic group this may be valid evidence for adaptive cultural coevolution (Mace & Pagel 1994). Nor is this because cultural traits within a particular domain are not transmitted vertically; indeed, some domains may exhibit huge conservatism, as with the consistency of kinship traits across language families (Jones 2003). Rather, because of ethnogenesis, the true trees of different domains may be different. Correcting for shared language (as is typically done in the application of phylogenetic methods to human cultures) may not be useful to control for shared origins in a domain that is not structured by language.
At this time, however, we lack a general framework for grappling with the most fundamental aspects of this problem (Borgerhoff Mulder et al. 2006). In particular, we need to address the following three questions: (i) Does horizontal transmission between societies produce a signal that is distinct from vertical transmission in comparative data? (ii) Which methods offer the most powerful means to detect horizontal transmission (or ethnogenesis)? (iii) What methods are most appropriate for investigating correlations between traits when both horizontal and vertical transmission occur? This latter question is particularly important, as it forms the basis for applying the comparative method to test adaptive hypotheses.
Some efforts have been made to address these questions, particularly with regard to methods developed from within anthropology (Dow 1984, 1993, 2007) and from biology (Borgerhoff Mulder et al. 2001; Mace & Holden 2005; Fortunato et al. 2006). But we also need to evaluate the different methods, which raises a fourth question: (iv) How can we evaluate the strengths and weaknesses of different methods when we usually lack information about the actual patterns of vertical and horizontal transmission in real-world data?
An answer to this last question again comes from comparative biology. Biologists typically investigate the appropriateness of a new comparative method by taking a phylogeny, simulating the evolution of traits down the tree and calculating statistical tests on these data (e.g. Martins & Garland 1991; Purvis et al. 1994; Nunn 1995; Diaz-Uriarte & Garland 1996; Harvey & Rambaut 2000). The statistical test might measure the correlation between traits, the degree to which more closely related species share similar trait values, measures of tree ‘balance’ or any other statistical or phylogenetic measure of interest. By generating many such artificial datasets with known characteristics—such as the model of evolution or the degree to which the traits are correlated—it becomes possible to evaluate the statistical properties of a particular method. Hence, these methods have played a pivotal role in assessing phylogeny-based comparative methods and making decisions about when and how they should be applied to biological data.
This paper has three major sections. First, we introduce a simulation model that was developed to assess methods for cross-cultural research (Nunn et al. 2006). Second, we review one set of results from the model involving the appropriateness of independent contrasts under conditions of horizontal transmission. Lastly, we apply the simulation model to a new question in which we evaluate whether two commonly used phylogenetic metrics of ‘treeness’—namely, the consistency index (CI) and the retention index (RI)—detect phylogenesis in cross-cultural datasets. These indices have been used in cross-cultural research (e.g. Tehrani & Collard 2002; Collard et al. 2006). Our simulations show that factors other than horizontal trait transmission are significant determinants of the CI and RI. The simulations also reveal that using external information on the historical relationships among societies (e.g. language; Mace & Pagel 1994) can improve the performance of tree-consistency measures in cross-cultural research.
2. The simulation model
(a). The basic framework
The purpose of a simulation model in phylogenetics research is to generate artificial datasets under known models of trait evolution, and then to assess how well a particular method can recover the parameters of the evolutionary model. To evaluate comparative methods in the context of cross-cultural comparisons, Nunn et al. (2006) developed a spatially explicit simulation approach to investigate trait evolution in relation to phylogeny and geography. Phylogeny in this case refers to the historical relationships among societies, such as a branching pattern indicated with a linguistic tree (e.g. Gray & Jordan 2000; Gray & Atkinson 2003), while geography is represented as a matrix of geographical distances among societies.
Nunn et al.'s (2006) simulation approach is derived from previous simulation protocols that have been used to test phylogenetic comparative methods in biology (e.g. Martins & Garland 1991; Purvis et al. 1994; Nunn 1995; Diaz-Uriarte & Garland 1996; Harvey & Rambaut 2000). Nunn et al. (2006) augmented this basic procedure with a spatial context that also allows for horizontal transmission of the simulated traits among neighbouring societies. Moreover, including spatial context required a procedure to generate links between geography and phylogeny, which is important because these two factors will tend to covary in real-world datasets—i.e. closely related societies are often in close spatial proximity. To achieve this, they developed an adaptive radiation model (e.g. Price 1997; Harvey & Rambaut 2000) in which mother populations produce daughter populations that tend to settle in nearby open niches, as described below.
Nunn et al. (2006) constructed their spatially explicit model of cultural trait evolution using the computer package Matlab (v. 6.5, Mathworks, Inc.). The model can be viewed as a metapopulation represented as a two-dimensional lattice (or matrix) with a ‘hard’ edge, such that societies on the edge of the lattice are not connected to societies on the opposite edge. Thus, the model assumes a geographically delimited area such as an island, a continent or an area bounded by impassable mountain ranges or bodies of water. The model has non-overlapping generations (discrete time) and each cell of the lattice is treated as a distinct society (discrete space).
The model examines the evolution of one or more traits, represented as X1, X2, … , Xn. These traits can be continuously varying—such as body mass—or they can be discrete—such as categorizations of the mating system. The user must also make assumptions about how traits transfer when they are horizontally donated. Traits can transfer as a group, which might be expected if the traits are functionally linked (and thus show correlated evolution). At the other extreme, traits can transfer independently. An intermediate position is also possible, with sets of traits moving stochastically as a function of the correlation between them (i.e. ‘stochastic yoking’; Nunn et al. 2006). In all cases, when trait X1 moves from society A to society B, the value of X1 in B is replaced by the value of X1 in A. As in most applications of the comparative method to real anthropological data, societies are assumed to exhibit no intra-societal variation; this is an assumption that could be relaxed in future research using the simulation model. Vertical transmission occurs by default when descendent societies inherit trait values of their ancestors across generations, including through speciation events.
(b). Initializing and running the model along discrete time steps
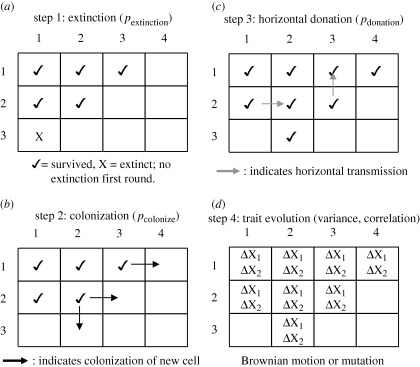
Figure 1 provides an overview of the simulation processes part way through a simulation run on a small 3 × 4 matrix. The parameters used in the simulation are summarized in table 1. The simulation begins with an empty matrix and a single society on the left-most column in the middle row of the matrix (e.g. row = 2, column = 1 in figure 1). Extinction, colonization of empty cells, horizontal trait donation and trait evolution occur sequentially and stochastically in discrete generations (i.e. time steps). Thus, in each generation, the following events can occur for each society, in the order described below.
Figure 1.
Simulation procedure. A simplified version of the simulation procedure using a 3-row by 4-column spatial matrix partway through a simulation run. The following stochastic processes occur in sequence for each generation in the simulation: (a) step 1: extinction of filled cells; (b) step 2: colonization of empty cells; (c) step 3: horizontal transmission among neighbouring societies; and (d) step 4: trait evolution. Empty cells indicate unfilled niches in the spatial model.
Table 1.
Parameters that can be varied in the simulation model.
| parameters | description |
|---|---|
| R, C | number of rows and number of columns in spatial matrix |
| pextinction | probability that a society goes extinct per time step |
| pdonation | probability that a society donates a trait to an adjacent society per time step |
| pcolonize | probability that a society colonizes an adjacent open cell per time step |
| r | correlation between continuously varying traits |
| N | number of traits simulated |
| pevolution | rate of evolutionary change (variance of trait change for continuous traits, mutation rate for discrete traits) |
Step 1: Extinction. Societies go extinct based on a user-defined probability of extinction (pextinction). Higher extinction rates increase the number of empty cells, and thus provide new niches for neighbouring societies to fill; this is important because it affects the degree to which diversification events occur close to the tips of the tree (see Nee et al. 1994; Nunn et al. 2006). To avoid the possibility of an entirely empty matrix, the probability of extinction is set to zero when only one cell of a matrix was occupied, including the first generation of a simulation run.
Step 2: Colonization. Neighbouring cells available for colonization are identified as empty cells on the ‘flat’ sides of a given society's cell (rather than cells attached by their corners). Thus, a society may possess a maximum of four neighbours, with societies on the edges of the matrix having fewer neighbours. Societies that colonize adjacent cells are treated as distinct societies in the next generation. Societies colonize neighbouring cells with probability pcolonize, and as this occurs, the evolutionary relationships are recorded as a bifurcating tree. The program updates branch lengths by one unit in each generation of the simulation. When a society colonized more than one cell in a generation, the relationship among societies was randomly resolved with short branch lengths (=0.001).
Step 3: Horizontal trait donation. Trait values spread among neighbours based on a user-defined probability that a society donates traits to one of its neighbours (pdonation). The values of traits in the recipient are replaced by values from the donor. Nunn et al. (2006) focused on results with traits transferred as a pair during horizontal transmission (i.e. if X1 moves, so does X2), applying the probability of trait donation to the paired movement of traits rather than independently for each trait (an issue that is discussed below and by Currie et al. 2010). For a society with more than one neighbour, more than one horizontal donation could take place in a single generation. To deal with this possibility, the probability that at least one donation event takes place for a potential recipient society was calculated using the binomial theorem based on the number of neighbours. If the condition for trait donation was met for a recipient society, the society donating the trait was randomly assigned from among recipient's neighbours. This means that the rate of horizontal transmission increases with the number of neighbours (as expected in real-world data) and that the spatial configuration of societies will impact the overall rate of horizontal transmission (e.g. by influencing the number of societies on edges of the matrix, as these will have fewer neighbours). Transfers of traits among all societies in the matrix were implemented simultaneously in a given generation after all cells were examined for possible trait transfer.
Step 4: Trait evolution. Evolutionary change in the traits occurs at the end of each generation. For continuously varying traits, one common model of trait evolution involves Brownian motion (Felsenstein 1985), with the user identifying variance in trait change per generation and the degree of correlation between characters. These changes are calculated for each society and added to existing trait values (indicated by ‘ΔX’ in figure 1). For discretely varying traits with two states, trait change can be modelled as a mutation rate that reflects the probability of switching between states at each time step. When simulating discrete traits, we assume that the probability of gains (0 to 1) equals the probability of losses (1 to 0).
(c). Output from a simulation run: the ‘true’ tree
When constructing a simulation program, the user is essentially playing ‘god’, and this allows him or her to decide what data to collect. Along with data on the traits, their geographical distribution and the simulation parameters, the simulation program also retains the history of splitting by the societies. This reflects the true phylogeny in the sense of the actual splitting of lineages and their times to last common ancestors. While we acknowledge that the true phylogeny is never known for real data, the true tree from the model can be viewed as analogous to a tree that is generated from independent data, such as linguistic data (Mace & Pagel 1994; Gray & Atkinson 2003; Gray et al. 2010). We will see that having independent information on historical relationships can often provide deeper insights into cultural transmission and evolution.
3. Horizontal transmission and correlated evolution
(a). Results
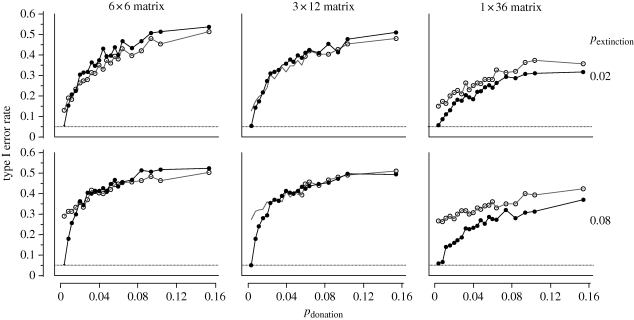
Nunn et al. (2006) used the simulation framework to investigate the effects of horizontal transmission on phylogenetically independent contrasts, which is a method that can be used to study whether two or more traits show correlated evolution (Felsenstein 1985; Harvey & Pagel 1991). Briefly, independent contrasts are calculated as differences in trait values among lineages that share a most recent common ancestor that are standardized for evolutionary time. Nunn et al. (2006) investigated whether an increasing probability of horizontal transmission reduces the statistical performance of independent contrasts, focusing on type I errors (incorrectly rejecting true null hypotheses of no association between traits). The authors thus simulated a wide range of values for the probability of trait donation (pdonation) to neighbours (in addition to other parameters, see Nunn et al. 2006). Most of the simulations were concentrated within a range of donation probabilities from 0 to 0.06. By simulating evolution with pdonation = 0, only vertical transmission occurred; thus, under this parameter setting, phylogeny-based methods were expected to produce results that match previous simulation studies that investigated the statistical performance of independent contrasts for biological traits (Martins & Garland 1991; Purvis et al. 1994). At the highest probability of donation (pdonation = 0.15) and four neighbours, the actual probability of receiving a trait from one or more neighbours in a given generation equalled 0.48 (based on the binomial theorem, see above). Such high rates are unlikely in most real-world datasets, but are not unreasonable, as suggested by examples such as the rapid spread of horses among New World peoples (Roe 1955), or the movement of Islam across many parts of west (Trimmingham 1970) and east Africa (Ensminger 1997). Simulations were run for 60 generations, and analyses were conducted only on full matrices of 36 societies (see Nunn et al. 2006 for details).
When considering traits that were entirely uncorrelated, Nunn et al. (2006) found that type I error rates increased with increasing probability of horizontal transmission (figure 2). It is often useful to compare methods that incorporate phylogenetic information with analyses that do not take this information into account. This comparison revealed that non-phylogenetic tests have higher type I errors than when controlling for phylogeny using independent contrasts, especially at low levels of horizontal transmission. In some simulations, type I errors for non-phylogenetic tests were lower than for analyses based on independent contrasts, but error rates were still extremely high (figure 2).
Figure 2.
Horizontal transmission and independent contrasts. Horizontal dotted line indicates expected type I error rate (α = 0.05). Plots show how the probability of trait donation (pdonation) affects type I error rates for independent contrasts (black circles) and in non-phylogenetic analyses (circles) for pextinction = 0.02 (top) and 0.08 (bottom row). There was no correlation between trait changes in these simulations of continuously varying characters (r = 0). Different plots reflect different combinations of spatial configuration (i.e. matrix dimensions) and probability of extinction (pextinction), and values plotted reflect the proportion of results in a given simulation run (n = 1000) in which a significant association was found between traits X and Y. Simulations were run with colonization probability (pcolonize) of 0.96 and a trait change variance of 0.02.
(b). Application and further considerations
Simulation results are dependent on the assumptions that were made in simulating and analysing the data; thus, it is important to critically evaluate these assumptions. Of particular importance in this regard, Nunn et al. (2006) assumed that traits are donated as a pair. This is a useful starting point, as it is analogous to the paired transmission of traits during vertical transmission and it means that both traits have the same evolutionary and geographical history. Currie et al. (2010) used results based on a different assumption, namely that traits are transmitted independently during donation events, which is also a reasonable assumption for many human cultural traits. In contrast to the patterns in figure 2, they found that type I error rates are not elevated by horizontal transmission. Thus, independent contrasts is a valid method when the traits are transmitted vertically or, for situations in which horizontal transmission occurs, when we have reason to believe that the traits are transmitted independently. The independence of traits will need to be determined on a case-by-case basis.
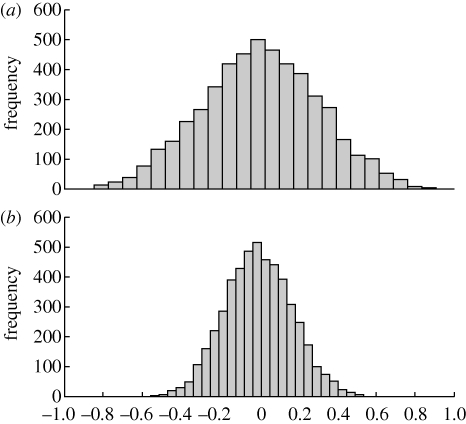
As to why the higher type I error rates emerge under paired transmission of traits, it could be due to one of two factors. First, the paired transmission could artificially create a correlation where none exists simply by the maintenance of traits with similar values across data points. This involves biased estimates of the correlation coefficient, and we would expect the correlation between the traits from the simulation to be centred above zero. Second, it could be that violations of the assumptions of independent contrasts, including an incorrect topology and model of evolution, create distributions of statistics that are too wide, as found in previous phylogenetic simulations (Martins & Garland 1991; Symonds 2002). In fact, we have good reason to believe that the assumptions of independent contrasts are violated with regard to the tree topology. More specifically, horizontal transmission under paired transmission results in a tree for the traits that differs from the true tree of societal splitting; yet the contrasts are calculated on the true tree. Consistent with this expectation, we find that paired horizontal transmission does not create bias, but does alter the distribution of statistics from that expected (figure 3; F4999,4999 = 3.02, p < 0.0001). Thus, it is the underlying assumption violations about the tree that cause the elevation in type I error rates, rather than bias originating from the design of the simulation model.
Figure 3.
Causes of higher type I error rates. Distribution of correlation coefficients statistics for (a) paired and (b) unpaired transmission of continuously varying traits across 5000 simulations. Paired transmission of traits during horizontal transmission events does not result in biased parameter estimates (mean value for paired transmission = 0.0064, versus −0.002 for unpaired transmission). Instead, the elevated type I error rates for paired compared with unpaired trait transmission result from a wider expected distribution of statistics (s.d. of 0.2996 and 0.1724 for paired and unpaired transmission, respectively). Results are from a 6 × 6 matrix with probability of extinction of 0.08, horizontal donation of 0.01, no correlation among the traits, variance of trait change of 0.02 and 60 generations.
4. Inferring vertical transmission based on levels of homology
Given the sensitivity of independent contrasts to some forms of horizontal transmission, it is critical to identify whether the degree of vertical transmission in cultural datasets is sufficient for applying phylogenetic methods. In this section, we use the simulation model to assess whether methods designed to detect homology in biological datasets can be used to assess the degree of vertical transmission in cultural data. Importantly, for what follows, the tree topology might be the most parsimonious tree for the cultural data matrix in question (hereafter called the ‘parsimony’ tree), or it could be a tree based upon separately analysed genetic or linguistic data (hereafter called the true tree, subject to the caveats given above). For a simulation study of Mantel tests to quantify vertical and horizontal signal, see Nunn et al. (2006).
(a). Two measures of homology
Episodes of independent evolutionary change (also known as evolutionary convergence or homoplasy) are expected to reduce phylogenetic signal in a dataset. Horizontal transmission is also expected to reduce the phylogenetic signal in the data, effectively resulting in more homoplasy. The CI and the RI are two statistics used in analyses of discrete biological data to assess phylogenetic signal. Both can be calculated for individual characters on a given tree, or as used here, an ‘ensemble’ metric calculated across multiple characters. The CI is measured as m/s, with m being the minimum number of parsimony steps possible for a given character on a tree completely congruent with it, and s being the minimum steps required on a given tree topology (Kluge & Farris 1969; Naylor & Kraus 1995). Thus, for a two-state character, m = 1; for a three-state character, m = 2 and so on. Thus, as the level of homoplasy increases for a character or character set, s goes up but m remains constant, and the CI is reduced. In principle, a CI of 1.0 reflects complete character-state homology on a tree, while a CI of 0.0 indicates a total absence of homology; in reality, however, the CI has an effective lower bound of about 0.38 (Archie 1990). CI values are not a simple reflection of character evolution alone, however, as they are known to decrease dramatically as a function of the number of taxa (Archie 1990). Additionally, the ensemble CI decreases, though less extremely, as character number increases (Archie 1989, 1990; Sanderson & Donoghue 1989). The RI is measured as (g − s)/(g − m). The parameters s and m in this equation are the same as in the CI calculation, while g indicates the highest number of parsimony steps that a character could possibly exhibit on any tree for the number of taxa in question (Farris 1989; Naylor & Kraus 1995). The RI in principle ranges from 0 to 1, with 1 indicating that all state distributions are homologous. As compared with the CI, the RI is substantially less sensitive to the number of taxa studied (Archie 1990).
The logic underlying the application of the RI and CI to cultural data is that if cultural traits are consistently transmitted from parent to daughter populations, we should find that cultural traits produce good trees with high levels of homology. Thus, a high RI or CI is taken as consistent with some degree of vertical transmission, which in turn is interpreted to indicate the preponderance of phylogenesis over ethnogenesis in the data.
These statistics are among the most commonly used in anthropology to infer the degree of vertical transmission in cultural datasets. For example, Collard et al. (2006) calculated the RI for 21 biological datasets and 20 cultural datasets (to which they added one additional published RI statistic, giving 21 cultural datasets). They reasoned that if a bifurcating tree model can account for cultural data, the RIs of cultural data should be similar to those of biological data, and this is exactly what they found: the average RI of the biological data was 0.61, while the average RI of the cultural data was 0.59. In another study that used the CI, Tehrani & Collard (2002) tested whether horizontal or vertical descent best describes decorative characteristics of Turkmen textiles. Their primary goal was to test the assertion that ethnogenesis is the predominant transmission mode for human cultural traits (e.g. Moore 1994). Using design characters from the period before Russian domination of the Turkmen, the CI was calculated at 0.68, suggesting that most of the characters are shared through common descent. Relatively similar patterns (with perhaps slightly more ethnogenesis) were found in the period following conquest of Central Asia by Tsarist Russians (Tehrani & Collard 2002). For additional examples, see Jordan & Shennan (2003) and Lycett et al. (2007).
(b). Concerns about using the consistency index and retention index with cultural data
How well do the CI and RI detect vertical and horizontal transmission, and can they provide definitive evidence for either? Several important issues concerning the use of the CI and RI have yet to be resolved. First, these statistics were designed to assess the degree of homology in biological data, and increases in rates of evolution can lead to lower homology, i.e. a lower CI or RI. We might thus expect that traits will show less tree-like signal when the rate of evolution is high, even if no horizontal transmission occurs. This means that low values of the CI and RI do not necessarily indicate horizontal transmission; they could instead indicate that evolutionary rates are high.
Second, if a large number of traits in the analysis are borrowed as a cultural ‘package’ (Boyd et al. 1997; Jones 2003), then parsimony inference will tend to produce a tree with low homoplasy. Such traits will tend to show a strong tree-like structure, albeit one that differs from the history of other cultural or genetic traits and despite possibly extensive borrowing. Indeed, some authors (e.g. O'Brien et al. 2001) have pointed out that cultural packages would still have a true tree-like structure to themselves that is not eroded by diffusion and cultural ‘hybridization’. If we have an independently derived tree, it becomes possible to evaluate the CI and RI for the traits on this true tree, and this should help to discern instances of borrowing.
Lastly, both statistics lack a firm statistical framework for deciding on the statistical importance of a particular value. For example, is a CI of 0.5 for a set of traits significantly higher or lower than expected, and does it constitute evidence for horizontal or vertical transmission? All that can be done is to compare values with those obtained from biological traits or systems that have better understood properties (Collard et al. 2006).
In summary, we can imagine scenarios where a low CI occurs in the absence of horizontal transmission, and other scenarios where a high CI occurs with high levels of horizontal transmission. Taking the low CI first, this could simply reflect homoplasy, i.e. the independent innovation of cultural traits, rather than horizontal transmission, owing to high rates of cultural evolution. A high CI might reflect lower rates of evolution, for example if the number of societies chosen is small and relatively homogeneous with respect to the traits in question. Moreover, horizontal transmission can produce a high CI on a tree inferred from traits that are borrowed as a package (Borgerhoff Mulder et al. 2006); in such a case, the phylogenetic signal will remain, but the true tree will differ from the tree that is reconstructed from cultural traits. Lastly, even if we can overcome these problems, how should we decide whether a given value is statistically significant, and should it be compared with the null hypothesis of maximal homoplasy or perfect homology?
(c). Applying the simulation model to study the consistency index and retention index
In light of these concerns, we altered the simulation model to test whether the RI and CI can detect horizontal transmission (Matlab 7.0, Mathworks, Inc.). We also investigated other variables that might influence the calculation of the CI and RI (table 2). Thus, in addition to rates of trait donation to neighbours, we included rates of evolution, as a higher rate of evolution should increase homoplasy in the data (thus reducing the CI and RI). Similarly, we examined variation in the rate of extinction, the number of societies, the number of traits and the rate of colonization into empty niches. In all of these simulations, the discretely varying traits underwent independent horizontal transmission (i.e. unpaired transmission).
Table 2.
Parameter ranges investigated in CI and RI simulations. (n.a., not applicable.)
| parameter | parameter values (range) |
|---|---|
| maximum number of societiesa | 36–100 societies, arranged as square lattices (i.e. number of rows = number of columns) |
| pextinction | 0.001–0.1 |
| pdonation | 0.001–0.1 |
| pcolonize | 0.1–0.95 |
| r | n.a. |
| N | 100–1000 |
| pevolution | 0.001–0.1 |
aThis is a maximum because some cells may be empty at the end of a simulation. The actual number of taxa ranged from 21 to 100.
In terms of output, we focused on four measures. The first two represent the CI and RI as typically calculated in cross-cultural research. Specifically, we used the trait data from the simulation model to generate phylogenies using parsimony. For this, we modified the simulation program so that it produced NEXUS files (Maddison et al. 1997), which we then analysed in PAUP* v. 4.0 Beta 10 (Swofford 2003). For each NEXUS file, we excluded parsimony uninformative characters and performed a heuristic search. We then selected at random one of the most parsimonious trees, and we calculated the CI and RI on this tree; these were ensemble statistics, calculated across the entire dataset from a simulation run. For the other two output measures, we calculated the CI and RI on the tree recorded from the simulation (i.e. the true tree). We expected that this might improve our ability to detect the signal of horizontal transmission, which should create differences between the parsimony tree and the true tree recorded in the simulation.
We faced two challenges in generating and analysing the data: the first involved effectively exploring the six-dimensional space of parameters that we varied, and the second involved analysing the output in a way that can account for possible interactions among variables. To explore parameter space, we used Latin hypercube sampling, which is a type of stratified Monte Carlo sampling that has been used in epidemiological modelling and is more efficient in this context than random sampling regimes (Seaholm et al. 1988; Blower & Dowlatabadi 1994; Rushton et al. 2000). Our Latin hypercube sample drew values from the range of values in table 2 10 times with 500 samples in each set, giving a total of 5000 simulation runs. To analyse the output, we used regression trees (De'ath & Fabricius 2000). This statistical method produces graphical output in the form of a decision tree that predicts the outcome of a simulation with particular parameter values. Methodologically, it repeatedly splits the data into homogeneous groups according to the six parameters used in our simulation. The advantages of regression tree analysis in the context of analysing simulation output are many, including its ability to deal with nonlinear effects, higher order interactions and ease of interpreting the graphical output. In addition, it does not rely on p-values, which can be highly significant with the large sample sizes used here, yet give little explanatory value. Regression trees were calculated using the Statistics Toolbox in Matlab. We split impure nodes when the number of observations for that node was 1000. After creating an initial tree using the simulation output, we used 10-fold cross-validation to identify the pruning level with the minimal cost (De'ath & Fabricius 2000), identified as the tree with the minimum error rate. Using this pruned tree, we calculated the percentage of variance explained by comparing predicted and observed values.
(d). Results
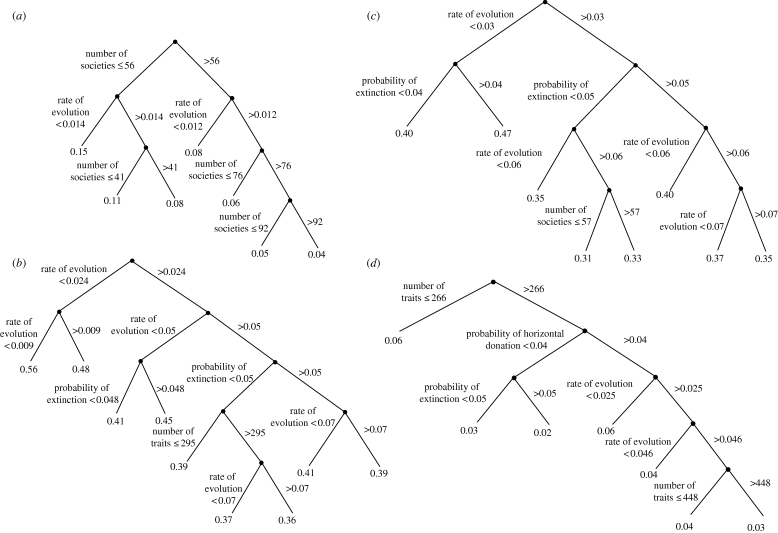
We first examined the regression trees for the CI and RI calculated from the simulated data. As shown in figure 4a, the primary predictor of the CI was the number of societies, with increasing number of societies leading to a lower CI (Archie 1990). The rate of evolution occurs at the second level in the regression tree, with higher rates of evolution leading to a lower CI. The model explained 83 per cent of the variation in CI scores, and horizontal transmission was not included in the regression tree. Figure 4b shows the regression tree for the RI, which is thought to be less sensitive to the number of taxa. In this case, the rate of evolution was the primary factor influencing the RI, with a higher rate of evolution leading to a lower RI (i.e. more homoplasy). The model accounted for 77 per cent of the variation in RI scores, and horizontal transmission was not included in the regression tree.
Figure 4.
Regression tree analyses. Regression trees for the (a) CI and (b) RI using phylogenies from the parsimony analyses of the simulated cultural trait data; (c) RI using phylogenies from the ‘true tree’ saved in the simulation program; and (d) the difference in the RI between the parsimony and ‘true’ trees.
We also examined the simulation parameters from the simulations that yielded the top 1 per cent of RI values (i.e. the 50 highest RIs in our simulation). The highest 1 per cent of RI scores ranged from 0.62 to 0.85. Relative to the values simulated, we found that these were from simulations with low rates of evolution (median = 0.003), a lower probability of donation (median = 0.01) and higher extinction rates (median = 0.08); importantly, however, four of the simulations with the highest RIs had values of horizontal transmission greater than 0.05. Other variables did not show an obvious association with the highest RIs. On the whole, this suggests that an RI above about 0.60 is usually indicative of a high degree of vertically transmission (phylogenesis) and a low degree of horizontal transmission (ethnogenesis).
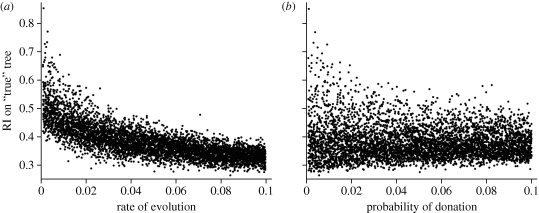
Next, we examined whether the CI and RI are better able to detect horizontal transmission when using the true tree generated in the simulation (as compared with the parsimony tree). The results for the CI revealed that the number of taxa was again the primary variable on the regression tree, with tertiary levels involving rates of evolution. This model explained 55 per cent of the variation, and the probability of donation was not included in the regression tree. Results for the RI are shown in figure 4c. The regression tree model explained 66 per cent of the variation and revealed that rates of evolution and extinction were the primary factors influencing RI values. Again, horizontal transmission was not included in the regression tree models. The importance of evolutionary rate is further illustrated in the bivariate plots in figure 5, which show that rates of evolution explain more of the variation in the RI than does probability of trait donation.
Figure 5.
The RI on the true tree. The RI declines with increasing rate of evolution (a), whereas the RI shows only a weak relationship with the probability of horizontal donation (b). Based on 5000 simulations.
We again examined the parameters of the simulations that yielded the top 1 per cent of RIs calculated from the true tree, which ranged from 0.58 to 0.85. In this case, we found that the top RI values were associated with very low rates of evolution (median = 0.004), and also generally high rates of extinction (median = 0.08) and low rates of horizontal transmission (as compared with the range of values used in our simulations, table 2). In the latter case, the median probability of horizontal donation was 0.009, with only one of 50 simulations having a value greater than 0.05.
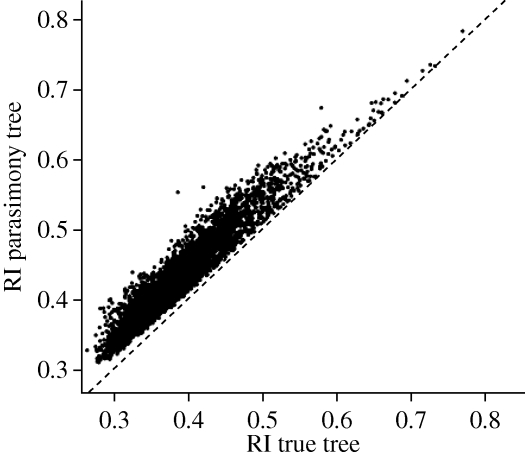
We found that the RI for the parsimony tree was always higher than the RI on the true tree, as expected given that the simulated data were used to both generate the parsimony tree and to calculate the RI (figure 6). We also found that increasing horizontal transmission is associated with a greater divergence between the RI calculated from the parsimony tree (RIparsimony_tree) and the RI calculated from the true tree (RItrue_tree), with the RI for the true tree tending to decline relative to the RI for the parsimony tree as horizontal transmission increases (b = −0.251, F1,4998 = 964, p < 0.0001). We therefore re-ran the analyses using the difference in RIs, calculated as RIparsimony_tree − RItrue_tree. The resulting regression tree model explained 57 per cent of the variation in RI differences, and for the first time in any of the models, the probability of horizontal transmission was included in the regression tree (figure 4d). Higher probabilities of donation tended to lead to a greater divergence in the RI scores between the trees, at least when the number of traits exceeded 266. Additional variables were included in the model, and this result emphasizes the importance of having external information on the history of societies, such as a linguistic tree (Mace & Pagel 1994).
Figure 6.
The RI on the parsimony tree and true tree. The RI is higher on the parsimony tree, as compared with the true tree. The dashed line indicates equal values and thus goes through the origin. Based on 5000 simulations.
Given that the simulations with the highest RI for both the parsimony tree and the true tree were characterized by low rates of evolution and low horizontal transmission, it seems relatively safe to conclude that a high RI is consistent with low horizontal transmission and a high degree of vertical transmission of cultural trait variation. Conversely, it is difficult to conclude that a low RI is indicative of horizontal transmission, as this is also consistent with high rates of evolution. As a last test to probe the effect of horizontal transmission, we examined the distribution of RIs when horizontal transmission takes place to the RIs in which all other parameters are identical, but no horizontal transmission was allowed. We used the RI from the true tree. On average, the RI was lower when horizontal transmission occurred (mean RIp–donation=0 = 0.411, mean RIp–donation>0 = 0.381), and in a paired t-test of the data, we found a significant effect (t = −22.4, p < 0.0001). Nonetheless, this difference was in the same direction for only 63 per cent of the pairs. In a general linear model that tested for an independent effect of horizontal transmission on the RI, we found that higher rates of horizontal transmission depressed the calculation of the RI (table 3).
Table 3.
Predictors of RI when horizontal transmission is allowed.
| term | estimate | t-statistic | p-value |
|---|---|---|---|
| intercept | 0.20 | 89.7 | <0.0001 |
| RIno_horizontal | 0.47 | 95.5 | <0.0001 |
| pdonation | −0.24 | −12.8 | <0.0001 |
Results from a general linear model: RIhorizontal = intercept + β1(RIno_horizontal) + β2(pdonation). R2 for the full model is 0.79. Results are based on the RI calculated using the ‘true’ tree.
(e). Applying the results to real-world data
In the new results presented here, we found that rates of evolution have a bigger impact on the RI and CI than do rates of horizontal transmission. Because the CI is sensitive to the number of societies, the RI is a better measure to use in cross-cultural research (see also Archie 1989, 1990). On a more positive note, however, a high RI is almost always associated with relatively low rates of horizontal transmission, especially when it is calculated on a tree that reflects the true evolutionary history of a set of societies. Thus, in future work, it would be worthwhile to use phylogenetic methods to compare trees calculated from cultural traits with trees obtained using other information, such as genetic or linguistic data, with the prediction that these trees will differ to a greater extent as horizontal transmission increases.
Our simulation results have several implications for the use of homology measures in studies of cultural traits. First, our results confirm that high CI and RI values (for example, greater than 0.60) are usually indicative of low horizontal transmission and thus the distribution of cultural variation being due to phylogenesis, which is consistent with previous uses of the metrics (O'Brien et al. 2001; Tehrani & Collard 2002; Collard et al. 2006). Second, low values were not consistently associated with high levels of horizontal transmission because the CI and RI are heavily influenced by other factors, such as the rate of character evolution and the extinction rate among societies. Thus, while high values may indicate phylogenesis, low values are uninformative. Lastly, low rates of horizontal transmission, and thus a high fidelity of cultural inheritance across generations, only rarely produced a high CI or RI. This may indicate that these metrics provide very low power to detect the prevalence of phylogenesis in cultural evolution. Caution should therefore be taken before adopting these metrics as the primary hypothesis test for a study of trait transmission.
Our simulation shows that further caution is needed if differences in RI values across the time depth of a tree are to be used to test whether cultural or genetic change is responsible for an observed distribution of traits. A recent study of chimpanzee behavioural data across populations found lower RI values on a tree with deeper evolutionary divergences than on two trees of more closely related chimpanzee populations (Lycett et al. 2007). While substantial evidence exists for chimpanzee social learning capabilities and traditional behaviours (Whiten et al. 1999), the RI-based test conducted by Lycett et al. (2007) is insufficient to rule out genetic inheritance of the behavioural variations because our simulation shows that the RI is very sensitive to the rate of evolution. Assuming the behavioural variations to be genetically inherited and not learned, then a sufficiently high evolutionary rate, which could be produced by selection, would also cause a reduction of RI on a tree with deeper phylogenetic separation. This effect would be analogous to the potential for saturation of a fast evolving gene with homoplasy when comparing distantly related organisms. While we might agree that genetic inheritance without social learning seems implausible for the behaviours in question, the point remains that a test based solely on the RI is insufficient.
5. Conclusions
This is an exciting time for cross-cultural research. Increasing availability of linguistic and genetic data is providing new opportunities to examine trait evolution in a historical context. Simultaneously, methodological developments in evolutionary biology and anthropology are providing the tools to examine cultural trait data in a more rigorous way. The success of this enterprise will depend on how well the methods work for particular types of data and under different evolutionary conditions. Cross-cultural studies have increasingly relied on phylogenetic methods to study correlated trait evolution, reconstruct ancestral states and detect vertical or horizontal transmission. To date, however, few studies have quantitatively examined whether phylogenetic methods are appropriate for cross-cultural research. We showed how simulation approaches can be used in this endeavour, specifically to test methods, to identify the conditions under which they fail and even to explore new approaches, such as comparing the RI calculated for a parsimony tree with the RI for a tree that reflects the true history of societal splitting.
Acknowledgements
Sasha Langley played a major role in developing parts of the computer code used in this study, particularly involving the tree structures. We thank two anonymous referees for helpful comments. This paper was supported in part through NSF grants BCS-0132927 and 0323793 and Harvard University.
Footnotes
One contribution of 14 to a Theme Issue ‘Cultural and linguistic diversity: evolutionary approaches’.
References
- Archie J. W.1989Homoplasy excess ratios: new indices for measuring levels of homoplasy in phylogenetic systematics and a critique of the consistency index. Syst. Zool. 38, 253–269 (doi:10.2307/2992286) [Google Scholar]
- Archie J. W.1990Homoplasy excess statistics and retention indices: a reply to Farris. Syst. Zool. 39, 169–174 (doi:10.2307/2992454) [Google Scholar]
- Billing J., Sherman P. W.1998Antimicrobial functions of spices: why some like it hot. Q. Rev. Biol. 73, 3–49 (doi:10.1086/420058) [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds O. R. P., et al. 2007The delayed rise of present-day mammals. Nature 446, 507–512 (doi:10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- Blower S. M., Dowlatabadi H.1994Sensitivity and uncertainty analysis of complex-models of disease transmission—an HIV model as an example. Int. Stat. Rev. 62, 229–243 (doi:10.2307/1403510) [Google Scholar]
- Borgerhoff Mulder M., George-Cramer M., Eshleman J., Ortolani A.2001A study of East African kinship and marriage using phylogenetically controlled comparison. Am. Anthropol. 103, 1059–1082 (doi:10.1525/aa.2001.103.4.1059) [Google Scholar]
- Borgerhoff Mulder M., Nunn C. L., Towner M. C.2006Macroevolutionary studies of cultural trait transmission. Evol. Anthropol. 15, 52–64 (doi:10.1002/evan.20088) [Google Scholar]
- Boyd R., Borgerhoff Mulder M., Durham W. H., Richerson P. J.1997Are cultural phylogenies possible? In Human by nature: between biology and the social sciences (eds Weingart P., Mitchell S. D., Richerson P. J., Maasen S.), pp. 355–386 Mahwah, NJ: Erlbaum [Google Scholar]
- Burton M. L., White D. R.1984Sexual division of labor in agriculture. Am. Anthropol. 86, 568–583 (doi:10.1525/aa.1984.86.3.02a00020) [Google Scholar]
- Collard M., Shennan S. J., Tehrani J. J.2006Branching, blending, and the evolution of cultural similarities and differences among human populations. Evol. Hum. Behav. 27, 169–184 (doi:10.1016/j.evolhumbehav.2005.07.003) [Google Scholar]
- Currie T. E., Greenhill S. J., Mace R.2010Is horizontal transmission really a problem for phylogenetic comparative methods? A simulation study using continuous cultural traits. Phil. Trans. R. Soc. B 365, 3903–3912 (doi:10.1098/rstb.2010.0014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- De'ath G., Fabricius K. E.2000Classification and regression trees: a powerful yet simple technique for ecological data analysis. Ecology 81, 3178–3192 (doi:10.1890/0012-9658(2000)081[3178:CARTAP]2.0.CO;2) [Google Scholar]
- Diaz-Uriarte R., Garland T.1996Testing hypotheses of correlated evolution using phylogenetically independent contrasts: sensitivity to deviations from Brownian motion. Syst. Biol. 45, 27–47 (doi:10.1093/sysbio/45.1.27) [Google Scholar]
- Dow M. M.1984A biparametric approach to network autocorrelation. Sociol. Methods Res. 13, 201–217 (doi:10.1177/0049124184013002002) [Google Scholar]
- Dow M. M.1993Saving the theory: on chi-square tests with cross-cultural survey data. Cross Cult. Res. 27, 247–276 (doi:10.1177/106939719302700305) [Google Scholar]
- Dow M. M.2007Galton's problem as multiple network autocorrelation effects—cultural trait transmission and ecological constraint. Cross Cult. Res. 41, 336–363 [Google Scholar]
- Ensminger J.1997Transaction costs and Islam: explaining conversion in Africa. J. Inst. Theor. Econ. 153, 4–29 [Google Scholar]
- Farris J.1989The retention index and the rescaled consistency index. Cladistics 5, 417–419 (doi:10.1111/j.1096-0031.1989.tb00573.x) [DOI] [PubMed] [Google Scholar]
- Felsenstein J.1985Phylogenies and the comparative method. Am. Nat. 125, 1–15 (doi:10.1086/284325) [Google Scholar]
- Fortunato L., Holden C., Mace R.2006From bridewealth to dowry? A Bayesian estimation of ancestral states of marriage transfers in Indo-European groups. Hum. Nat. 17, 355–376 (doi:10.1007/s12110-006-1000-4) [DOI] [PubMed] [Google Scholar]
- Gordon R. G. (ed.) 2005Ethnologue: languages of the world. Dallas, TX: SIL International; See http://www.ethnologue.com [Google Scholar]
- Gray R. D., Bryant D., Greenhill S. J.2010On the shape and fabric of human history. Phil. Trans. R. Soc. B 365, 3923–3933 (doi:10.1098/rstb.2010.0162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R. D., Atkinson Q. D.2003Language-tree divergence times support the Anatolian theory of Indo-European origin. Nature 426, 435–439 (doi:10.1038/nature02029) [DOI] [PubMed] [Google Scholar]
- Gray R. D., Jordan F. M.2000Language trees support the express-train sequence of Austronesian expansion. Nature 405, 1052–1055 (doi:10.1038/35016575) [DOI] [PubMed] [Google Scholar]
- Hartung J.1982Polygyny and the inheritance of wealth. Curr. Anthropol. 23, 1–12 (doi:10.1086/202775) [Google Scholar]
- Harvey P. H., Pagel M. D.1991The comparative method in evolutionary biology. Oxford Series in Ecology and Evolution Oxford, UK: Oxford University Press [Google Scholar]
- Harvey P. H., Rambaut A.2000Comparative analyses for adaptive radiations. Proc. R. Soc. Lond. B 355, 1599–1605 (doi:10.1098/rstb.2000.0721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden C. J.2002Bantu language trees reflect the spread of farming across sub-Saharan Africa: a maximum-parsimony analysis. Proc. R. Soc. Lond. B 269, 793–799 (doi:10.1098/rspb.2002.1955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles M. E., Matisoo-Smith E., Gray R. D., Penny D.2003Untangling oceanic settlement: the edge of the knowable. Trends Ecol. Evol. 18, 531–540 (doi:10.1016/S0169-5347(03)00245-3) [Google Scholar]
- Jones D.2003Kinship and deep history: exploring connections between culture areas, genes, and languages. Am. Anthropol. 105, 501–514 (doi:10.1525/aa.2003.105.3.501) [Google Scholar]
- Jordan P., Shennan S. J.2003Cultural transmission, language, and basketry traditions amongst the California Indians. J. Anthropol. Archaeol. 22, 43–74 (doi:10.1016/S0278-4165(03)00004-7) [Google Scholar]
- Kluge A. G., Farris J. S.1969Quantitative phyletics and evolution of anurans. Syst. Zool. 18, 1–32 [Google Scholar]
- Lipo C. P., O'Brien M. J., Collard M., Shennan S. (eds) 2006Mapping our ancestors. New Brunswick, NJ: Aldine Transaction [Google Scholar]
- Lycett S. J., Collard M., McGrew W. C.2007Phylogenetic analyses of behavior support existence of culture among wild chimpanzees. Proc. Natl Acad. Sci. USA 104, 17 588–17 592 (doi:10.1073/pnas.0707930104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace R., Holden C. J.2005A phylogenetic approach to cultural evolution. Trends Ecol. Evol. 20, 116–121 (doi:10.1016/j.tree.2004.12.002) [DOI] [PubMed] [Google Scholar]
- Mace R., Pagel M.1994The comparative method in anthropology. Curr. Anthropol. 35, 549–564 (doi:10.1086/204317) [Google Scholar]
- Mace R., Pagel M.1995A latitudinal gradient in the density of human languages in North America. Proc. R. Soc. Lond. B 261, 117–121 (doi:10.1098/rspb.1995.0125) [Google Scholar]
- Mace R., Holden C. J., Shennan S. (eds) 2005The evolution of cultural diversity: a phylogenetic approach. London, UK: UCL Press [Google Scholar]
- Maddison D. R., Swofford D. L., Maddison W. P.1997Nexus: an extensible file format for systematic information. Syst. Biol. 46, 590–621 (doi:10.2307/2413497) [DOI] [PubMed] [Google Scholar]
- Martins E. P., Garland T.1991Phylogenetic analyses of the correlated evolution of continuous characters: a simulation study. Evolution 45, 534–557 (doi:10.2307/2409910) [DOI] [PubMed] [Google Scholar]
- Mesoudi A., Whiten A., Laland K. N.2004Perspective: is human cultural evolution Darwinian? Evidence reviewed from the perspective of the origin of species. Evolution 58, 1–11 (doi:10.1554/03-212) [DOI] [PubMed] [Google Scholar]
- Moore J. H.1994Putting anthropology back together again: the ethnogenetic critique of cladistic theory. Am. Anthropol. 96, 925–948 (doi:10.1525/aa.1994.96.4.02a00110) [Google Scholar]
- Moore C. C., Romney A. K.1994Material culture, geographic propinquity, and Linguist affiliation on the north coast of New-Guinea—a reanalysis. Am. Anthropol. 96, 370–392 (doi:10.1525/aa.1994.96.2.02a00050) [Google Scholar]
- Moore J. L., Manne L., Brooks T., Burgess N. D., Davies R., Rahbek C., Williams P., Balmford A.2002The distribution of cultural and biological diversity in Africa. Proc. R. Soc. Lond. B 269, 1645–1653 (doi:10.1098/rspb.2002.2075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moylan J. W., Borgerhoff Mulder M., Graham C. M., Nunn C. L., Hakansson T.2006Cultural traits and Linguistic trees: phylogenetic signal in East Africa. In Mapping our ancestors (eds Lipo C. P., O'Brien M. J., Collard M., Shennan S.), pp. 33–52 New Brunswick, NJ: Aldine Transaction [Google Scholar]
- Murdock G. P., White D.1969Standard cross-cultural sample. Ethnology 8, 329–369 (doi:10.2307/3772907) [Google Scholar]
- Naroll R.1961Two solutions to Galton problem. Phil. Sci. 28, 15–39 (doi:10.1086/287778) [Google Scholar]
- Naylor G., Kraus F.1995The relationship between s and m and the retention index. Syst. Biol. 44, 559–562 (doi:10.1093/sysbio%2F44.4.559) [Google Scholar]
- Nee S., Holmes E. C., May R. M., Harvey P. H.1994Extinction rates can be estimated from molecular phylogenies. Phil. Trans. R. Soc. Lond. B 344, 77–82 (doi:10.1098/rstb.1994.0054) [DOI] [PubMed] [Google Scholar]
- Nunn C. L.1995A simulation test of Smith's ‘degrees of freedom’ correction for comparative studies. Am. J. Phys. Anthropol. 98, 355–367 (doi:10.1002/ajpa.1330980308) [DOI] [PubMed] [Google Scholar]
- Nunn C. L., Barton R. A.2001Comparative methods for studying primate adaptation and allometry. Evol. Anthropol. 10, 81–98 (doi:10.1002/evan.1019) [Google Scholar]
- Nunn C. L., Borgerhoff Mulder M., Langley S.2006Comparative methods for studying cultural trait evolution: a simulation study. Cross Cult. Res. 40, 177–209 (doi:10.1177/1069397105283401) [Google Scholar]
- O'Brien M. J., Darwent J., Lyman R. L.2001Cladistics is useful for reconstructing archaeological phylogenies: Palaeoindian points from the southeastern United States. J. Archaeol. Sci. 28, 1115–1136 (doi:10.1006/jasc.2001.0681) [Google Scholar]
- Pagel M., Mace R.2004The cultural wealth of nations. Nature 428, 275–278 (doi:10.1038/428275a) [DOI] [PubMed] [Google Scholar]
- Pagel M. D., May R. M., Collie A. R.1991Ecological aspects of the geographical-distribution and diversity of mammalian-species. Am. Nat. 137, 791–815 (doi:10.1086/285194) [Google Scholar]
- Price T.1997Correlated evolution and independent contrasts. Phil. Trans. R. Soc. Lond. B 352, 519–529 (doi:10.1098/rstb.1997.0036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A., Gittleman J. L., Luh H.1994Truth or consequences: effects of phylogenetic accuracy on two comparative methods. J. Theor. Biol. 167, 293–300 (doi:10.1006/jtbi.1994.1071) [Google Scholar]
- Roe F. G.1955The Indian and the horse. Norman, OK: University of Oklahoma Press [Google Scholar]
- Rushton S. P., Lurz P. W. W., Gurnell J., Fuller R.2000Modelling the spatial dynamics of parapoxvirus disease in red and grey squirrels: a possible cause of the decline in the red squirrel in the UK? J. Appl. Ecol. 37, 997–1012 (doi:10.1046/j.1365-2664.2000.00553.x) [Google Scholar]
- Sanderson M. J., Donoghue M. J.1989Patterns of variation in levels of homoplasy. Evolution 43, 1781–1795 (doi:10.2307/2409392) [DOI] [PubMed] [Google Scholar]
- Seaholm S. K., Ackerman E., Wu S. C.1988Latin hypercube sampling and the sensitivity analysis of a Monte–Carlo epidemic model. Int. J. Biomed. Comput. 23, 97–112 (doi:10.1016/0020-7101(88)90067-0) [DOI] [PubMed] [Google Scholar]
- Sutherland W. J.2003Parallel extinction risk and global distribution of languages and species. Nature 423, 276–279 (doi:10.1038/nature01607) [DOI] [PubMed] [Google Scholar]
- Swofford D. L.2003PAUP*: phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates [Google Scholar]
- Symonds M. R. E.2002The effects of topological inaccuracy in evolutionary trees on the phylogenetic comparative method of independent contrasts. Syst. Biol. 51, 541–553 (doi:10.1080/10635150290069977) [DOI] [PubMed] [Google Scholar]
- Tehrani J., Collard M.2002Investigating cultural evolution through biological phylogenetic analyses of Turkmen textiles. J. Anthropol. Archaeol. 21, 443–463 (doi:10.1016/S0278-4165(02)00002-8) [Google Scholar]
- Terrell J. E., Hunt T. L., Gosden C.1997The dimensions of social life in the Pacific—human diversity and the myth of the primitive isolate. Curr. Anthropol. 38, 155–195 (doi:10.1086/204604) [Google Scholar]
- Trimmingham J. S.1970A history of Islam in west Africa. London, UK: Oxford University Press [Google Scholar]
- Tylor E. B.1889On a method of investigating the development of institutions applied to the law of marriage and descent. J. R. Anthropol. Inst. 18, 245–272 [Google Scholar]
- White D. R., Burton M. L., Dow M. M.1981Sexual division of labor in African agriculture—a network auto-correlation analysis. Am. Anthropol. 83, 824–849 (doi:10.1525/aa.1981.83.4.02a00040) [Google Scholar]
- Whiten A., Goodall J., McGrew W. C., Nishida T., Reynolds V., Sugiyama Y., Tutin C. E. G., Wrangham R. W., Boesch C.1999Cultures in chimpanzees. Nature 399, 682–685 (doi:10.1038/21415) [DOI] [PubMed] [Google Scholar]