Abstract
Differences between individuals in the copy-number of whole genes have been found in every multicellular species examined thus far. Such differences result in unique complements of protein-coding genes in all individuals, and have been shown to underlie adaptive phenotypic differences. Here, we review the evidence for copy-number variants (CNVs), focusing on the methods used to detect them and the molecular mechanisms responsible for generating this type of variation. Although there are multiple technical and computational challenges inherent to these experimental methods, next-generation sequencing technologies are making such experiments accessible in any system with a sequenced genome. We further discuss the connection between copy-number variation within species and copy-number divergence between species, showing that these values are exactly what one would expect from similar comparisons of nucleotide polymorphism and divergence. We conclude by reviewing the growing body of evidence for natural selection on copy-number variants. While it appears that most genic CNVs—especially deletions—are quickly eliminated by selection, there are now multiple studies demonstrating a strong link between copy-number differences at specific genes and phenotypic differences in adaptive traits. We argue that a complete understanding of the molecular basis for adaptive natural selection necessarily includes the study of copy-number variation.
Keywords: duplication, copy-number variation, paralogue, natural selection, humans, Drosophila
1. Introduction
The sequencing of whole genomes has revealed large numbers of polymorphisms in every species examined. When either fully outbred individuals are sequenced (e.g. Mikkelsen et al. 2005) or multiple inbred lines from the same species are sequenced (e.g. Begun et al. 2007), millions of single-nucleotide polymorphisms (SNPs) and small insertion/deletion (indel) polymorphisms are found. Because of their ubiquity and the ease with which they are genotyped, these types of variation have been the focus of most population-level studies.
However, in recent years it has been revealed that copy-number variants—large regions of the genome that differ in copy number between individuals within a species owing to duplication or deletion events—are an important source of genetic variation. Indeed, copy-number variants (CNVs; sometimes also called ‘copy-number polymorphisms’ or CNPs) have been shown to be widespread in a variety of organisms, including humans (Sebat et al. 2004; Conrad et al. 2006; McCarroll et al. 2006; Redon et al. 2006), mice (Graubert et al. 2007; She et al. 2008), chimpanzees (Perry et al. 2006, 2008), rhesus macaques (Lee et al. 2008), cows (Liu et al. 2010), dogs (Chen et al. 2009; Nicholas et al. 2009), chickens (Griffin et al. 2008), maize (Springer et al. 2009), Arabidopsis thaliana (Ossowski et al. 2008), fruitflies (Dopman & Hartl 2007; Emerson et al. 2008), Caenorhabditis elegans (Maydan et al. 2010) and Saccharomyces cerevisiae (Carreto et al. 2008). Though it is often harder to experimentally identify and genotype CNVs relative to SNPs and indels, many are big enough to encompass whole genes and are therefore more likely to affect organismal fitness.
The exact number of genic differences between individuals owing to CNVs is often a hard number to pin down; this is due to a number of factors, including the genomic resolution of individual experiments, whether the overlap with genes is partial or complete, and the fact that many studies report the total number of variants found and not the average number of pairwise differences between individuals. Owing to the biomedical focus of most studies, the best data on CNVs come from humans. These studies have revealed that a sizable proportion—0.2 per cent (six megabases)—of the human genome varies in copy number between two individuals (McCarroll et al. 2008). Earlier low-resolution studies vastly overestimated the size of CNVs and, therefore, had highly inflated estimates (as demonstrated in Kidd et al. 2008 and McCarroll et al. 2008). Considering only protein-coding genes, studies show that any two humans are likely to differ at CNVs completely encompassing approximately 105 genes (as calculated from the Yoruban samples in Conrad et al. 2010). Similar numbers of genic CNVs can be found in every species examined, with much larger counts if all CNVs that partially overlap genes are also counted (e.g. approx. 367 genes in humans based on unrelated Yoruban individuals in Conrad et al. 2010).
The importance of this result cannot be over-emphasized: any two individual genomes taken from nature, in any species, will have dozens to hundreds of differences in their total number of functional genes. Because CNVs are due to both duplications and deletions, these differences will be due to newly arising duplications in some genes and deletions in others. And these copy-number differences are not confined to large, multi-gene families or some other subset of genes thought to be unimportant for fitness—single-copy genes can be duplicated or deleted in any individual, though selection against such deletions is probably much stronger (see below). What is more, most estimates of polymorphism owing to CNVs were derived using methods that will fail to detect all gene copy-number polymorphisms. For example, there are many well-known examples of segregating pseudogenes, including the large number of polymorphic olfactory receptor pseudogenes found in humans (e.g. Menashe et al. 2003). Because the differences between functional and non-functional olfactory receptors are due to single-nucleotide changes or small indels, they will not be detected by most CNV experiments. Therefore, even these counts of functional genic differences among individuals are underestimates.
In this paper, we review recent studies that have increased our understanding of the mutational mechanisms that form CNVs as well as the degree to which CNVs are impacted by natural selection and drift. We then discuss how these evolutionary forces result in copy-number differences among individuals and eventually differences between species. However, we first begin with a discussion of the methods used to detect CNVs.
2. CNV detection methods
There are two general categories of methods used to detect CNVs and regions with overlapping CNVs (CNVRs). The first (‘hybridization-based mapping’) uses the fact that any region duplicated or deleted in a sample individual will show an excess or deficit, respectively, of DNA that is highly similar to that region relative to the reference genome. These methods are therefore aimed at detecting these localized differences in relative DNA content. The second category of methods (‘paired-end mapping’) does not detect the duplications and deletions directly, but instead detects length differences in the size of captured fragments from a sample relative to the reference genome. Fragments that appear too large must contain insertions or duplications, while those that are too small must contain deletions. Other methods, such as quantitative PCR and fluorescent in situ hybridization, can be used to verify CNVs but are not useful for the discovery process.
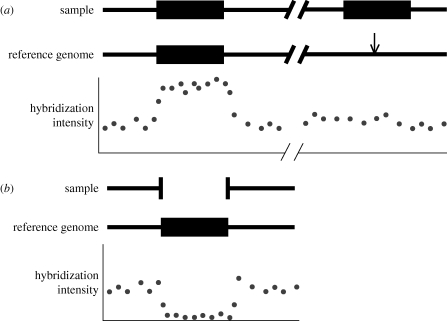
Methods focused on comparisons of relative DNA content (i.e. hybridization-based mapping) were first performed using microarrays (Sebat et al. 2004; Conrad et al. 2006; McCarroll et al. 2006; Redon et al. 2006). This method detects differences in copy number by allowing fluorescently labelled DNA from a sample individual to hybridize to an array designed from different regions of the genome. The first such methods used cDNA or BAC-based arrays, though custom oligonucleotide arrays that are designed to have probes covering as much of the genome as possible are now most commonly used. In either case, regions with elevated hybridization intensity are inferred to correspond to sequences with one or more highly similar duplicate copies present in the sample but absent in the reference—hereafter referred to as duplications relative to the reference genome (figure 1a). Similarly, regions with lower hybridization intensity correspond to sequences absent in the sample individual but present in the reference genome, referred to as deletions relative to the reference genome (figure 1b). This method is useful for detecting deletions and duplications, and is the source of the vast majority of CNV data; reliable genotyping of homozygotes and heterozygotes for copy-number differences is also possible (e.g. McCarroll et al. 2008). A drawback of hybridization-based methods is that while they can detect regions of the reference genome that have a duplicate copy, this data cannot be used to determine where the duplicate copy resides in the genome (see §3). Finally, hybridization-based methods cannot detect CNVs lying in poorly assembled regions of the reference genome that cannot be probed, or highly repetitive CNVs such as transposable elements that may not be represented on the array. Depending on the emphasis on specificity versus sensitivity, the experimental platform used, and the length of CNVs, error rates when using this method can range from over 25 per cent to less than 1 per cent (Redon et al. 2006; Emerson et al. 2008; McCarroll et al. 2008; Conrad et al. 2010).
Figure 1.
Detecting duplications and deletions relative to a reference genome using hybridization intensities. (a) When a region of the genome has one copy in the reference genome but two copies in the sample (black rectangles), DNA from both paralogues in the sample hybridize to probes corresponding to the only copy in the reference, resulting in a spike in hybridization intensity at these probes (illustrated by the elevated intensities directly below the copy in the reference). The location of the additional copy present in the sample genome is denoted with an arrow in the reference genome. (b) When a region of the genome has one copy in the reference genome (black rectangle) but no copies in the sample, hybridization intensity is significantly diminished at probes corresponding to the sequence missing from the sample.
With the advent of next-generation sequencing technologies (e.g. Illumina or SOLiD), CNVs are now detectable in genome resequencing projects by finding regions with unusually high or low read depth. This method is analogous to array-based hybridization methods and is characterized by many of the same advantages and drawbacks. The use of sequence-capture arrays (e.g. Burbano et al. 2010) even allows targeted sequencing of specific genomic regions, which means that these technologies can also be used as genotyping platforms for CNVs.
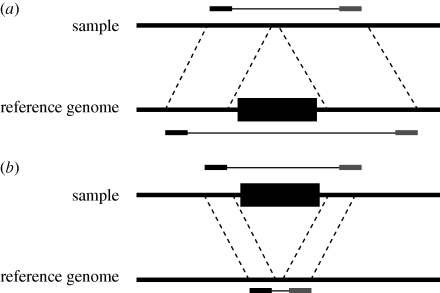
The second, and often more experimentally challenging, method used to detect CNVs involves sequencing the paired ends of DNA fragments collected from an individual and then ‘mapping’ these end sequences to a reference genome using BLAST or some other fast alignment tool. While the methods for creating and collecting these fragments differ in important ways, the key idea is that the sequenced endpoints are a known, fixed distance apart in the sample. If the portion of the genome spanned by the two end sequences is larger than the expected size of the fragment, then the individual probably harbours a deletion relative to the reference at that locus (figure 2a). On the other hand, if the spanned portion of the genome is smaller than expected, then the individual is inferred to have a stretch of sequence at that locus that is absent in the reference genome (figure 2b). This extra sequence is likely to be a duplicated copy of DNA, though there are other possibilities (see below). Thus, unlike hybridization-based mapping, paired-end mapping detects the location of the sequence that is the result of the duplication (the ‘daughter’ locus), rather than just the location of the sequence that it is copied from (the ‘parent’ locus; figure 2b).
Figure 2.
Detecting insertions and deletions using paired-end mapping data. The ends of a DNA fragment from a sample individual are mapped to a reference genome. In both of these illustrations, a depiction of the DNA fragment appears above its location in the sample chromosome. The black and grey ends correspond to the unique sequenced ends of the fragment, and the expected length of the sequence is shown above the sample chromosome. Dashed lines indicate homologous regions in the two genomes, and the location of the black and grey ends below the reference chromosome corresponds to their mapped locations. (a) If the portion of the reference genome spanned by the fragment ends is larger than expected, then the sample genome probably contains a deletion relative to the reference. (b) If the length of the region spanned by the locations of the end sequences in the reference genome is smaller than expected, then an insertion is inferred to be present in the sample genome.
There are two main methods used for paired-end mapping, one using next-generation sequencing technologies and the other using fosmid or other clone-based technologies. Paired-end mapping using, for instance, Illumina sequencing relies on the fact that different libraries can be constructed with insert sizes ranging from 150 bp to 10 kb, with little variation in length within an insert-size class. The main advantage of this method is that millions of paired-end sequences can be generated in a single run, and CNVs are often supported by many independent pairs of reads. There are two significant disadvantages of next-generation paired-end methods. First, the insert size is quite limited, such that only small duplications will be contained in the end-sequenced DNA fragments. Second, there is no way to capture the inserted DNA and to sequence it; this means that the identity of the insert is not known, and therefore the ‘parental’ locus is also unknown. To get around both of these problems, fosmid-based methods capture much longer stretches of DNA in semi-permanent clone libraries maintained in bacterial cells (e.g. Tuzun et al. 2005). While fosmid methods are still somewhat limited in their insert sizes (up to approx. 40 kb), the insert can be sequenced and mapped to the reference genome, and therefore the identity and location of both the parent and daughter copies can be revealed. Clone-based methods are much more time-consuming in general, not least because the paired ends must be sequenced by the Sanger method. The error rate of paired-end methods has been measured at below 20 per cent (Tuzun et al. 2005), though, as with hybridization-based methods, this depends on thresholds that can be adjusted based on preferences for specificity versus sensitivity.
Both paired-end methods have advantages relative to hybridization-based methods. Differences in the distance and orientation of paired-end reads between the sample and the reference genome can also be used to detect larger duplications, inversions, transposable element insertions and other types of ‘structural variants’ invisible to hybridization-based methods (e.g. Korbel et al. 2007). In addition, deletions in the reference assembly—which cannot normally be detected using hybridizaton-based methods (see below)—can be found using paired-end mapping, and insertion sites can be found with high resolution. However, paired-end mapping is more expensive and time-consuming than hybridization-based methods, and has its own limitations. Like hybridization-based mapping, paired-end mapping can usually only identify CNVs when paired-end sequences map unambiguously to a reference genome. And because most duplication events are detected as insertions between the paired ends, even with the larger insert sizes afforded by fosmids, paired-end approaches are likely to significantly underestimate the number and average length of polymorphic duplications relative to a reference genome. For now, both hybridization-based and paired-end methods for detecting CNVs offer complementary insights into the nature of these polymorphisms. It is also worth noting that both methods can be combined via next-generation paired-end resequencing.
3. Mutational mechanisms of duplication and deletion
There are a number of mutational mechanisms that will result in duplication and/or deletion of stretches of DNA: replication slippage, non-allelic homologous recombination (NAHR), non-homologous end joining (NHEJ) and retrotransposition. At least in humans, NAHR appears to be the most common mechanism generating CNVs, followed closely by NHEJ and replication slippage, and more distantly by retrotransposition (Kidd et al. 2008; Conrad et al. 2010). There may also be variation across the genome in the dominance of one mechanism versus the others (e.g. Cardoso-Moreira & Long 2010).
Replication slippage is perhaps the simplest mechanism by which CNVs are formed. Many smaller variants such as variable-number tandem repeats are caused by slippage, and it appears that occasional stretches greater than 5 kb in length can be added and subsequently subtracted by this mechanism (calculated from Conrad et al. 2010). The mechanism apparently responsible for the largest proportion of known CNVs is NAHR. NAHR occurs when previously duplicated sequences that are still highly similar to one another recombine; because non-allelic sequences have recombined, this process will result in both a duplication and a deletion when recombination occurs between homologous chromosomes or between sister chromatids, or only deletions when recombination occurs on the same chromatid (Turner et al. 2008a). The commonly cited mechanism of unequal crossing over is actually driven by NAHR between duplicated sequences located in close proximity. When new duplicates are formed, they themselves can become the substrate for additional mutations, thereby increasing the local mutation rate (though the magnitude of this increase is not clear). Thus, NAHR hotspot formation may be a runaway process (Eichler 2001). While duplication caused by NAHR requires highly similar sequences at the breakpoints of the mutation, a related recombination repair pathway, NHEJ, requires little or no sequence homology, and can result in both deletion and insertion of DNA into double-strand breaks (Hastings et al. 2009). Retrotransposition is another common mutational mechanism that, unlike NAHR and NHEJ, results only in new duplications. Retrotransposed duplicate genes result from the reverse-transcription of mRNA into cDNA, which is then inserted into a new genomic position. If methods used to look for differential hybridization are applied only to exonic sequences, these ‘retroCNVs’ are detectable (e.g. Conrad et al. 2010).
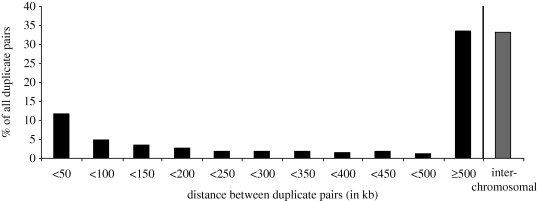
All of the mechanisms mentioned above can result in polymorphic duplications (though not all result in deletions). Many of these duplications are tandem, meaning the daughter copy is located very near to the parental copy. However, there is evidence to suggest that many of these CNVs can be dispersed duplications in which the two paralogues are located on different chromosomes or far apart on the same chromosome (Conrad et al. 2010; Schrider & Hahn 2010). This is not surprising, since a large proportion of fixed duplicates in humans—and a small but significant number in fruitflies—lie on different chromosomes (figure 3; Bailey et al. 2002; McGrath et al. 2009; Meisel et al. 2009), and these fixed dispersed duplicates probably arose as polymorphic dispersed duplicates. Since hybridization-based methods and next-generation paired-end methods generally locate only one paralogue, it is possible that the other paralogue is not located very close by—it may even lie on a different chromosome.
Figure 3.
Distances between recent gene duplicates in the human genome formed after the human–macaque split (from data presented in McGrath et al. 2009).
4. CNVs are detected relative to a reference genome
If one wishes to detect CNVs in a certain species, a reference genome for that species is required—all of the detection methods described earlier involve observing a difference in the copy number of a locus in a sample compared with the reference genome. Importantly, one should keep in mind that this reference genome is (usually) just the genome of a single individual. Therefore, a ‘duplication’ detected in a sample individual may actually be a deletion of a previously duplicated sequence, with the reference genome having the deletion allele and the sample individual not having the mutation (electronic supplementary material, figure S1a). Likewise, ‘deletions’ relative to a reference genome may in fact be novel duplication alleles present in the reference genome but absent in the sample (electronic supplementary material, figure S1b). However, the majority of detected duplications ‘relative to the reference’ are in fact due to duplication events, and most deletions relative to the reference are actual deletions (Emerson et al. 2008; Schrider & Hahn 2010). These observations are consistent with the population-genetic expectation that the probability of finding a derived allele in the reference genome is simply equal to the population frequency of the derived allele, which is 25 per cent on average in an idealized population.
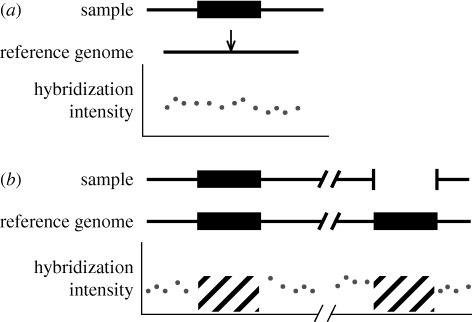
Since the reference genome is the sequence of a random individual, it may also contain deletion alleles at otherwise single-copy loci where other individuals do not. In these cases, sample individuals contain a sequence that is completely absent from the reference, which may not necessarily be homologous to any sequence in the reference genome, and which therefore would be impossible to detect via hybridization-based methods; the same is true of novel insertions in a sample that are not present in the reference genome (figure 4a). A similar argument can be made for duplication alleles present in the reference genome: if, for technical reasons, duplicated regions of the reference genome are not queried in hybridization-based experiments (and they often are not), then these CNVs will not be detected (figure 4b). In total, expectations from a Wright–Fisher population suggest that approximately 25 per cent of all CNVs will not be detected by hybridization-based methods because the derived allele is present in the reference genome.
Figure 4.
Hybridization-based methods will not detect either deletions in the reference or highly similar duplications in the reference. As in figure 1, sample hybridization intensities are shown under the corresponding regions of the reference genome. (a) If a deletion allele is present in the reference genome (the location is shown by the arrow), then an array designed from the reference will not be able to probe this sequence in sample individuals. (b) If a duplication allele is present in the reference genome, arrays designed from the reference will probably not probe these repetitive regions (shown as diagonal black lines). Because of ambiguous sequence mapping, next-generation sequencing methods will also have difficulty detecting variants in these regions.
The inability to detect sequences not present in the reference genome, or to detect changes in copy number of sequences having more than one copy in the reference genome—at least using hybridization-based methods—leads to a pernicious ascertainment bias (Emerson et al. 2008): a larger fraction of high-frequency deletion and duplication alleles will be missed relative to low-frequency deletion and duplication alleles. For example, if a derived deletion allele reaches a population frequency of p = 0.90, there is a 90 per cent chance that the derived state will be present in the reference genome and therefore that it will go undetected; likewise, there is only a 10 per cent chance of missing a deletion allele at frequency 0.10. If a duplication allele reaches a population frequency of p = 0.90, there is also a 90 per cent chance that it is found in the reference genome and therefore that it could go undetected.
Summing over all CNVs that are deletions or duplications segregating in a population, these calculations imply that a survey using hybridization-based methods will miss 90 per cent of all alleles at frequency 0.90, 80 per cent at frequency 0.80, 70 per cent at 0.70, etc. (there is no bias for paired-end methods since both derived insertions and deletions can be detected). If we were to plot the allele frequency spectrum for deletion or duplication alleles without accounting for this bias, we would find a strong skew towards low-frequency alleles (electronic supplementary material, figure S2). Because methods for inferring natural selection often use the allele frequency spectrum (see below), it is important to correct for this bias. A straightforward way to correct for ascertainment bias is to divide the number of counts in each frequency bin by 1 − p; this simple correction accounts for the proportion of derived alleles that are missed in the first place (electronic supplementary material, figure S2; cf. Emerson et al. 2008).
These arguments assume that the reference genome is constructed from a single haploid genome or a highly inbred individual, as is the case with Drosophila melanogaster. In cases where the reference genome is constructed from one or more outbred diploid individuals, expectations are more complicated (electronic supplementary material, figure S2). Regardless of the details of the correction, it is clear that many studies have failed to detect a large portion of common CNVs in humans and other organisms because of this bias.
5. Copy-number variation results in copy-number divergence
While the sheer number of polymorphic whole-gene duplications and deletions may at first seem surprising, this level of variation should have been predictable: there are thousands of very young duplicated genes found in every eukaryotic reference genome (e.g. Lynch & Conery 2000; Gu et al. 2002), and closely related species differ substantially in gene copy number (Demuth et al. 2006; Hahn et al. 2007a,b). Because polymorphism is a sine qua non of evolutionary divergence, results from comparative genomics have always implied this level of within-species variation.
To demonstrate that CNVs are a phase of molecular evolution like any other polymorphism, it is useful to consider the expected amounts of variation and divergence in a particular species. In an idealized Wright–Fisher population at equilibrium, the amount of polymorphism is expected to be 4Neμ, where Ne is the effective population size and μ is the neutral mutation rate. Comparing two species, the amount of divergence is expected to be 2tμ, where t is the time since the most recent common ancestor of the species. Therefore, regardless of the type of mutation considered, the ratio of polymorphism to divergence is 4Ne/2t. There should also be a constant ratio between polymorphism and divergence even if the equilibrium assumptions do not hold.
Table 1 gives the values of coding polymorphism calculated for both SNPs and CNVs within humans, and divergence between humans and chimpanzees. Because the ratio of polymorphism to divergence appears to be quite similar for the two types of variation, these estimates suggest that CNVs fix at a rate comparable to SNPs. There are a number of things to be cautious of in this comparison, including the fact that it lumps multiple mutational mechanisms, as well as duplications and deletions, into one ‘CNV’ category; the problem of undetected CNVs owing to ascertainment biases; and the fact that values in each cell are based on different methodologies. The genomic instability associated with NAHR may also change the relationship between polymorphism and divergence, as has been observed previously (Newman et al. 2005). Nonetheless, the comparison at least provides quantitative support for previous estimates of interspecific divergence owing to gene gain and loss (e.g. Demuth et al. 2006).
Table 1.
Human polymorphism and divergence for nucleotide and copy-number variation. Nucleotide data are expressed per site and copy-number data are expressed per gene; divergence data are calculated from pairwise comparisons with chimpanzees.
| polymorphism | divergence | |
|---|---|---|
| nucleotide | 0.0009a | 0.0123b |
| copy number | 0.0038c | 0.064d |
6. Evidence for natural selection on CNVs
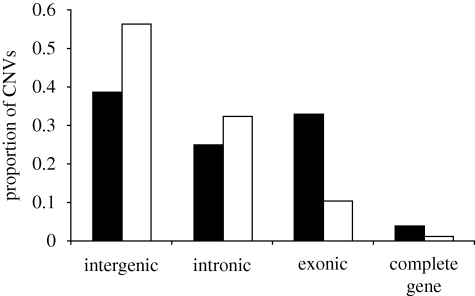
A number of studies have attempted to make inferences about the selective forces acting on different types of CNVs (e.g. duplications versus deletions) or even individual CNVs. Several of these studies have suggested that deletions tend to be under stronger purifying selection than duplications. This assertion is supported in part by the data presented in figure 5, where we use CNVs collected in Emerson et al. (2008) to show that there is a deficit of deletions in coding sequences relative to duplications, as well as a deficit of deletions in exons relative to introns. A deficit of genic deletions has also been observed in humans (Conrad et al. 2006, 2010; Redon et al. 2006). This implies that deletions in coding sequences are more deleterious than duplications of these sequences and are therefore more likely to be removed by purifying selection. Interestingly, Emerson et al. (2008) also found that duplications in Drosophila are very likely to be under purifying selection. This finding may be due primarily to strong purifying selection acting on duplications within but not wholly encompassing genes, as tandem duplications contained within genes would probably disrupt the coding sequence or impact splicing (Emerson et al. 2008). But even increases in the copy number of whole genes can be deleterious when CNVs contain dosage-sensitive genes (Schuster-Bockler et al. 2010), and CNVs occurring outside of coding regions can also cause gene expression changes (Stranger et al. 2007), which could result in selection against intergenic CNVs.
Figure 5.
Proportion of duplications and deletions residing within exons, introns, intergenic regions or encompassing complete genes (redrawn from Emerson et al. 2008). Black bars, duplications; white bars, deletions.
The frequency spectrum of alleles can also be used to infer that one type of polymorphism is under stronger selection than another. For example, non-synonymous SNPs are on average at lower frequencies than synonymous SNPs, implying that non-synonymous mutations are being kept at low frequency by stronger purifying selection (Eyre-Walker & Keightley 2007). An examination of the allele frequency spectra of CNVs in humans (Locke et al. 2006) concluded that deletions are on average confined to lower frequencies than are duplications. Provided the ascertainment bias against variants present in the reference genome has an equal effect on both duplications and deletions, these observations lend further support to the claim that deletions are under stronger purifying selection than duplications. The finding by Emerson et al. (2008) that duplications are under purifying selection was based on a comparison of allele frequencies of duplications and synonymous SNPs. Specifically, it was found that duplications were slightly but significantly skewed towards the lower frequency, even after correcting for ascertainment bias, indicating weak purifying selection. Similar lines of evidence have been used to suggest that selection is stronger against large CNVs (Itsara et al. 2009), presumably because they are more likely to affect functional DNA. These findings, taken together with the deficit of CNVs in coding regions, imply that strong natural selection quickly eliminates most copy-number changes of functional sequence—deletions in particular—with the observed variants that remain being slightly deleterious, neutral or advantageous.
Although CNVs as a whole may be under purifying selection, a growing number of studies have shown that individual CNVs and whole-gene families polymorphic in copy number are under positive selection. In order to detect cases of positive selection, recent studies have adapted methods previously used to detect selection on nucleotide changes. Tests based on the fact that selective sweeps will result in extended haplotype homozygosity (Sabeti et al. 2002; Voight et al. 2006) have been used to detect CNVs probably under recent positive selection (e.g. Conrad et al. 2010). A number of studies have detected candidate CNVs under selection by examining allele frequency differentiation between populations (Redon et al. 2006; Perry et al. 2007; Xue et al. 2008; Conrad et al. 2010) or by examining differentiation at the ends of a cline (Turner et al. 2008b). For example, Perry et al. (2007) found that the number of copies of the human salivary amylase gene, AMY1, is typically higher in populations with high-starch diets than in those with low-starch diets, and that this difference is probably due to adaptive natural selection. Another example comes from the human UGT2B17 gene, the enzyme product of which metabolizes steroids and foreign compounds, and has a polymorphic deletion that may be experiencing balancing selection in Europeans and positive selection in East Asians (Xue et al. 2008); the deletion allele has been associated with several phenotypes that are the possible targets of selection (Xue et al. 2008).
In addition to methods only considering within-species variation, several studies have attempted to use methods comparing polymorphism to divergence in order to detect selection on CNVs. The McDonald–Kreitman (MK) test (McDonald & Kreitman 1991) has been used to compare the ratio of polymorphism to divergence in copy-number variant genes of a particular function to the ratio of polymorphism to divergence of intergenic CNVs between humans and chimpanzees (Perry et al. 2008). Zhang (2007) used a similar variant of the MK test to compare the number of polymorphic and fixed functional OR genes with the numbers of OR pseudogenes polymorphic and fixed between humans and chimps. Though neither of these studies found statistical support to reject the null hypothesis, there are some important caveats when using the MK test and related methods (e.g. the HKA test; Hudson et al. 1987). These methods all assume that the neutral mutation rate does not change over time; this assumption probably does not hold for changes in copy number because each additional gene contributes independently to the overall probability of change in the number of copies.
One of the areas in which future studies of natural selection on CNVs may make the most impact is the implications such research has for the various models for the maintenance of gene duplicates (reviewed in Hahn 2009). Different models make explicit predictions about the role of adaptive natural selection in the maintenance of gene duplicates, and whether selection is acting on the duplicative mutation itself or on post-fixation nucleotide changes. The clearest examples may be in cases where there is selection for increased dosage of protein products (e.g. Amy1): selection requires no change in the underlying sequence and is simply acting to increase the total number of identical copies in an individual. Accumulating evidence for this form of selection—or for differences in protein function or gene expression for segregating duplicates—may move research on gene duplication from purely comparative to more mechanistic population-genetic studies.
7. Conclusions
It is now clear that individuals differ in the number of functional genes contained within their genomes. Although the technologies used to detect these differences can be computationally and technically challenging, they offer researchers a much richer view of molecular variation. As this variation has been found to underlie multiple adaptive phenotypes—and as new examples appear all the time—understanding the molecular basis for phenotypic differences will begin to require an accounting of all types of mutations, not just single-nucleotide differences.
Acknowledgements
We thank J. J. Emerson and A. Kern for very helpful discussions. The authors are supported by National Science Foundation grants DBI-0543586 and DBI-0845494.
References
- Bailey J. A., Gu Z. P., Clark R. A., Reinert K., Samonte R. V., Schwartz S., Adams M. D., Myers E. W., Li P. W., Eichler E. E.2002Recent segmental duplications in the human genome. Science 297, 1003–1007 (doi:10.1126/science.1072047) [DOI] [PubMed] [Google Scholar]
- Begun D. J., et al. 2007Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5, e310 (doi:10.1371/journal.pbio.0050310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbano H. A., et al. 2010Targeted investigation of the Neandertal genome by array-based sequence capture. Science 328, 723–725 (doi:10.1126/science.1188046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M. M., Long M.2010Mutational bias shaping fly copy number variation: implications for genome evolution. Trends Genet. 26, 243–247 (doi:10.1016/j.tig.2010.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreto L., Eiriz M. F., Gomes A. C., Pereira P. M., Schuller D., Santos M. A. S.2008Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics 9, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. K., Swartz J. D., Rush L. J., Alvarez C. E.2009Mapping DNA structural variation in dogs. Genome Res. 19, 500–509 (doi:10.1101/gr.083741.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. F., Andrews T. D., Carter N. P., Hurles M. E., Pritchard J. K.2006A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 38, 75–81 (doi:10.1038/ng1697) [DOI] [PubMed] [Google Scholar]
- Conrad D. F., et al. 2010Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (doi:10.1038/nature08516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth J. P., De Bie T., Stajich J. E., Cristianini N., Hahn M. W.2006The evolution of mammalian gene families. PLoS ONE 1, e85 (doi:10.1371/journal.pone.0000085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman E. B., Hartl D. L.2007A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 104, 19 920–19 925 (doi:10.1073/pnas.0709888104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E. E.2001Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 17, 661–669 (doi:10.1016/S0168-9525(01)02492-1) [DOI] [PubMed] [Google Scholar]
- Emerson J. J., Cardoso-Moreira M., Borevitz J. O., Long M.2008Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320, 1629–1631 (doi:10.1126/science.1158078) [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D.2007The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8, 610–618 (doi:10.1038/nrg2146) [DOI] [PubMed] [Google Scholar]
- Graubert T. A., et al. 2007A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 3, e3 (doi:10.1371/journal.pgen.0030003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D., et al. 2008Whole genome comparative studies between chicken and turkey and their implications for avian genome evolution. BMC Genomics 9, 168 (doi:10.1186/1471-2164-9-168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z. L., Cavalcanti A., Chen F. C., Bouman P., Li W. H.2002Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 19, 256–262 [DOI] [PubMed] [Google Scholar]
- Hahn M. W.2009Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100, 605–617 (doi:10.1093/jhered/esp047) [DOI] [PubMed] [Google Scholar]
- Hahn M. W., Demuth J. P., Han S. G.2007aAccelerated rate of gene gain and loss in primates. Genetics 177, 1941–1949 (doi:10.1534/genetics.107.080077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., Han M. V., Han S. G.2007bGene family evolution across 12 Drosophila genomes. PLoS Genet. 3, e197 (doi:10.1371/journal.pgen.0030197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P. J., Lupski J. R., Rosenberg S. M., Ira G.2009Mechanisms of change in gene copy number. Nat. Rev. Genet. 10, 551–564 (doi:10.1038/nrg2593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kreitman M., Aguade M.1987A test of neutral molecular evolution based on nucleotide data. Genetics 116, 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A., et al. 2009Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 84, 148–161 (doi:10.1016/j.ajhg.2008.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd J. M., et al. 2008Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (doi:10.1038/nature06862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J. O., et al. 2007Paired-end mapping reveals extensive structural variation in the human genome. Science 318, 420–426 (doi:10.1126/science.1149504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. S., Gutierrez-Arcelus M., Perry G. H., Vallender E. J., Johnson W. E., Miller G. M., Korbel J. O., Lee C.2008Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum. Mol. Genet. 17, 1127–1136 (doi:10.1093/hmg/ddn002) [DOI] [PubMed] [Google Scholar]
- Liu G. E., et al. 2010Analysis of copy number variations among diverse cattle breeds. Genome Res. 20, 693–703 (doi:10.1101/gr.105403.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke D. P., et al. 2006Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 79, 275–290 (doi:10.1086/505653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S.2000The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (doi:10.1126/science.290.5494.1151) [DOI] [PubMed] [Google Scholar]
- Maydan J. S., Lorch A., Edgley M. L., Flibotte S., Moerman D. G.2010Copy number variation in the genomes of twelve natural isolates of Caenorhabditis elegans. BMC Genomics 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S. A., et al. 2006Common deletion polymorphisms in the human genome. Nat. Genet. 38, 86–92 (doi:10.1038/ng1696) [DOI] [PubMed] [Google Scholar]
- McCarroll S. A., et al. 2008Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 40, 1166–1174 (doi:10.1038/ng.238) [DOI] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M.1991Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (doi:10.1038/351652a0) [DOI] [PubMed] [Google Scholar]
- McGrath C. L., Casola C., Hahn M. W.2009Minimal effect of ectopic gene conversion among recent duplicates in four mammalian genomes. Genetics 182, 615–622 (doi:10.1534/genetics.109.101428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R. P., Han M. V., Hahn M. W.2009A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol. Evol. 2009, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I., Man O., Lancet D., Gilad Y.2003Different noses for different people. Nat. Genet. 34, 143–144 (doi:10.1038/ng1160) [DOI] [PubMed] [Google Scholar]
- Mikkelsen T. S., Hillier L. W., Eichler E. E., Zody M. C., Jaffe D. B., Yang S. P., et al. 2005Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (doi:10.1038/nature04072) [DOI] [PubMed] [Google Scholar]
- Newman T. L., Tuzun E., Morrison V. A., Hayden K. E., Ventura M., McGrath S. D., Rocchi M., Eichler E. E.2005A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 15, 1344–1356 (doi:10.1101/gr.4338005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas T. J., Cheng Z., Ventura M., Mealey K., Eichler E. E., Akey J. M.2009The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res. 19, 491–499 (doi:10.1101/gr.084715.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R. M., Lanz C., Warthmann N., Weigel D.2008Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18, 2024–2033 (doi:10.1101/gr.080200.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., et al. 2006Hotspots for copy number variation in chimpanzees and humans. Proc. Natl Acad. Sci. USA 103, 8006–8011 (doi:10.1073/pnas.0602318103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., et al. 2007Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 (doi:10.1038/ng2123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., et al. 2008Copy number variation and evolution in humans and chimpanzees. Genome Res. 18, 1698–1710 (doi:10.1101/gr.082016.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., et al. 2006Global variation in copy number in the human genome. Nature 444, 444–454 (doi:10.1038/nature05329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C., et al. 2002Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837 (doi:10.1038/nature01140) [DOI] [PubMed] [Google Scholar]
- Schrider D. R., Hahn M. W.2010Lower linkage disequilibrium at CNVs is due to both recurrent mutation and transposing duplications. Mol. Biol. Evol. 27, 103–111 (doi:10.1093/molbev/msp210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B., Conrad D., Bateman A.2010Dosage sensitivity shapes the evolution of copy-number varied regions. PLoS ONE 5, e9474 (doi:10.1371/journal.pone.0009474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., et al. 2004Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (doi:10.1126/science.1098918) [DOI] [PubMed] [Google Scholar]
- She X. W., Cheng Z., Zollner S., Church D. M., Eichler E. E.2008Mouse segmental duplication and copy number variation. Nat. Genet. 40, 909–914 (doi:10.1038/ng.172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N. M., et al. 2009Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5, e1000734 (doi:10.1371/journal.pgen.1000734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J. E., Hahn M. W.2005Disentangling the effects of demography and selection in human history. Mol. Biol. Evol. 22, 63–73 (doi:10.1093/molbev/msh252) [DOI] [PubMed] [Google Scholar]
- Stranger B. E., et al. 2007Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–853 (doi:10.1126/science.1136678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. J., Miretti M., Rajan D., Fiegler H., Carter N. P., Blayney M. L., Beck S., Hurles M. E.2008aGermline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 40, 90–95 (doi:10.1038/ng.2007.40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Levine M. T., Eckert M. L., Begun D. J.2008bGenomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179, 455–473 (doi:10.1534/genetics.107.083659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., et al. 2005Fine-scale structural variation of the human genome. Nat. Genet. 37, 727–732 (doi:10.1038/ng1562) [DOI] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X. Q., Pritchard J. K.2006A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (doi:10.1371/journal.pbio.0040072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y. L., et al. 2008Adaptive evolution of UGT2B17 copy-number variation. Am. J. Hum. Genet. 83, 337–346 (doi:10.1016/j.ajhg.2008.08.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.2007The drifting human genome. Proc. Natl Acad. Sci. USA 104, 20 147–20 148 (doi:10.1073/pnas.0710524105) [DOI] [PMC free article] [PubMed] [Google Scholar]