Abstract
We are now reaching the stage at which specific genetic factors with known physiological effects can be tied directly and quantitatively to variation in phenology. With such a mechanistic understanding, scientists can better predict phenological responses to novel seasonal climates. Using the widespread model species Arabidopsis thaliana, we explore how variation in different genetic pathways can be linked to phenology and life-history variation across geographical regions and seasons. We show that the expression of phenological traits including flowering depends critically on the growth season, and we outline an integrated life-history approach to phenology in which the timing of later life-history events can be contingent on the environmental cues regulating earlier life stages. As flowering time in many plants is determined by the integration of multiple environmentally sensitive gene pathways, the novel combinations of important seasonal cues in projected future climates will alter how phenology responds to variation in the flowering time gene network with important consequences for plant life history. We discuss how phenology models in other systems—both natural and agricultural—could employ a similar framework to explore the potential contribution of genetic variation to the physiological integration of cues determining phenology.
Keywords: phenology, genetic architecture, life-history evolution, seasonal timing, local adaptation
1. Introduction
Within the last 50 years, drastic, directional shifts have occurred in the seasonal timing of many natural events including bud burst, flowering and migration (Fitter & Fitter 2002; Walther et al. 2002; Parmesan & Yohe 2003; Lehikoinen et al. 2004; Parmesan 2006; Bertin 2008; van Buskirk et al. 2009). The observed changes correspond in general to patterns of human-induced climate change (Rosenzweig et al. 2008). Advancing timing of spring events, alteration of range limits and clines and changing phenology in urban versus rural areas have all been demonstrated to mirror recent changes in temperature and growing season length (Roetzer et al. 2000; Bradshaw & Holzapfel 2001; Parmesan & Yohe 2003; Primack et al. 2004; Menzel et al. 2006a; Miller-Rushing et al. 2006; Miller-Rushing & Primack 2008). Shifts in seasonal timing are obvious indicators of climate change not only to scientists but also to the general public, and farming practices have already begun to adapt to altered climate patterns (Menzel et al. 2006a,b). As a result, the changing timing of biotic and abiotic indicators of season has recently received widespread popular and scientific coverage (e.g. Post et al. 2009).
Nonetheless, organisms' patterns of response are neither uniform nor universal, and the underlying causes of some common patterns remain mysterious. For example, in temperate environments, spring phenological events have advanced far further and more consistently than autumn events (Lehikoinen et al. 2004; Bertin 2008; van Buskirk et al. 2009; but see Ibáñez et al. 2010). Even within a given community, different species have shown contrasting long-term responses to directional climate change as well as to inter-annual variation in climate (Miller-Rushing & Primack 2008; Willis et al. 2008; Primack et al. 2009).
Under changing climates, the magnitude and flexibility of species phenological responses have many important consequences. Species responsiveness to year-to-year climate variation has been linked to long-term persistence versus local extinction in both bird and plant communities (Moller et al. 2008; Willis et al. 2008; Davis et al. 2010). Timing mismatches that are attributed to climate change have resulted in disrupted trophic interactions and altered competitive dynamics within community assemblages (Durant et al. 2005; Post & Forchhammer 2008; van der Jeugd et al. 2009; Singer & Parmesan 2010). Emphasis has been placed on understanding species tolerances to novel combinations of environmental factors (Williams & Jackson 2007), but the basis of this tolerance will probably depend on the way in which species respond phenologically to different environmental variables individually and in combination. Thus, to understand the basis of observed changes (or stasis) in phenological timing, and to make predictions for future responses, it will be necessary to have an understanding of the mechanisms underlying phenological response.
Plants serve as ideal model organisms in which to examine the mechanistic bases of seasonal response and adaptation. Plants come in many different life forms and inhabit a broad variety of geographical and seasonal habitats. And yet, plants are (for the most part) sessile and ectothermic, so they must cope with the climatic conditions into which they are dispersed. Nonetheless, plants can control the climatic conditions they experience during critical life stages through phenological control of dormancy, quiescence and/or the timing of developmental transitions. Several phenological traits in plants are of great economic importance and have been the object of extensive study; for instance, the timing of flowering and fruiting in cereals has been studied intensively because these plants supply the majority of food calories to the human population. Accumulated understanding of genetic and environmental influences on development, multiple seasonal traits and a rich history of manipulative experiments make plants prime candidates for studying how evolution has shaped phenology as a function of different external cues.
Understanding the genetic and physiological mechanisms that plants use for the timing of seasonal responses may allow us to predict phenological responses to no-analogue climates that will become increasingly common with anthropogenic climate change (Williams et al. 2007), as well as the capacity for adaptation under these scenarios. Such an understanding will also inform breeding strategies by highlighting signalling pathways and conditions under which sensitivities to different environmental factors are exposed. Thus, a more mechanistic understanding of phenology has become of major interest within the fields of conservation, ecology, evolution and agronomy, among others.
Here, we examine what is known about the seasonal cues to which plants respond, and the importance of these cues for appropriate timing of plant life-history events. Focusing on recent advances in uncovering the genetic mechanisms underlying seasonal traits, we elaborate on common themes and genetic architectures of plant responses. Finally, we explore genetically informed models of plant development and life history that link genetic architecture and sensitivity to differences in phenological response with geographical and temporal variation in climate.
2. Seasonal cues regulating plant phenology
Timing developmental events to coincide with favourable seasonal conditions is critical for plant growth, survival and reproduction. Spring, summer and autumn are characterized by different combinations of environmental cues (figure 1), and plant traits expressed in these seasons are subjected to distinct selection pressures. For example, early establishment in spring can provide competitive advantages, but not if it exposes delicate growing tissues to late frosts (Howe et al. 2003). Cues that precede or anticipate seasonal changes are particularly important because plant responses involve cellular, metabolic, morphological or developmental changes that require time to complete. Plants make use of several cues that serve as reliable indicators of season and thus resource availability, of which light and temperature are usually most important in temperate plant species. The environmental sensitivity of many plant life cycles reflects these different life-history strategies in both natural and agricultural settings.
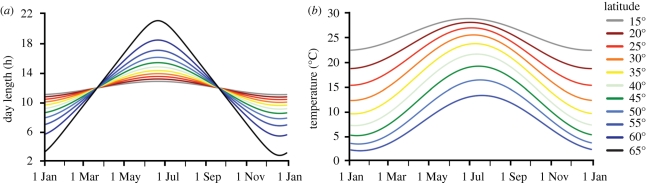
Figure 1.
(a) Seasonal variation in photoperiod (from Ham 2004) from 15° to 65° latitude and (b) daily average temperatures (from Charles-Edwards et al. 1986) from 15° to 55° latitude. The yearly range in photoperiod increases with latitude, and the amplitude of photoperiod increases more rapidly towards the poles. In general, the lag between daily temperature and photoperiod cycles increases with latitude.
Temperature is a seasonal cue that cycles annually in temperate climates following patterns of day length and insolation (figure 1b). Ambient temperature also directly affects growth and development rates. Such rates typically increase with ambient temperatures up to some optimum or maximum, and then decline as warming continues. In temperate environments, however, optimum ambient temperatures for growth are rarely exceeded (e.g. Schaber & Badeck 2002). Many plants also respond to cold temperature cues, typically referred to as chilling or vernalization effects. For sensitive traits, passage through a cold season accelerates the subsequent pace of development (Henderson et al. 2003). For traits that respond to chilling, changes in seasonal temperature can have complex effects on phenology when the generally promotive effects of increasing temperature oppose the influence of reduced vernalization (see below). Plant life-cycle events that occur in spring often rely on vernalization as well as photoperiod and/or warming temperature cues. In these traits, response to increasing day length (or ambient temperature) is greatly amplified following prolonged exposure to cold that serves as an indication that winter has passed (Harrington et al. 2010).
Light quantity contributes to plant growth and development, but day length can also serve as an important developmental cue. Decreasing day lengths are reliable cues of the impending end of the growing season and winter onset for many temperate biomes; increasing day length indicates the arrival of spring (figure 1a). Bud-set timing is more influenced by declining day lengths that indicate the approach of autumn than by low temperatures per se (Bohlenius et al. 2006; Savolainen et al. 2007), most probably because declining photoperiods are a more reliable indicator of the end of the growing season. Day length can also serve as an important cue for the appropriate timing of flowering and fruiting with respect to seasonal patterns of temperature and precipitation. Spring-flowering, Mediterranean-adapted plants (e.g. barley, wheat) often accelerate development in response to lengthening days, which allows them to complete their life cycle before the hot, dry conditions of summer. In tropical plants such as sorghum, the shortening days of late summer can serve as a cue signalling the end of summer and the onset of the autumn monsoon rains, which are favourable for grain filling (Dingkuhn et al. 2008). Owing to its dependable annual cycle, plants use day length as an important cue of season, and the genes involved in response to photoperiodic events are anciently conserved (see below).
Precipitation affects both plant survival and growth and can also show strong seasonal patterns. In seasonally dry communities, the initiation of seasonal growth (as measured by greenness at a landscape scale) closely tracks the onset of rains (Zhang et al. 2006). It is unclear whether moisture in itself serves as an anticipatory cue, or whether plants use other seasonal cues to become competent to respond to precipitation once it arrives. Pre-formation of leaves or other organs whose emergence depends on permissive moisture conditions may allow plants to get a ‘jump start’ when favourable conditions arrive (Damascos et al. 2005). Whether or not precipitation serves as a cue, water availability may determine the length of the growing season and thus can have important effects on the relationship between phenology and fitness (Franks et al. 2007).
Day length and temperature can serve as reliable cues of seasonal conditions across a broad range of temperate climates and geographical scales, and plants use both cues to appropriately time important life-cycle events; however, the temperatures and photoperiods indicative of season vary. Even under current climate conditions, the appropriate cue of a favourable seasonal environment in one location may not be the same in another (figure 1 and table 1). For instance, the characteristic day length three weeks prior to autumn frost falls precipitously with latitude. The underlying geographical distribution of relevant seasonal environmental cues and resources can serve as an important driver of locally adapted phenological responses. That is, given geographical variation in the seasonal availability of different resources, plants might be expected to and often do show distinct phenologies in different habitats. And yet, many plant species have broad distributions. How do species adapt to this variation within their range? When is it advantageous to have populations with rigid seasonal responses, and when is it advantageous to respond plastically to environmental cues? Understanding the physiological and genetic basis of phenology can help to answer these questions.
Table 1.
General, large-scale patterns in the seasonal distribution of temperatures. (Compiled from information in Landsberg (1941), Sellers (1965), Akin (1990) and Linacre (1992).)
| winter temperature | summer temperature | yearly temperature range | diurnal temperature range | seasonal lag | |
|---|---|---|---|---|---|
| proximity to equator | significantly warmer | slightly warmer | reduced | reduced | reduced |
| proximity to ocean | significantly warmer | slightly warmer | reduced | reduced | increased |
| northern versus southern hemisphere | slightly cooler | significantly warmer | increased | ||
| proximity to forest | slightly cooler | significantly cooler | reduced | reduced | |
| increase in altitude | slightly cooler | significantly cooler | variable, generally reduced | variable | reduced |
3. Genetic basis of phenological response
(a). Flowering time gene network in Arabidopsis thaliana
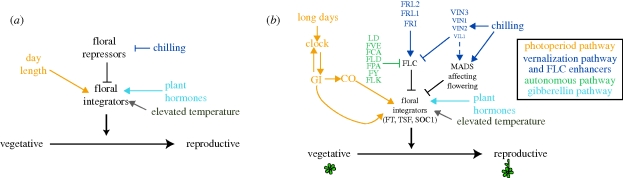
The converging genetic signalling pathways mediating environmental response of flowering time have been particularly well studied in the model annual plant Arabidopsis thaliana (figure 2). Arabidopsis thaliana integrates the environmental signals of long days, growing degree days and winter chilling, all of which speed the rate of development towards flowering. Under long days, the photoperiod pathway promotes flowering via the transcriptional regulator CONSTANS (CO; Koornneef et al. 1991; Lee & Amasino 1995) and its upstream activator GIGANTEA (GI; Mizoguchi et al. 2005). These signals activate floral integrator genes including FLOWERING LOCUS T (FT), TWIN SISTER OF FT (TSF) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1; Kim et al. 2005; Yamaguchi et al. 2005; Kobayashi & Weigel 2007), which in turn promotes the transition from vegetative to reproductive development. Higher ambient temperatures speed the accumulation of growing degree days and also promote flowering (Granier et al. 2002; Blazquez et al. 2003; Welch et al. 2003; Lempe et al. 2005; Balasubramanian et al. 2006b).
Figure 2.
(a) General diagram of environmentally sensitive flowering time pathways in plants. (b) Simplified diagram of the network of the environmentally sensitive flowering time pathways in Arabidopsis thaliana. See text for details.
The ability of floral integrator genes to respond to inductive signals is mediated by a suite of repressor genes, notably the MADS-box transcription factor FLOWERING LOCUS C (FLC) and related MADS-box genes such as SHORT VEGETATIVE PHASE (SVP), FLOWERING LOCUS M (FLM or MAF1) and MADS AFFECTING FLOWERING 2–5 (MAF2-5; Alexandre & Hennig 2008). FLC is activated by genes such as FRIGIDA (FRI; Geraldo et al. 2009) and its relatives FRIGIDA-LIKE1 and 2 (FRL1, FRL2; Michaels et al. 2004; Schlappi 2006), and is repressed by genes in the ‘autonomous’ or ‘endogenous’ pathway (Baurle & Dean 2006). Autonomous pathway genes are identified as those that affect flowering regardless of the environment (or in all environments), and this pathway is thought to be sensitive to internal or endogenous signals of developmental stage. Attenuation of floral repressors can also be achieved through prolonged winter chilling (vernalization) that induces expression of VERNALIZATION-INSENSITIVE-3 (VIN3), which initiates stable epigenetic repression of FLC via the vernalization pathway (Sung & Amasino 2004; Finnegan & Dennis 2007). Deficiencies in FLC activators such as FRI remove the vernalization requirement for flowering, and under laboratory conditions result in a ‘rapid-cycling’ life history (Johanson et al. 2000; Michaels et al. 2003; Boss et al. 2004; Lempe et al. 2005; Moon et al. 2005; Shindo et al. 2005; Searle et al. 2006; Schmitz et al. 2008). Through this complex network of converging pathways, Arabidopsis plants balance different seasonal cues in order to time flowering appropriately (Boss et al. 2004; Wilczek et al. 2009; see below).
Historically, A. thaliana genes underlying variation in flowering time have been discovered and described from forward genetic screens carried out in controlled conditions using a combination of mutagenized and transformed lines, recombinant inbred lines (RILs) and naturally occurring variation in wild-collected accessions (Alonso-Blanco et al. 2005, 2009; Engelmann & Purugganan 2006). The genetic basis of flowering time has received a great deal of attention both because of its agronomic relevance and also because there is enormous variation in flowering time exhibited in the laboratory and in natural populations (Koornneef et al. 2004). Explicit genetic models based on gene expression profiles and interactions have successfully modelled behaviour and feedback integration of the Arabidopsis circadian clock (Locke et al. 2005, 2006; Zeilinger et al. 2006; Salazar et al. 2009) and time to flowering as a function of temperature and day length in various mutant lines (Dong 2003; Welch et al. 2003, 2005). Much interest has been focused on working out the signalling pathways involved using both experimental and modelling approaches, but understanding the synthesis by floral integrator genes of signals from these pathways and their relative importance in different environments has proved more difficult.
The contributions of the different flowering time candidate genes and pathways under complex combinations of environmental factors have begun to be explored only recently. Field studies have largely validated the described roles of these flowering time genes, but several studies in natural conditions or with natural populations have highlighted the conditions (both environmental and genetic) in which variation in these signalling pathways is expressed. For instance, studies with field-sown RILs have demonstrated both site- and season-specific quantitative trait loci (QTLs) (Weinig et al. 2002; Malmberg et al. 2005). Recent controlled environment studies have included more complex temperature and photoperiod interactions, and perhaps as a result have uncovered more subtle environment-dependent pleiotropic, epistatic and dominance effects of known flowering time genes (Li et al. 2006; Scarcelli et al. 2007; Scarcelli & Kover 2009). Together, these studies have highlighted how a combination of field and controlled environment studies can be used to explore genetic determination of phenological traits and their role in adaptation to environment.
(b). Genetic architecture of seasonal sensitivity in plants
Among important plant seasonal responses, the timing of flowering, fruit or grain production, bud burst and bud set have all been extensively studied in crop and forestry species (Cooper & Hammer 1996). The genetic basis of these traits is important for plant breeding and improvement strategies (Cooper & Hammer 1996; Hammer et al. 2006) as well as for predicting responses to changing climates (Davis et al. 2005; Aitken et al. 2008). Extensive quantitative genetic studies have demonstrated that most of these phenological traits have a heritable genetic basis (reviewed in Cooper & Hammer 1996; Howe et al. 2003; Savolainen et al. 2007). In temperate species, the loci involved appear to respond to one or more of a combination of factors including endogenous developmental status, day length and chilling (figure 2; Colasanti & Coneva 2009). We focus here on phenology in temperate regions because of the greater seasonality in these areas, and the greater amount of data available. Since the timing of seasonal events in plant life cycles has important fitness consequences in natural habitats, the genetic architecture of response bears the signature of past evolution and depends on habitat, type of signal and life history of species considered. Seasonal traits in many plant species have a similar architecture of underlying sensitivity to environmental factors involving integration of temperature, day length and chilling cues (see above; Howe et al. 2003; Cockram et al. 2007b; Savolainen et al. 2007).
More recently, specific genes involved in phenology and seasonal traits have been described in several plant species (see review in Alonso-Blanco et al. 2009). These genes have been characterized through complementary approaches that include identifying the causal loci of QTL through positional cloning as well as identification and characterization of orthologues of known flowering-time genes from model species, particularly Arabidopsis. The genetic module involved in photoperiod integration is remarkably ancient, which may have advantages for understanding the functional basis and manipulation of day length responses across a wide range of important plant species. Several photoperiod genes identified initially through quantitative genetic studies have since been revealed to be orthologous to genes in the photoperiod pathway in Arabidopsis (e.g. from rice, barley, wheat). The CONSTANS gene family in particular is involved in photoperiodic response in all plants studied, including the bryophyte Physcomitrella (Zobell et al. 2005), and is even implicated in the photoperiodic response of growth and starch accumulation in the green alga Chlamydomonas (Serrano et al. 2009). However, expression of CO in planta has different effects on phenology depending on the downstream targets. In both Oryza sativa (rice) and A. thaliana, CO and its rice orthologue Hd1 combine signals from the diurnal clock oscillator with the outputs of photoreceptors to measure day length as per the external coincidence model (Hayama & Coupland 2004). However, whereas high levels of CO upregulate the floral integrator FT in A. thaliana, expression of rice Hd1 represses the FT rice orthologue Hd3a, thus producing a short-day flowering response (Hayama et al. 2003).
Meristem identity (i.e. vegetative versus floral) and floral integrator genes are also highly conserved, particularly FT, which has been shown to act as a mobile signal acting in the manner of ‘florigen’ in rice (Tamaki et al. 2007), tomato (Lifschitz et al. 2006) and Arabidopsis (Corbesier et al. 2007), with similar roles suggested in several other plant species including grasses, sunflowers, poplar and morning glory (Turck et al. 2008; Kikuchi et al. 2009; Blackman et al. 2010). Related genes that, like FT, carry a phosphatidyl ethanolamine-binding protein domain appear to be involved in the determination of phenological traits and/or onset of reproduction over evolutionary time dating all the way to bryophytes (Hedman et al. 2009) and spruce (Gyllenstrand et al. 2007). Despite conservation of involvement, the details of the environmental sensitivity of these integrator genes and their interaction with other floral/seasonal trait network genes may differ by species (Nemoto et al. 2003).
There appears to be greater variation in the genes underlying the response to chilling, although lifting of repression following chilling is a common response among temperate plants. While nonetheless variably involved, MIKCC-type MADS box genes including FLC orthologues are implicated in integrating vernalization cues and repressing flowering or growth pre-chilling across a broad range of plant taxa including sugar beet, citrus and peach (Kim et al. 2007; Reeves et al. 2007; Li et al. 2009; Zhang et al. 2009). In cereals, however, not all genes involved in vernalization sensitivity fall in this family (Trevaskis et al. 2007; Greenup et al. 2009). PEP1, an FLC orthologue in the perennial species Arabidopsis alpine, is involved in both flowering time as well as the resumption of vegetative growth after chilling, which is accompanied by a ‘resetting’ of the epigenetic state of this gene (Wang et al. 2009). Arabidopsis alpina is one of the few species for which we understand how genes involved in non-circadian environmental sensing reversibly shift their sensitivity within a single individual plant. (In the annual A. thaliana, the epigenetic resetting of FLC occurs during embryo development; Sheldon et al. 2008.) Understanding these resetting processes will be critical to determining the mechanistic bases of annually recurring traits in perennial species as well as in different traits that use the same genes as environmental reporters.
Plant traits that are expressed in different seasonal environments appear to have distinct genetic bases, which might provide greater response flexibility (both in short and evolutionary time frames) if they are responding to different cues. On the other hand, genes involved in diverse developmental events that occur in the same season may be jointly regulated, as has been recently described for cold tolerance and vernalization responses in cereals (Galiba et al. 2009). Autumn traits in trees such as timing of bud set and cold hardening also appear to be genetically correlated, but there is little relationship between these traits and bud burst in the spring (Howe et al. 2003; Savolainen et al. 2007). Additionally, in tree species such as poplar, autumn traits may have greater heritability and greater standing genetic variation than spring traits, which respond more plastically to local environmental conditions (e.g. Hall et al. 2007; Luquez et al. 2008). Oddly, results of those quantitative genetic studies that find little correlation between different seasonal traits often contrast with functional studies of individual loci that are implicated in several seasonal responses. For example, the poplar orthologue of FT is involved in both bud set (autumn) and flowering (spring) and shows a strong north–south cline in the timing and environmental sensitivity of activity (Bohlenius et al. 2006). This seeming paradox remains to be resolved, but may involve shifting upstream or downstream interactions, with the relevant genetic variation identified in quantitative genetic studies located in these interacting genes (Ingvarsson et al. 2006, 2008).
To summarize, many plant species and traits show similar genetic architecture of seasonal response, as might be expected given the commonality of cues indicating season. We are still in the early stages of identifying and characterizing the individual genes underlying sensitivity to photoperiod, temperature and developmental state in most plant species. Work to date has shown that many of these environmental-sensing modules involve orthologous genes or similar gene family members even across deep evolutionary divides, which suggests that a unified understanding of the genetic basis of phenology may be a tractable goal.
(c). Intraspecific variation in sensitivity to environmental cues
Where phenology contributes to local adaptation, we might expect to see genetic differentiation in response across the native range of species. Combinations of light and temperature that correspond to the beginning and end of the growing season vary geographically (table 1 and figure 1), and clinal patterns in environmental sensitivity may allow species to respond appropriately to local seasonal cues (see also Chuine 2010). For example, autumn frosts arrive earlier in the north where day lengths before the equinox are also longer. Most probably reflecting this latitudinal trend, many tree species show marked clines in the critical short-day length that induces bud set at both the phenotypic and allelic level, resulting in bud set at earlier calendar dates and longer days in more northern populations (Bohlenius et al. 2006; Hall et al. 2007; Luquez et al. 2008). Within species or genera, chilling cues may become subordinate to photoperiod cues in the timing of bud burst closer to the tropics and/or the temperatures required to fulfil chilling requirements may rise (Borchert et al. 2005; Wilkie et al. 2008; Colasanti & Coneva 2009). In Arabidopsis, clines in vernalization response (Lempe et al. 2005; Shindo et al. 2005; Stinchcombe et al. 2005), light sensitivity (Maloof et al. 2001; Stenoien et al. 2002), circadian clock period (Michael et al. 2003) and day length sensitivity (Balasubramanian et al. 2006a; but see Samis et al. 2008) have all been identified. Overall developmental rates or response to endogenous cues may also shift with climate. A broad survey of Arabidopsis accessions revealed that lines from cooler native climates were more responsive to warmer temperatures (Hoffmann et al. 2005); more rapid flowering under warmer temperatures might reflect the need to complete growth in a shorter growing season (cf. Bradshaw & Holzapfel 2008). In the northeast USA, there is apparent community-level, landscape-scale differentiation in the environmental cues influencing bud burst, although this may involve both changes in intraspecific variation and species-level shifts in plant assemblage composition (Fisher et al. 2007). The elucidation of clines or lack of clines in phenological traits and in the genes underlying these traits can give important information about the past selective history of species (Joost et al. 2007) as well as their ability to respond to changing environments (Aitken et al. 2008).
The selection of varieties within crop species provides an interesting counterpoint to the natural clines set up over evolutionary time through natural selective events. Many crop plants including wheat and barley have both ‘spring’ and ‘winter’ varieties. At a phenotypic level, these are distinguished by a loss of vernalization sensitivity in spring-sown varieties. More recent work has revealed that this change typically involves only a few loci and alleles in wheat and barley (Cockram et al. 2007a,b, 2009; Jones et al. 2008). Photoperiod sensitivity also plays an important role in the seasonal timing of grain production, but this usually marks differences in varieties sown in the same season but in different geographical regions. As one moves north, sensitivity to photoperiod results in yield losses in the Mediterranean-adapted winter wheat (Worland et al. 1998; Cockram et al. 2007a). In the south, flowering and heading under lengthening days allow plants to avoid the dry conditions of summer, but in the north, the lower temperatures and wetter summers mean that early flowering induced by lengthening days results in flowering at smaller size and with less productivity. Land races of both barley and wheat show strong south-to-north clines of declining photoperiod sensitivity and allele frequency of the major mutation that causes this insensitivity (Turner et al. 2005; Cockram et al. 2007a; Lister et al. 2009). Soybean, a short-day plant, exhibits a similar cline in which the flowering time of northern landraces is relatively insensitive to day length, but also—in contrast to wheat and barley—uniformly rapid (Zhang et al. 2008). Interestingly, this pattern of decreased photoperiod sensitivity in northern populations in agricultural settings is in direct contrast to the common pattern of many natural species such as Arabidopsis that show increased photoperiod sensitivity and more extreme critical day lengths in northern populations (see also above; Ray & Alexander 1966; Griffith & Watson 2006). The factors that drive selection of ideally suited local crop varieties and how these might differ from natural seasonal selection pressures remain an interesting topic to explore further.
4. Phenology and life-history variation in arabidopsis thaliana
(a). Linking genetic sensitivity to the timing of flowering
Arabidopsis thaliana's well understood flowering time network and extensive natural variation in flowering time genes make it an ideal model in which to explore the link between genetic factors and phenological response. Moreover, this weedy annual species inhabits a wide range of climates across its native range in Europe and central Asia and exhibits distinct geographical patterns in its seasonal life history. According to field observations, at high latitudes, A. thaliana behaves as a winter annual, germinating in autumn, overwintering as a rosette and flowering in the spring soon after snowmelt (Petipas et al. in preparation). Similarly, in Mediterranean climates close to its southern range limit, Arabidopsis overwhelmingly germinates in autumn and flowers in spring (Montesinos et al. 2009). However, these two winter annual life histories differ dramatically in total length. Populations in Oulu, Finland, near the Arctic Circle germinate in September and flower in May (Petipas et al. in preparation), while those near the Mediterranean coast in Spain germinate in November and flower in February (Wilczek et al. 2009). In northwestern European locations including the UK and Germany, autumn germinants display diverse flowering times spanning from later autumn to mid-spring (Thompson 1994; Wilczek et al. 2009). In these climates, rapid-cycling life histories have also been observed in which individuals germinate in spring or summer and flower within one to two months of emergence (Thompson 1994; Wilczek et al. 2009). Field studies have demonstrated that both the germination timing of A. thaliana (reviewed in Donohue 2009) and genetic differences in integration of environmental signals after germination can contribute to spatial and temporal variability of life-history expression; however, the complex interplay of environmental and genetic factors underlying life-history variation in natural populations has remained largely unexplored.
Studies that sample geographical and seasonal variation in climate can help inform the relative contribution of environmental inputs and genetic sensitivity that underlies the observed diversity of natural responses. Using climate and phenology data from Arabidopsis lines grown in a multinational European field study, we created a genetically informed photothermal model of development that successfully explained over 90 per cent of the variation in the timing of flowering in wild-type plants and mutant lines carrying disruptions to the gene pathways involved in environmental sensing (Wilczek et al. 2009). This model, the first to predict quantitatively the integration of these pathways in a field setting, thus provides a powerful tool for examining the balance of genetic and environmental factors in determining life history in complex natural environments. We focus here specifically on four genotypes harbouring mutations affecting day length and vernalization response. The Columbia ecotype (Col, wild-type) is a widely studied accession from northwest Europe that is rapid-cycling in laboratory studies and carries a natural lesion in the locus FRI that mediates the strength of the vernalization response (figure 2b). We also studied a line in the Col background into which a natural functional FRI allele from the Spanish ecotype Sf-2 had been introgressed (Col FRI), resulting in greater initial floral repression and a more pronounced vernalization response (Lee & Amasino 1995). Comparisons between Col and Col FRI revealed the effects of vernalization sensitivity on the pre-flowering vegetative interval. Understanding the allelic effect of FRI is of special interest because both null and functional alleles at the FRI locus are found in natural populations, and chamber studies have suggested that FRI is involved in determining life-history variation in the wild (Johanson et al. 2000; Boss et al. 2004; Lempe et al. 2005; Shindo et al. 2005). We also considered a mutant line (Col FRI vin3) that does not respond developmentally to winter chilling, and by comparing this line with its control (Col FRI), we were able to assess the contribution of vernalization signals to developmental rate and timing of flowering. Finally, we grew a mutant in the day-length-sensing photoperiod pathway (Col gi), with which it was possible to quantify the importance of long-day signals.
Plantings of these lines in five European common gardens revealed important differences in the sensitivity of flowering time to genetic perturbation depending on site and season of growth, with most of this variation in response captured by our photothermal model. For example, in the mild oceanic climate of Norwich, UK, where wild Arabidopsis cohorts germinate naturally throughout much of the year, a wide range of genotypes are expected to flower rapidly when they germinate in spring and summer (Wilczek et al. 2009). Using 2 years of on-site weather data to simulate reaction norms of life history with our developmental model, we predicted that later autumn germinants would overwinter in the vegetative state and transition to reproductive growth (bolt) at similar times in the spring. Thus, in a narrow germination window in the early autumn, bolting time was exceptionally sensitive to both small changes in genetic background and germination timing. Outside of this window, the effects of genotype on bolting time were muted and no single allelic change resulted in a major life-history conversion. The predicted sensitivity window was observed in field plantings of Col and Col FRI, where the presence of a functional FRI allele caused a life-history conversion only during a limited portion of the year, with the exact timing depending on the climate at the growth site (Wilczek et al. 2009). Thus, genetically informed developmental modelling approaches can be used to highlight the environmental signals that influence life history in different environments, and the sensitivity of life-history variation to genetic variation in signalling pathways.
Because the photothermal model uses detailed plant-level measurements of temperature, initial explorations of life history were limited to the five sites and 2 years for which we had such data (Wilczek et al. 2009). In order to generalize the photothermal model to other years and locations, we needed to convert the widely available temperature data (daily maxima and mimima at 1.5 m) to hourly, ground-level temperatures. Therefore, we developed a set of simple conversions that captured over 99 per cent of the variation in bolting-time-relevant temperatures across our five sites (appendix A). Clearly, the exact correspondence of simulated life-history data to finer-scale geographical patterns will depend largely on the accuracy and precision of the underlying climate models (for future climate scenarios) and the grain of recorded air temperature data (for spatial extrapolation). Our simulations of the timing of reproduction using available climate models nevertheless broadly reproduce the patterns we observed in our European field plantings (see below). Here, we expand our analysis temporally via projected future climate scenarios and spatially across the native range in order to illustrate large-scale patterns of Arabidopsis life history under changing climates.
(b). Contributions of vernalization sensitivity to life history of Arabidopsis thaliana under changing climates
Our first example is a temporal analysis of patterns of life-history variation under current and projected climates in Norwich, UK, where natural A. thaliana cohorts are seen throughout the year. Comparing on-site temperature data from Norwich in 2006–2008 against projected temperatures for current and end-of-century future climates under the A1B scenario (NOAA GFDL 2004; Delworth et al. 2006) revealed some startling differences. We found that while real and simulated minimum temperatures were in general agreement for the current time period, real air temperature maxima were consistently higher than even simulated future maxima. The years 2006–2008 were all considerably warmer than the 1961–2000 regional average (UK National Weather Service 2009); an alternate explanation for our high measured temperatures is that the microclimate at our site is warmer than that of the 2.5° × 2.5° geographical grid cell that includes Norwich. Owing to the warm temperature maxima experienced by field plants in 2006–2008, bolting responses of those plants mimic more closely the responses of plants to projected future climates (figure 3).
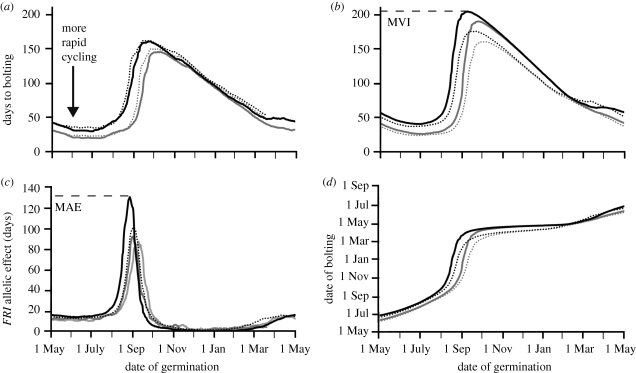
Figure 3.
(a) Days to bolting as a function of germination date from May 2006 to May 2007 and May 2007 to May 2008 for genotypes (Col, Col FRI) differing in vernalization sensitivity. Predicted development time calculated using on-site plant-level measurements of temperature in Norwich, UK. Data for 2006–2007 reproduced from Wilczek et al. (2009). Solid grey line, Col 2006; dotted grey line, Col 2007; solid black line, FRI 2006; dotted black line, FRI 2007. (b) Days to bolting as a function of germination date in projected current (average for 2004–2009) and future (2094–2099) climates for genotypes (Col, Col FRI) differing in vernalization sensitivity. A dashed line shows the maximum vegetative interval (MVI) for Col FRI under current conditions. Predicted development time was based on 6 year average time to bolting using simulated 2004–2009 temperatures and for 6 year average using simulated 2094–2099 temperatures in Norwich, UK. Temperature projections from the A1B scenario. Solid grey line, Col present; dotted grey line, Col future; solid black line, FRI present; dotted black line, FRI future. (c) The effect of functional FRI on flowering time under current measured climates, as well as projected current and future climates, using data from (a) and (b). A dashed line shows the maximum allelic effect (MAE) of FRI under current conditions. Solid light grey line, 2006; solid dark grey line, 2007; solid black line, present; dotted black line, future.(d) Date of bolting as a function of germination date in projected current (average for 2004–2009) and future (2094–2099) climates for genotypes (Col, Col FRI) differing in vernalization sensitivity. Data replotted from (b). Key to plots as for (b).
Simulations of bolting as a function of germination date under warmer future climates at this site indicated that the impact of FRIGIDA-mediated vernalization sensitivity will remain qualitatively similar to its life-history effect in cooler climates. A functional FRI allele increased the amount of time elapsed between germination and flowering (figure 3a) or the predicted maximum vegetative interval (MVI) both in current and future climates. Further, the predicted magnitude of the FRI allele effect was not much altered in future climates for late spring and late summer germinants (figure 3c). In contrast, for FRI functional plants germinating from late January through early April, a decrease in late winter chilling actually delayed bolting in warmer, as compared with cooler, climates (figure 3b). However, climate change did cause quantitative shifts in both the seasonal timing and magnitude of life-history transitions. For cohorts germinating throughout much of the late autumn and winter, FRI had no allelic effect on flowering time, yet under projected future climates, the flowering of late autumn germinants occurred earlier in the spring for both Col and Col FRI genotypes (figure 3d). Additionally, we observed a shift in the timing of the sensitivity window, where the change from rapid-cycling, autumn-flowering behaviour to spring-flowering behaviour occurred at a later germination date in warmer climates and with loss of FRIGIDA function (figure 3b).
The maximum allelic effect (MAE) of FRI on flowering time (figure 3c) is a measure of the largest potential effect on the life history of a given perturbation of gene function. The MAE of FRI not only decreased in magnitude under warmer climates, but also the germination dates on which the MAE occurred shifted later in the autumn season. The attenuation of the influence of FRI under climates with less winter chilling may seem counterintuitive, given that prior to chilling FRI functional plants develop more slowly and that FRI has the strongest effect in constant warm conditions in controlled environment studies (Lempe et al. 2005; Balasubramanian et al. 2006b; Shindo et al. 2006). The weakening of chilling cues that would equalize the developmental rates of the two genotypes was counterbalanced as warmer temperatures accelerated overall development, leading to decreased MVI in both genetic backgrounds. Thus, regardless of the maintenance or even exaggeration of relative differences in developmental rates between the two genotypes with warming climates, in most cases (with the exception of some spring germinants, figure 3b), their difference in flowering time as measured in days decreased (figure 3c).
Based on our projections in Norwich, we find that FRIGIDA genotype alone is grossly insufficient to explain life-history variation at this site in both current and future climates. We would not expect variation in FRI status to account completely for rapid-cycling versus winter annual life histories in co-occurring populations that have been observed in the UK, despite the fact that FRI has a measurable effect on flowering time throughout most of the year (figure 3c). Instead, the length of time spent as a vegetative individual prior to flowering depended largely on the timing of germination (figure 3b), with genotype playing a major role in flowering phenology only for a subset of autumn-germination cohorts (figure 3c). What emerges in this analysis of response to climate change at a single location is a complex set of relationships between genetic sensitivity, seasonal distribution of environmental cues and the timing of reproduction that can nonetheless be precisely predicted and mapped when the physiological bases of these developmental processes are understood.
(c). Genetic sensitivity and life-history variation across the native range of Arabidopsis thaliana
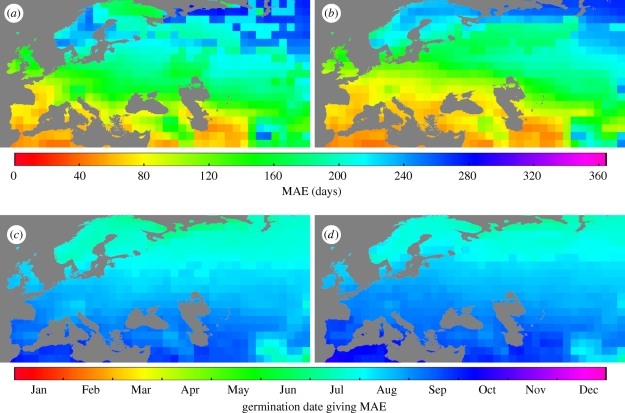
What then drives variation in life history in this species? What is the effect of the candidate locus FRIGIDA on life-history expression across a range of different seasonal climates? How conserved is the pattern of sensitivity of life-cycle length to germination timing under different climates? To address these questions, we expanded our phenological analysis of FRI to a broader geographical scale encompassing much of the species's native range in Europe and central Asia. From simulations of time to bolting (length of vegetative interval) as a function of germination date under current and future projected climates, we found that early summer germinants of both Col and Col FRI showed very little variation in life-cycle length (appendix A; electronic supplementary material, movies S1 and S2). These rapid-cycling cohorts transitioned from vegetative to reproductive growth within two months of germination regardless of geographical location or genotype, although FRI functional plants generally required about 10 more days to reach bolting. As our simulations progressed through summer and into autumn-germination cohorts, the seasonal transition from rapid-cycling to spring-flowering life history as a function of germination date occurred in a wave from north to south, with the onset of this wave coming earlier in FRI functional plants and in current (versus future) climates.
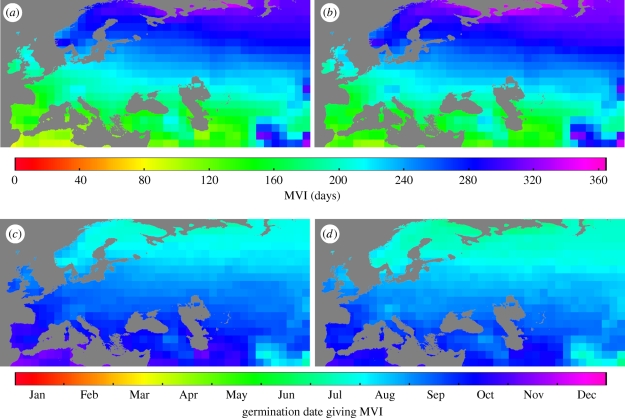
To characterize geographical patterns in life-cycle variation, we determined the MVI between germination and bolting for each genotype, and the date of germination on which life-cycle length was maximized (figures 3b and 4). Since the minimum life-history length achieved by summer germinants was similar everywhere, the MVI is representative of the magnitude of seasonal variation in life history at a given location; the date on which this transition to spring-flowering occurs gives an indication of the germination season in which the window of life-history sensitivity occurs. We found that more northern populations had the greatest variation in life-cycle length (figure 4a,b). Depending on location (and genetic background to a lesser extent), the MVI varied from less than 3 months to over a year. Despite transitioning to spring-flowering behaviour in earlier germination cohorts (figure 4c,d), autumn germinants in northern populations flowered later in the spring (electronic supplementary material, movies S1 and S2) so that both the length of the life cycle and the seasonal timing of flowering showed geographical structure. Under projected future climates, these patterns were largely maintained but shifted slightly northwards (electronic supplementary material, figure S1).
Figure 4.
Current geographical patterns of (a,b) the maximum vegetation interval (MVI) between germination and bolting, and (c,d) the germination date on which the MVI occurs in A. thaliana accessions that differ in vernalization sensitivity. (a,c) Col ecotype (Col) and (b,d) Col ecotype with functional FRIGIDA (Col FRI). Estimates of bolting time are based on a 6 year average from 2004 to 2009 under the A1B projection scenario.
In order to better understand the effect of FRI on life history in current and future climates, we looked at the maximum life-history effect owing to allelic variation in FRI and the germination date on which this MAE is achieved. We used these values to assess the magnitude and timing of the life-history transition caused by changes in the strength of vernalization response. The magnitude of the difference in life-cycle length between ecotypes that differ in FRI functionality, and thus vernalization response, decreased with latitude (figure 5a) and, generally, with warming climates within sites (figure 5b).
Figure 5.
Geographical patterns in the maximum difference in bolting time (maximum allelic effect, MAE) between A. thaliana ecotypes that differ at the FRIGIDA locus under (a) current (2004–2009) and (b) projected (2094–2099) climates. The time of year at which this life-history transition window occurred also showed geographical variation in both (c) current and (d) projected climates across the native range of the species.
Other perturbations to the environmentally sensitive flowering pathways in A. thaliana revealed distinct geographical and temporal patterns of life history. Complete genetic insensitivity to vernalization signals, such as that found in Col FRI vin3 plants, resulted in transition to spring-flowering behaviour at even earlier summer and autumn germination dates accompanied by greater increases in the MVI (electronic supplementary material, figure S2 and movie S3). The maximum effect of the VIN3 locus was in general smaller than at the FRI locus and showed a stronger oceanic to continental gradient, even while the timing of this life-history transition showed a clear latitudinal pattern (electronic supplementary material, figure S4). Col and Col FRI plants differed in developmental rate prior to exposure to chilling cues, while Col FRI and Col FRI vin3 plants diverged after chilling had occurred. Thus, changes in vernalization sensitivity can have disparate effects on life-history expression depending on details of the response mechanism (i.e. genetic basis), seasonal timing of germination and geographical location.
The effects of the photoperiod-sensing pathway on flowering time also varied strikingly with site and season. At lower latitudes, both plants that are insensitive to the accelerating effects of long days (Col gi) and plants with the sensitive wild-type background transitioned to spring-flowering at similar germination dates (electronic supplementary material, figure S3). As a result, life history of autumn-germinating cohorts was more sensitive to variation in vernalization pathways than in photoperiod-response pathways. At more northern latitudes, however, photoperiod-insensitive individuals had a longer MVI that was reached in earlier germinating cohorts (electronic supplementary material, figure S3 and movie S4). The MAE of the GI mutation was small towards the more southern (but extratropical, approx. 35–40° N) latitudes we explored (electronic supplementary material, figure S5). Thus, as might have been expected a priori, we found a greater overall importance of photoperiod sensitivity for life history in the north; however, the seasonal timing of the photoperiod sensitivity effect was less expected. One might predict the MAE of complete photoperiod insensitivity to occur in spring- or early summer-germinating cohorts, which would experience the longest days. This was true only at our southernmost locations, where the MAE, maximum day length and number of days per year above the critical long-day length were also smallest. Moving north into the regions where the maximum life-history effect of GI increased (to levels comparable to that of FRI), the timing of the MAE shifted to later autumn-germinating cohorts. At northern latitudes, longer days occur earlier in the spring and contribute more to the importance of photoperiod effects in the flowering behaviour of winter annuals. The unexpected influence of photoperiod pathway disruption for seedlings germinating in shortening, non-inductive days would be difficult to understand without a detailed photothermal model of development.
(d). Model limitations and future extensions
This photothermal model of Arabidopsis development can be used to describe seasonal and geographical patterns of genetic sensitivity in flowering time and to project these patterns into predicted climates. Nonetheless, there are several clear extensions to the model presented here that would further enhance our understanding of phenological response to novel climates in Arabidopsis.
Our model demonstrates the exquisite sensitivity of life history to seasonal timing, and yet for most habitats, we know little about the factors determining the season in which natural Arabidopsis cohorts occur. For instance, our model of Arabidopsis phenology is at present silent about the factors that influence the timing of germination, even though recent studies have shown that certain genes in the flowering time network are also involved in seed dormancy and germination behaviour (Heschel et al. 2008; Chiang et al. 2009). Both maternal environmental factors such as temperature (Schmuths et al. 2006) and genetic factors (Alonso-Blanco et al. 2003; Holdsworth et al. 2008; Chiang et al. 2009) affect germination behaviour in this species. Despite such advances, we still know relatively little about the natural seasonal and climatic conditions that are permissive for germination (see also review in Donohue 2009). For instance, the warmer temperatures at the southern range limit confer a longer window in which summer and early autumn germinants could complete their life cycle prior to winter; however, drier summers at these sites (in concert with warmer temperatures, which can induce secondary dormancy in this species) may mean that germination during this window is impossible despite the permissive photothermal conditions for vegetative development (Montesinos et al. 2009). Without a detailed understanding of these factors, we cannot know what portion of the possible life-history variation predicted by the model will be expressed in nature.
The genetic and physiological bases of several later life-history stages, and their effects on the coordination of the complete life cycle, likewise remain to be explored. Our model estimates the timing of bolting, which is the appearance of the floral meristem that signals the switch from vegetative to reproductive growth. Although bolting time is used interchangeably with flowering time in much of the Arabidopsis developmental literature, the two events do not coincide and may respond to different environmental cues or be under distinct selection pressures. Elucidating the physiological and developmental responses of each life stage may lead to a better understanding of how and why some habitats support multiple natural cohorts of Arabidopsis per year while others have only a winter annual generation. For example, studies of insect life cycles have shown that the combination of seasonally distributed environmental signals and life-history stages with distinct sensitivities can be sufficient to generate multiple cohorts and synchronize the life cycles of individuals within a population across years (Jenkins et al. 2001; Powell & Logan 2005). Future models that explore the sensitivity, phenology and coordination of distinct life stages in A. thaliana will contribute further to the goal of creating a complete life-history map (cf. Donohue 2009).
Finally, we must understand the relationship between seasonal expression of phenology and fitness. Such information is necessary to explore how selection will act on the environmental sensitivity of phenological traits in changing environments (cf. Davis et al. 2005; Aitken et al. 2008). Phenological transitions can have direct fitness consequences, but selection for timing traits may vary with the environment. In Arabidopsis, inappropriate early bolting in winter annuals can lead to decreased fitness, but earlier bolting is advantageous in spring germinants under field conditions (Korves et al. 2007). Selection experiments under contrasting CO2 or simulated seasonal conditions have identified distinct genetic responses in Arabidopsis that account for evolutionary changes in flowering time depending on the selection environment (Springer et al. 2008; Scarcelli & Kover 2009). The fitness consequences of dormancy characteristics differ between seeds dispersed in autumn and those in spring, and seasonal QTL involved in dormancy response and fitness can also be identified (Donohue et al. 2005; Huang et al. 2010). Thus, knowledge at the genetic level of the basis of phenological traits, the amount of natural variation in these traits, the effect of season on expression of these traits and their fitness consequences in seasonal environments will be necessary to achieve more accurate predictions of the integrated life-history responses of plants to novel environments.
5. Prospects for understanding phenology in changing climates
Process-based phenology models that link mechanism to responsiveness provide an important step forward in predicting plant behaviour and life history under future climate scenarios (cf. Chuine & Beaubien 2001; Morin et al. 2007). In particular, models that integrate and balance the importance of different environmental cues should obviate some of the problems associated with predictions of behaviour under no-analogue climates of the future (Williams & Jackson 2007; Williams et al. 2007). In Arabidopsis, even a simple model using only day length and thermal inputs can explain a great deal of the observed phenological behaviour in a variety of genetic backgrounds, seasons and geographical locations (Wilczek et al. 2009).
Despite a wide diversity of physiologically motivated thermal, photothermal and hydrothermal models of development for various plant species and phenological traits, most existing approaches have focused on generating separate and separately parametrized models for every species and variety considered (Cooper et al. 1995; Cooper & Hammer 1996). The environmentally driven response of plant timing traits modelled using these approaches can be thought of as simulated reaction norms. The Arabidopsis model presented here represents a step forward in understanding the genetic influence on phenology because it both ties parameters that mediate environmental response to known genes in environmental-sensing pathways and also scales developmental rates according to pathway sensitivity. Models of phenology that are sensitive to changes in allele strength at candidate loci or changes in pathway strength in genetic networks can be used to understand the magnitude of phenotypic response as a function of both environmental conditions and the genetic variation sampled. With this approach, we can also generate reaction norms in phenology space along axes of genetic pathway sensitivity. For instance, the predicted timing of life-history sensitivity as a function of genetic pathway function can inform strategies to optimize flowering to specific times in different geographical areas, as well as illustrating how planting dates and ploughing schedules should be shifted to expose genetic variation in sensitivity to different environmental variables. Thus, such trajectories in genetic sensitivity can inform plant breeding strategies for novel climates and will also be critical to understanding potential evolutionary responses to changing climates.
At the moment, there are few other plant species or traits for which the genetic basis of phenology is so well understood at the molecular level. Nonetheless, the approach of modelling general pathway sensitivity, balance and integration should be possible in any species where the basic genetic architecture of environmental response is known (figure 2)—a category that already includes several crop and forestry species. Numerous plant studies provide a rich source of phenological data from complex (and semi-natural) field environments (e.g. Betancourt & Schwartz 2005), in which individuals experience a range of temperature and photoperiod cues in combination. Building on well-developed traditions in crop research that quantify influences of different environmental variables even when they covary (Cooper & Hammer 1996), future work would benefit from considering how perturbation in the sensitivities to environmental variables singly and in concert would affect phenology. Genetic research and screens can also be used to explore the loci and pathways that underlie these environmental sensitivities (Cooper et al. 2005; Welch et al. 2005; Hammer et al. 2006). From work to date, we can predict that for any particular network architecture, the exact balance of converging gene pathways in determining phenology will depend on genetic sensitivity to environmental cues, the input of relevant environmental factors and the seasonal timing of life history.
6. Conclusions
Even simple process-based models can be sufficient to explain a great deal of variation in phenology at a broad scale. Given the growing interest in phenology under changing climates and the relative paucity of detailed physiological data for many plant species, it may be valuable to explore a more general approach to physiological sensitivity of different seasonal environmental inputs with models that are more heuristic and tractable. Agronomy, forestry and other applied fields are rich in data and well-developed methods for exploring the physiological basis of phenology that can be directly transferred to natural systems. To date, a great deal of effort has been focused on understanding plasticity of response to environmental factors, and as a result, the basic seasonal cues to which most plant species and traits respond are fairly well understood. Examination of intraspecific variation in phenology within the framework of models that balance response to different environmental cues may help elucidate the genetic pathways involved as well as the amount of genetic variation in this sensitivity (see also Laurie et al. 2004; Welch et al. 2005). The magnitude and distribution of natural intraspecific variation in environmental response will inform whether adaptation to new climatic regimes will be genetically constrained or how it might be facilitated through natural or assisted migration. Future challenges that will help complete our understanding of phenology in changing environments include further explorations of the genetic basis of phenological traits, the integration of seasonal timing across different life stages and the resetting of developmental time in recurring seasonal traits.
Unlike many indicators of global change that are only detectable to scientists with specialized equipment, many seasonal shifts in abiotic and biotic phenological events are immediately obvious to a large audience. The timing of leafing out in deciduous forests, onset of different pollen seasons, date of first and last frost, bloom time of showy wildflowers, timing of migration of songbirds and butterflies and bird nesting are all undergoing rapid and obvious changes detectable on the scale of a human lifetime. In exploring the mechanistic basis of phenology, evolutionary biologists, ecologists and geneticists have the opportunity to prove the explanatory power of physiological and genetic models in both recreating observed responses and projecting responses to novel environments.
Acknowledgements
We thank R. Early for assistance with GIS data formats and J. Roe for helpful discussions about the structure of flowering time networks. We are grateful to members of the Brown University Autumn 2009 Graduate Seminar for many interesting discussions about phenology in changing climates. This work was supported by an NSF Frontiers in Integrative Biological Research programme grant EF-0425759.
Appendix A
A.1. Modified photothermal unit calculation
The model used to calculate photothermal unit accumulation, and thus the progression of development to bolting, was that described in Wilczek et al. (2009). The modified photothermal time uj of genotype j at time t, uj(t), was calculated from the hourly instantaneous rate 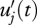 at which genotype j accumulated modified photothermal (developmental) time. This instantaneous rate was calculated as the product of temperature above threshold Θ, a photoperiod factor p and a vernalization effectiveness e,
at which genotype j accumulated modified photothermal (developmental) time. This instantaneous rate was calculated as the product of temperature above threshold Θ, a photoperiod factor p and a vernalization effectiveness e,
| A1 |
All inputs were calculated from temperature and photoperiod information by site, S. Temperature T(S; t), day length d(S; t) and chilling duration v(S; t) were all site-dependent functions of time representing the growth environment to which plants were exposed. The four genotypes in our simulations were isogenic for floral integrator genes (as well as other background loci), and thus all genotypes were assumed to bolt at a common threshold of modified photothermal time. However, the rate at which any given genotype accumulated developmental time depended both on environmental inputs (day length, temperature) and on its genetically determined sensitivity to these environmental factors. Full calculation details and genotype-specific parameters for day length and vernalization sensitivity can be found in Wilczek et al. (2009).
A.2. Converting daily maxima and minima to hourly air temperatures
The photothermal accumulation model depended on hourly plant-level temperature profiles, but only daily maxima and minima were available for most climate projection datasets. We therefore simulated hourly temperature profiles from daily maxima and minima based on equations modified from Cesaraccio et al. (2001)
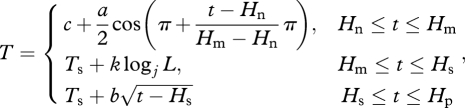 |
A2 |
where Hn, Hm and Hs are the time of dawn, daily maximum and dusk of that day, and t the hour. Local times of dawn and dusk were calculated using equations from Ham (2004). Additional intermediates were as follows: temperature at sunset Ts is estimated as Ts = Tm − s(Tm − Tp), where Tn is the day's minimum temperature, Tm the day's maximum temperature, Tp the next day's minimum temperature, and s is a parameter; the average daily increase c is the arithmetic mean of Tm and Tn, i.e. c = (Tm + Tn)/2; a is the amplitude of the increase Tm − Tn; k is Tm − Ts; the logarithmic base j is 1 + Hs − Hm, 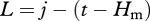 and
and
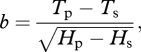 |
where Hp−Hs is the interval from sunset until the next dawn. Hm, the time of daily maximum temperature, is simulated as
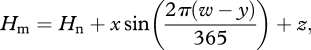 |
A3 |
where w is the day of year (doy) and x, y, z and s are parameters. Equation (A 2) differed from that in Cesaraccio et al. (2001) with respect to the calculation of temperature from the time of maximum until sunset, as inspection of data from weather stations at five field sites in Europe revealed a systematic bias in the estimation of temperature decline that could be better approximated using a log function (L. Burghardt 2010, unpublished data). Parameters were fit using real data from five European weather stations, spanning from 38° N to 65° N, at which we gathered real hourly air temperature profiles for at least 1 year at each site and compared these with hourly profiles simulated using measured daily maxima and minima (Wilczek et al. 2009). Final parameter values were set to x = 2.036391, y = 79.22015, z = 9.285504 and s = 0.227538.
A.3. Simulation of ground temperature from air temperature
Surface temperature Tg in kelvins was simulated based on Kelvin air temperature Ta as
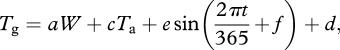 |
A4 |
where a, c, d, e and f are parameters, W (hour, doy) is clear sky irradiance as calculated in Ham (2004) and t is (fractional) time since midnight on 1 January in days. Values of parameters were fit empirically using data from the same five European weather stations to a = 0.004099, c = 0.920493, d = 22.466179, e = −1.861643, and f = 1.549941.
A.4. Accuracy of hourly ground temperature simulations from daily air temperature
Using the photothermal unit accumulation model, days to bolting (DTB) for the Col wild-type genotype was estimated for between 346 and 815 (average 644, median 662) germination dates from 2006 to 2008 at each of the five sites. DTB was calculated using either real hourly ground temperatures measured from on-site weather stations (Wilczek et al. 2009) or measured daily air temperature maxima and minima from these same stations and equations (A 2), (A 3) and (A 4), from which hourly ground temperature profiles were simulated. The correlation between DTB calculated from real hourly ground temperature data and from simulated hourly ground temperature profiles was greater than 0.99 in each of the sites. The slope of DTB predicted from real hourly ground versus simulated hourly ground temperatures was on average 1.00 between the five sites, with a range of 0.98–1.05. Thus, these hourly profile simulations can capture much of the flowering-relevant temperature variation across a broad geographical and seasonal range.
A.5. Estimating phenology in projected climate scenarios
We chose to explore flowering behaviour in an area of Europe and central Asia (11–86° E and 35–71° N) that encompasses much of A. thaliana's native contiguous range. The northern and eastern borders of this grid were set by the extreme locations of recent A. thaliana sampling efforts and range descriptions as well as online available herbaria records (Hoffmann 2002; Schmid et al. 2006; Beck et al. 2008; GBIF). However, even within this grid, there are probably large areas from which natural populations of A. thaliana are absent, particularly in northern Scandinavia and much of Siberia (Hoffmann 2002; Koornneef et al. 2004). In fact, the described northern range limit closely follows the pattern described by our model predictions of MVI for FRI populations under current climates. Arabidopsis is largely absent from areas in which the MVI exceeds 320 days (figure 4), suggesting that the inability to complete a winter annual life cycle may limit the maintenance of populations in these areas. Described populations of A. thaliana that occur outside (particularly south) of this grid occur largely at high elevations (e.g. in the Himalayas and northern Africa) or on oceanic islands, where the microclimate is not likely to be captured at the scale of available climate projections.
For the simulations of flowering time across the native range (approximated here by the area from 11° to 86° E and 35° to 71° N), we used data from the NOAA GFDL CM2.1 A1B_X1 climate scenario for 2001–2100 (NOAA GFDL 2004; Delworth et al. 2006). This scenario, which simulates temperatures under increasing CO2 concentrations up to 720 ppm in the year 2100, projects global daily temperature maxima and minima at a spatial resolution of 2.5° × 2.5°. For each day and geographical grid cell, hourly temperature profiles were simulated from the projected daily maxima and minima using equations (A 2), (A 3) and (A 4). We chose two 6 year time intervals, 2004–2009 and 2094–2099, inclusive, to represent current and future projected climates, respectively. For germination on each successive calendar day of the year, the required number of days to reach the developmental threshold for bolting as a function of simulated local temperature and photoperiod conditions was calculated for the Col, Col FRI, Col FRI vin3 and Col gi genotypes according to the model in Wilczek et al. (2009). For the two 6 year intervals, the average time to bolting (in days) and standard deviation of time to bolting were then calculated for each genotype for each germination day.
Footnotes
One contribution of 11 to a Theme Issue ‘The role of phenology in ecology and evolution'.
References
- Aitken S. N., Yeaman S., Holliday J. A., Wang T. L., Curtis-Mclane S.2008Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1, 95–111 (doi:10.1111/j.1752-4571.2007.00013.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin W. E.1990Global patterns: climate, vegetation and soils. Norman, OK: University of Oklahoma Press [Google Scholar]
- Alexandre C. M., Hennig L.2008FLC or not FLC: the other side of vernalization. J. Exp. Bot. 59, 1127–1135 (doi:10.1093/jxb/ern070) [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Bentsink L., Hanhart C. J., Blankestijn-de Vries H., Koornneef M.2003Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164, 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C., Mendez-Vigo B., Koornneef M.2005From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. Int. J. Dev. Biol. 49, 717–732 (doi:10.1387/ijdb.051994ca) [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Aarts M. G. M., Bentsink L., Keurentjes J. J. B., Reymond M., Vreugdenhil D., Koornneef M.2009what has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21, 1877–1896 (doi:10.1105/tpc.109.068114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., et al. 2006aThe PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat. Genet. 38, 711–715 (doi:10.1038/ng1818) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D.2006bPotent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2, 980–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurle I., Dean C.2006The timing of developmental transitions in plants. Cell 125, 655–664 (doi:10.1016/j.cell.2006.05.005) [DOI] [PubMed] [Google Scholar]
- Beck J. B., Schmuths H., Schaal B. A.2008Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol. Ecol. 17, 902–915 [DOI] [PubMed] [Google Scholar]
- Bertin R. I.2008Plant phenology and distribution in relation to recent climate change. J. Torrey Bot. Soc. 135, 126–146 [Google Scholar]
- Betancourt J. L., Schwartz M.2005Implementing a U.S. national phenology network. EOS Trans. AGU 86, 539–541 [Google Scholar]
- Blackman B., Strasburg J. L., Raduski A., Michaels S., Rieseberg L.2010The role of recently derived FT paralogs in sunflower domestication. Curr. Biol. 20, 629–635 (doi:10.1016/j.cub.2010.01.059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez M. A., Ahn J. H., Weigel D.2003A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33, 168–171 (doi:10.1038/ng1085) [DOI] [PubMed] [Google Scholar]
- Bohlenius H., Huang T., Charbonnel-Campaa L., Brunner A. M., Jansson S., Strauss S. H., Nilsson O.2006CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043 (doi:10.1126/science.1126038) [DOI] [PubMed] [Google Scholar]
- Borchert R., Robertson K., Schwartz M. D., Williams-Linera G.2005Phenology of temperate trees in tropical climates. Int. J. Biometeorol. 50, 57–65 (doi:10.1007/s00484-005-0261-7) [DOI] [PubMed] [Google Scholar]
- Boss P. K., Bastow R. M., Mylne J. S., Dean C.2004Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16, S18–S31 (doi:10.1105/tpc.015958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw W. E., Holzapfel C. M.2001Genetic shift in photoperiodic response correlated with global warming. Proc. Natl Acad. Sci. USA 98, 14 509–14 511 (doi:10.1073/pnas.241391498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw W. E., Holzapfel C. M.2008Genetic response to rapid climate change: it's seasonal timing that matters. Mol. Ecol. 17, 157–166 (doi:10.1111/j.1365-294X.2007.03509.x) [DOI] [PubMed] [Google Scholar]
- Cesaraccio C., Spano D., Duce P., Snyder R. L.2001An improved model for determining degree-day values from daily temperature data. Int. J. Biometeorol. 45, 161–169 [DOI] [PubMed] [Google Scholar]
- Charles-Edwards D. A., Doley D., Rimmington G. M.1986Modelling plant growth and development. North Ryde, Australia: Academic Press [Google Scholar]
- Chiang G. C. K., Barua D., Kramer E. M., Amasino R. M., Donohue K.2009Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 106, 11 661–11 666 (doi:10.1073/pnas.0901367106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuine I.2010Why does phenology drive species distribution? Phil. Trans. R. Soc. B 365, 3149–3160 (doi:10.1098/rstb.2010.0142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuine I., Beaubien E. G.2001Phenology is a major determinant of tree species range. Ecol. Lett. 4, 500–510 (doi:10.1046/j.1461-0248.2001.00261.x) [Google Scholar]
- Cockram J., Chiapparino E., Taylor S. A., Stamati K., Donini P., Laurie D. A., O'Sullivan D. M.2007aHaplotype analysis of vernalization loci in European barley germplasm reveals novel VRN-H1 alleles and a predominant winter VRN-H1/VRN-H2 multi-locus haplotype. Theor. Appl. Genet. 115, 993–1001 (doi:10.1007/s00122-007-0626-x) [DOI] [PubMed] [Google Scholar]
- Cockram J., Jones H., Leigh F. J., O'Sullivan D., Powell W., Laurie D. A., Greenland A. J.2007bControl of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot. 58, 1231–1244 (doi:10.1093/jxb/erm042) [DOI] [PubMed] [Google Scholar]
- Cockram J., Norris C., O'Sullivan D. M.2009PCR-based markers diagnostic for spring and winter seasonal growth habit in barley. Crop Sci. 49, 403–410 (doi:10.2135/cropsci2008.07.0398) [Google Scholar]
- Colasanti J., Coneva V.2009Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiol. 149, 56–62 (doi:10.1104/pp.108.130500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M., Hammer G. (eds) 1996Plant adaptation and crop improvement. Wallingford, CT: CAB International [Google Scholar]
- Cooper M., Podlich D. W., Smith O. S.2005 Gene-to-phenotype models and complex trait genetics. Aust. J. Agric. Res. 56, 895–918 (doi:10.1071/AR05154) [Google Scholar]
- Corbesier L., et al. 2007FT protein movement contributes to long-distance signaling in floral induction in Arabidopsis. Science 316, 1030–1033 (doi:10.1126/science.1141752) [DOI] [PubMed] [Google Scholar]
- Damascos M. A., Prado C., Ronquim C. C.2005Bud composition, branching patterns and leaf phenology in Cerrado woody species. Ann. Bot. 96, 1075–1084 (doi:10.1093/aob/mci258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. B., Shaw R. G., Etterson J. R.2005Evolutionary responses to changing climate. Ecology 86, 1704–1714 (doi:10.1890/03-0788) [Google Scholar]
- Davis C. C., Willis C. G., Primack R. B., Miller-Rushing A. J.2010The importance of phylogeny to the study of phenological response to global climate change. Phil. Trans. R. Soc. B 365, 3201–3213 (doi:10.1098/rstb.2010.0130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delworth T. L., et al. 2006GFDL's CM2 global coupled climate models. Part I: formulation and simulation characteristics. J. Climate 19, 643–674 (doi:10.1175/JCLI3629.1) [Google Scholar]
- Dingkuhn M., Kouressy M., Vaksmann M., Clerget B., Chantereau J.2008A model of sorghum photoperiodism using the concept of threshold-lowering during prolonged appetence. Eur. J. Agron. 28, 74–89 (doi:10.1016/j.eja.2007.05.005) [Google Scholar]
- Dong Z. S.2003Incorporation of genomic information into the simulation of flowering time in Arabidopsis thaliana. Manhattan, KS: Kansas State University [Google Scholar]
- Donohue K.2009Completing the cycle: maternal effects as the missing link in plant life histories. Phil. Trans. R. Soc. B 364, 1059–1074 (doi:10.1098/rstb.2008.0291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K., Dorn L., Griffith C., Kim E., Aguilera A., Polisetty C. R., Schmitt J.2005The evolutionary ecology of seed germination of Arabidopsis thaliana: variable natural selection on germination timing. Evolution 59, 758–770 [PubMed] [Google Scholar]
- Durant J. M., Hjermann D. O., Anker-Nilssen T., Beaugrand G., Mysterud A., Pettorelli N., Stenseth N. C.2005Timing and abundance as key mechanisms affecting trophic interactions in variable environments. Ecol. Lett. 8, 952–958 (doi:10.1111/j.1461-0248.2005.00798.x) [DOI] [PubMed] [Google Scholar]
- Engelmann K., Purugganan M.2006The molecular evolutionary ecology of plant development: flowering time in Arabidopsis thaliana. Adv. Bot. Res. 44, 507–526 (doi:10.1016/S0065-2296(06)44013-1) [Google Scholar]
- Finnegan E. J., Dennis E. S.2007Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 17, 1978–1983 (doi:10.1016/j.cub.2007.10.026) [DOI] [PubMed] [Google Scholar]
- Fisher J. I., Richardson A. D., Mustard J. F.2007Phenology model from surface meteorology does not capture satellite-based greenup estimations. Global Change Biol. 13, 707–721 (doi:10.1111/j.1365-2486.2006.01311.x) [Google Scholar]
- Fitter A. H., Fitter R. S. R.2002Rapid changes in flowering time in British plants. Science 296, 1689–1691 (doi:10.1126/science.1071617) [DOI] [PubMed] [Google Scholar]
- Franks S. J., Sim S., Weis A. E.2007Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl Acad. Sci. USA 104, 1278–1282 (doi:10.1073/pnas.0608379104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiba G., Vagujfalvi A., Li C. X., Soltesz A., Dubcovsky J.2009Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci. 176, 12–19 (doi:10.1016/j.plantsci.2008.09.016) [Google Scholar]
- GBIF. Data Portal, data.gbif.org . Accessed 2009-07-01.
- Geraldo N., Baurle I., Kidou S., Hu X. Y., Dean C.2009FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol. 150, 1611–1618 (doi:10.1104/pp.109.137448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C., Massonnet C., Turc O., Muller B., Chenu K., Tardieu F.2002Individual leaf development in Arabidopsis thaliana: a stable thermal-time-based programme. Ann. Bot. 89, 595–604 (doi:10.1093/aob/mcf085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A., Peacock W. J., Dennis E. S., Trevaskis B.2009The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 103, 1165–1172 (doi:10.1093/aob/mcp063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith T. M., Watson M. A.2006Is evolution necessary for range expansion? Manipulating reproductive timing of a weedy annual transplanted beyond its range. Am. Nat. 167, 153–164 [DOI] [PubMed] [Google Scholar]
- Gyllenstrand N., Clapham D., Kallman T., Lagercrantz U.2007A Norway spruce FLOWERING LOCUS T homolog is implicated in control of growth rhythm in conifers. Plant Physiol. 144, 248–257 (doi:10.1104/pp.107.095802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D., Luquez V., Garcia V. M., St Onge K. R., Jansson S., Ingvarsson P. K.2007Adaptive population differentiation in phenology across a latitudinal gradient in European aspen (Populus tremula, L.): a comparison of neutral markers, candidate genes and phenotypic traits. Evolution 61, 2849–2860 (doi:10.1111/j.1558-5646.2007.00230.x) [DOI] [PubMed] [Google Scholar]
- Ham J. M.2004Useful equations in micrometeorology. In Micrometeorology of agricultural systems (eds Hatfield J. L., Baker J. M.). Madison, WI: ASA-CSSA-SSSA [Google Scholar]
- Hammer G. L., Cooper M., Tardieu F., Welch S. M., Walsh B., van Eeuwijk F., Chapman S., Podlich D.2006Models for navigating biological complexity in breeding improved crop plants. Trends Plant Sci. 11, 587–593 (doi:10.1016/j.tplants.2006.10.006) [DOI] [PubMed] [Google Scholar]
- Harrington C. A., Gould P. J., St. Clair J. B.2010Modeling the effects of winter environment on dormancy release of Douglas-fir. For. Ecol. Manage. 259, 798–808 (doi:10.1016/j.foreco.2009.06.018) [Google Scholar]
- Hayama R., Coupland G.2004The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135, 677–684 (doi:10.1104/pp.104.042614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K.2003Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722 (doi:10.1038/nature01549) [DOI] [PubMed] [Google Scholar]
- Hedman H., Kallman T., Lagercrantz U.2009Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 70, 359–369 (doi:10.1007/s11103-009-9478-x) [DOI] [PubMed] [Google Scholar]
- Henderson I. R., Shindo C., Dean C.2003The need for winter in the switch to flowering. Annu. Rev. Genet. 37, 371–392 (doi:10.1146/annurev.genet.37.110801.142640) [DOI] [PubMed] [Google Scholar]
- Heschel M. S., Butler C. M., Barua D., Chiang G. C. K., Wheeler A., Sharrock R. A., Whitelam G. C., Donohue K.2008New roles of phytochromes during seed germination. Int. J. Plant Sci. 169, 531–540 (doi:10.1086/528753) [Google Scholar]
- Hoffmann M. H.2002Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J. Biogeog. 29, 125–134 (doi:10.1046/j.1365-2699.2002.00647.x) [Google Scholar]
- Hoffmann M. H., Tomiuk E., Schmuths H., Koch C., Bachmann K.2005Phenological and morphological responses to different temperature treatments differ among a world-wide sample of accessions of Arabidopsis thaliana. Acta Oecol. Int. J. Ecol. 28, 181–187 (doi:10.1016/j.actao.2005.03.010) [Google Scholar]
- Holdsworth M. J., Bentsink L., Soppe W. J. J.2008Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179, 33–54 (doi:10.1111/j.1469-8137.2008.02437.x) [DOI] [PubMed] [Google Scholar]
- Howe G. T., Aitken S. N., Neale D. B., Jermstad K. D., Wheeler N. C., Chen T. H. H.2003From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Can. J. Bot. Rev. 81, 1247–1266 (doi:10.1139/b03-141) [Google Scholar]
- Huang X., Schmitt J., Dorn L., Griffith C., Effgen S., Takao S., Koornneef M., Donohue K.2010The earliest stages of adaptation in an experimental plant population: strong selection on QTLS for seed dormancy. Mol. Ecol. 19, 1335–1351 (doi:10.1111/j.1365-294x.2010.04557.x) [DOI] [PubMed] [Google Scholar]
- Ibáñez I., Primack R. B., Miller-Rushing A. J., Ellwood E., Higuchi H., Lee S. D., Kobori H., Silander J. A.2010Forecasting phenology under global warming. Phil. Trans. R. Soc. B 365, 3247–3260 (doi:10.1098/rstb.2010.0120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson P. K., Garcia M. V., Hall D., Luquez V., Jansson S.2006Clinal variation in phyB2, a candidate gene for day-length-induced growth cessation and bud set, across a latitudinal gradient in European aspen (Populus tremula). Genetics 172, 1845–1853 (doi:10.1534/genetics.105.047522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson P. K., Garcia M. V., Luquez V., Hall D., Jansson S.2008Nucleotide polymorphism and phenotypic associations within and around the phytochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178, 2217–2226 (doi:10.1534/genetics.107.082354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins J. L., Powell J. A., Logan J. A., Bentz B. J.2001Low seasonal temperatures promote life cycle synchronization. Bull. Math. Biol. 63, 573–595 (doi:10.1006/bulm.2001.0237) [DOI] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C.2000Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347 (doi:10.1126/science.290.5490.344) [DOI] [PubMed] [Google Scholar]
- Jones H., et al. 2008Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the fertile crescent. Mol. Biol. Evol. 25, 2211–2219 (doi:10.1093/molbev/msn167) [DOI] [PubMed] [Google Scholar]
- Joost S., Bonin A., Bruford M., Després L., Conord C., Erhardt G., Taberlet P.2007A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol. Ecol. 16, 3955–3969 (doi:10.1111/j.1365-294X.2007.03442.x) [DOI] [PubMed] [Google Scholar]
- Kikuchi R., Kawahigashi H., Ando T., Tonooka T., Handa H.2009Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149, 1341–1353 (doi:10.1104/pp.108.132134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., He Y. H., Jacob Y., Noh Y. S., Michaels S., Amasino R.2005Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17, 3301–3310 (doi:10.1105/tpc.105.034645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., et al. 2007Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L. ssp pekinensis). Plant Cell Rep. 26, 327–336 (doi:10.1007/s00299-006-0243-1) [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Weigel D.2007Move on up, it's time for change—mobile signals controlling photoperiod-dependent flowering. Gen. Dev 21, 2371–2384 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C. J., Vanderveen J. H.1991A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Vreugdenhil D.2004Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55, 141–172 (doi:10.1146/annurev.arplant.55.031903.141605) [DOI] [PubMed] [Google Scholar]
- Korves T. M., Schmid K. J., Caicedo A. L., Mays C., Stinchcombe J. R., Purugganan M. D., Schmitt J.2007Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am. Nat. 169, E141–E157 [DOI] [PubMed] [Google Scholar]
- Landsberg H. Physical climatology. State College, PA: Gray Printing Company; 1941. [Google Scholar]
- Laurie D. A., Griffiths S., Dunford R. P., Christodoulou V., Taylor S. A., Cockram J., Beales J., Turner A.2004Comparative genetic approaches to the identification of flowering time genes in temperate cereals. Field Crops Res. 90, 87–99 (doi:10.1016/j.fcr.2004.07.007) [Google Scholar]
- Lee I., Amasino R. M.1995Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the Frigida gene. Plant Physiol. 108, 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehikoinen E., Sparks T. H., Zalakevicius M.2004Arrival and departure dates. In The effects of climate change on birds (eds Moller A. P., Fielder W., Berthold P.), pp. 1–31 New York, NY: Academic Press Ltd [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., Weigel D.2005Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1, 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Roycewicz P., Smith E., Borevitz J. O.2006Genetics of local adaptation in the laboratory: flowering time quantitative trait loci under geographic and seasonal conditions in Arabidopsis. PLoS ONE 1, e105 (doi:10.1371/journal.pone.0000105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. G., Reighard G. L., Abbott A. G., Bielenberg D. G.2009Dormancy-associated MADS genes from the EVG locus of peach Prunus persica (L.) Batsch have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60, 3521–3530 (doi:10.1093/jxb/erp195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J. P., Eshed Y.2006The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl Acad. Sci. USA 103, 6398–6403 (doi:10.1073/pnas.0601620103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linacre E.1992Climate, data and resources: a reference and guide. New York, NY: Routledge [Google Scholar]
- Lister D. L., et al. 2009Latitudinal variation in a photoperiod response gene in European barley: insight into the dynamics of agricultural spread from ‘historic’ specimens. J. Archaeol. Sci. 36, 1092–1098 (doi:10.1016/j.jas.2008.12.012) [Google Scholar]
- Locke J. C. W., Southern M. M., Kozma-Bognar L., Hibberd V., Brown P. E., Turner M. S., Millar A. J.2005Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1 (doi:10.1038/msb4100018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J. C. W., Kozma-Bognar L., Gould P. D., Feher B., Kevei E., Nagy F., Turner M. S., Hall A., Millar A. J.2006Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquez V., Hall D., Albrectsen B. R., Karlsson J., Ingvarsson P., Jansson S.2008Natural phenological variation in aspen (Populus tremula): the SwAsp collection. Tree Genet. Genomes 4, 279–292 (doi:10.1007/s11295-007-0108-y) [Google Scholar]
- Malmberg R. L., Held S., Waits A., Mauricio R.2005Epistasis for fitness-related quantitative traits in Arabidopsis thaliana grown in the field and in the greenhouse. Genetics 171, 2013–2027 (doi:10.1534/genetics.105.046078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof J. N., et al. 2001Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29, 441–446 (doi:10.1038/ng777) [DOI] [PubMed] [Google Scholar]
- Menzel A., et al. 2006aEuropean phenological response to climate change matches the warming pattern. Global Change Biol. 12, 1969–1976 (doi:10.1111/j.1365-2486.2006.01193.x) [Google Scholar]
- Menzel A., Von Vopelius J., Estrella N., Schleip C., Dose V.2006bFarmers' annual activities are not tracking the speed of climate change. Climate Res. 32, 201–207 [Google Scholar]
- Michael T. P., Salome P. A., McClung C. R.2003Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc. Natl Acad. Sci. USA 100, 6878–6883 (doi:10.1073/pnas.1131995100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., He Y. H., Scortecci K. C., Amasino R. M.2003Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl Acad. Sci. USA 100, 10 102–10 107 (doi:10.1073/pnas.1531467100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Bezerra I. C., Amasino R. M.2004FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc. Natl Acad. Sci. USA 101, 3281–3285 (doi:10.1073/pnas.0306778101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Rushing A. J., Primack R. B.2008Global warming and flowering times in Thoreau's concord: a community perspective. Ecology 89, 332–341 (doi:10.1890/07-0068.1) [DOI] [PubMed] [Google Scholar]
- Miller-Rushing A. J., Primack R. B., Primack D., Mukunda S.2006Photographs and herbarium specimens as tools to document phenological changes in response to global warming. Am. J. Bot. 93, 1667–1674 (doi:10.3732/ajb.93.11.1667) [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., et al. 2005Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270 (doi:10.1105/tpc.105.033464) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller A. P., Rubolini D., Lehikoinen E.2008Populations of migratory bird species that did not show a phenological response to climate change are declining. Proc. Natl Acad. Sci. USA 105, 16 195–16 200 (doi:10.1073/pnas.0803825105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos A., Tonsor S. J., Alonso-Blanco C., Xavier Pico F.2009Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS ONE 4, no. e7213. (doi:10.1371/journal.pone.0007213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Lee H., Kim M., Lee I.2005Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 46, 292–299 (doi:10.1093/pcp/pci024) [DOI] [PubMed] [Google Scholar]
- Morin X., Augspurger C., Chuine I.2007Process-based modeling of species' distributions: what limits temperate tree species' range boundaries? Ecology 88, 2280–2291 (doi:10.1890/06-1591.1) [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Kisaka M., Fuse T., Yano M., Ogihara Y.2003Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 36, 82–93 (doi:10.1046/j.1365-313X.2003.01859.x) [DOI] [PubMed] [Google Scholar]
- NOAA GFDL (US Department of Commerce/NOAA/Geophysical Fluid Dynamics Laboratory) 2004IPCC Fourth Assessment and US CCSP Projects. Princeton, NJ, USA. See http://data1.gfdl.noaa.gov/CM2.X/references/ (Accessed)
- Parmesan C.2006Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637–669 (doi:10.1146/annurev.ecolsys.37.091305.110100) [Google Scholar]
- Parmesan C., Yohe G.2003A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 (doi:10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- Petipas R. H., Wilczek A. M., Schmitt J., Savolainen O.In preparation Demography of Arabidopsis thaliana at its northern range limit in Finland. [Google Scholar]
- Post E., Forchhammer M. C.2008Climate change reduces reproductive success of an Arctic herbivore through trophic mismatch. Phil. Trans. R. Soc. B 363, 2369–2375 (doi:10.1098/rstb.2007.2207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post E., et al. 2009Ecological dynamics across the Arctic associated with recent climate change. Science 325, 1355–1358 (doi:10.1126/science.1173113) [DOI] [PubMed] [Google Scholar]
- Powell J. A., Logan J. A.2005Insect seasonality: circle map analysis of temperature-driven life cycles. Theor. Popul. Biol. 67, 161–179 (doi:10.1016/j.tpb.2004.10.001) [DOI] [PubMed] [Google Scholar]
- Primack D., Imbres C., Primack R. B., Miller-Rushing A. J., Del Tredici P.2004Herbarium specimens demonstrate earlier flowering times in response to warming in Boston. Am. J. Bot. 91, 1260–1264 (doi:10.3732/ajb.91.8.1260) [DOI] [PubMed] [Google Scholar]
- Primack R. B., Ibanez I., Higuchi H., Lee S. D., Miller-Rushing A. J., Wilson A. M., Silander J. A.2009Spatial and interspecific variability in phenological responses to warming temperatures. Biol. Conserv. 142, 2569–2577 (doi:10.1016/j.biocon.2009.06.003) [Google Scholar]
- Ray P. M., Alexander W. E.1966Photoperiodic adaptation to latitude in Xanthium strumarium. Am. J. Bot. 53, 806–816 (doi:10.2307/2440183) [Google Scholar]
- Reeves P. A., He Y. H., Schmitz R. J., Amasino R. M., Panella L. W., Richards C. M.2007Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics 176, 295–307 (doi:10.1534/genetics.106.069336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer T., Wittenzeller M., Haeckel H., Nekovar J.2000Phenology in central Europe—differences and trends of spring phenophases in urban and rural areas. Int. J. Biometeorol. 44, 60–66 (doi:10.1007/s004840000062) [DOI] [PubMed] [Google Scholar]
- Rosenzweig C., et al. 2008Attributing physical and biological impacts to anthropogenic climate change. Nature 453, 353–357 (doi:10.1038/nature06937) [DOI] [PubMed] [Google Scholar]
- Salazar J. D., Saithong T., Brown P. E., Foreman J., Locke J. C. W., Halliday K. J., Carre I. A., Rand D. A., Millar A. J.2009Prediction of photoperiodic regulators from quantitative gene circuit models. Cell 139, 1170–1179 (doi:10.1016/j.cell.2009.11.029) [DOI] [PubMed] [Google Scholar]
- Samis K. E., Heath K. D., Stinchcombe J. R.2008Discordant longitudinal clines in flowering time and phytochrome c in Arabidopsis thaliana. Evolution 62, 2971–2983 (doi:10.1111/j.1558-5646.2008.00484.x) [DOI] [PubMed] [Google Scholar]
- Savolainen O., Pyhajarvi T., Knurr T.2007Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 38, 595–619 (doi:10.1146/annurev.ecolsys.38.091206.095646) [Google Scholar]
- Scarcelli N., Kover P. X.2009Standing genetic variation in FRIGIDA mediates experimental evolution of flowering time in Arabidopsis. Mol. Ecol. 18, 2039–2049 (doi:10.1111/j.1365-294X.2009.04145.x) [DOI] [PubMed] [Google Scholar]
- Scarcelli N., Cheverud J. M., Schaal B. A., Kover P. X.2007Antagonistic pleiotropic effects reduce the potential adaptive value of the FRIGIDA locus. Proc. Natl Acad. Sci. USA 104, 16 986–16 991 (doi:10.1073/pnas.0708209104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaber J., Badeck F. W.2002Evaluation of methods for the combination of phenological time series and outlier detection. Tree Physiol. 22, 973–982 [DOI] [PubMed] [Google Scholar]
- Schlappi M. R.2006FRIGIDA LIKE 2 is a functional allele in Landsberg erecta and compensates for a nonsense allele of FRIGIDA LIKE 1. Plant Physiol. 142, 1728–1738 (doi:10.1104/pp.106.085571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K., Torjek O., Meyer R., Schmuths H., Hoffmann M. H., Altmann T.2006Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor. Appl. Genet. 112, 1104–1114 (doi:10.1007/s00122-006-0212-7) [DOI] [PubMed] [Google Scholar]
- Schmitz R. J., Sung S., Amasino R. M.2008Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 105, 411–416 (doi:10.1073/pnas.0710423104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuths H., Bachmann K., Weber W. E., Horres R., Hoffmann M. H.2006Effects of preconditioning and temperature during germination of 73 natural accessions of Arabidopsis thaliana. Ann. Bot. 97, 623–634 (doi:10.1093/aob/mcl012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I., He Y. H., Turck F., Vincent C., Fornara F., Krober S., Amasino R. A., Coupland G.2006The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912 (doi:10.1101/gad.373506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers W. D.1965Physical climatology. Chicago, IL: University of Chicago Press [Google Scholar]
- Serrano G., Herrera-Palau R., Romero J. M., Serrano A., Coupland G., Valverde F.2009Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr. Biol. 19, 359–368 (doi:10.1016/j.cub.2009.01.044) [DOI] [PubMed] [Google Scholar]
- Sheldon C. C., Hills M. J., Lister C., Dean C., Dennis E. S., Peacock W. J.2008Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl Acad. Sci. USA 105, 2214–2219 (doi:10.1073/pnas.0711453105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Aranzana M. J., Lister C., Baxter C., Nicholls C., Nordborg M., Dean C.2005Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138, 1163–1173 (doi:10.1104/pp.105.061309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Lister C., Crevillen P., Nordborg M., Dean C.2006Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20, 3079–3083 (doi:10.1101/gad.405306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. C., Parmesan C.2010Phenological asynchrony between herbivorous insects and their hosts: signal of climate change or pre-existing adaptive strategy? Phil. Trans. R. Soc. B 365, 3161–3176 (doi:10.1098/rstb.2010.0144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer C. J., Orozco R. A., Kelly J. K., Ward J. K.2008Elevated CO2 influences the expression of floral-initiation genes in Arabidopsis thaliana. New Phytol. 178, 63–67 (doi:10.1111/j.1469-8137.2008.02387.x) [DOI] [PubMed] [Google Scholar]
- Stenoien H. K., Fenster C. B., Kuittinen H., Savolainen O.2002Quantifying latitudinal clines to light responses in natural populations of Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 89, 1604–1608 (doi:10.3732/ajb.89.10.1604) [DOI] [PubMed] [Google Scholar]
- Stinchcombe J. R., Caicedo A. L., Hopkins R., Mays C., Boyd E. W., Purugganan M. D., Schmitt J.2005Vernalization sensitivity in Arabidopsis thaliana (Brassicaceae): the effects of latitude and FLC variation. Am. J. Bot. 92, 1701–1707 (doi:10.3732/ajb.92.10.1701) [DOI] [PubMed] [Google Scholar]
- Sung S. B., Amasino R. M.2004Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164 (doi:10.1038/nature02195) [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H. L., Yokoi S., Shimamoto K.2007Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036 (doi:10.1126/science.1141753) [DOI] [PubMed] [Google Scholar]
- Thompson L.1994The spatiotemporal effects of nitrogen and litter on the population-dynamics of Arabidopsis thaliana. J. Ecol. 82, 63–68 [Google Scholar]
- Trevaskis B., Hemming M. N., Dennis E. S., Peacock W. J.2007The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357 (doi:10.1016/j.tplants.2007.06.010) [DOI] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G.2008Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594 (doi:10.1146/annurev.arplant.59.032607.092755) [DOI] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R. P., Laurie D. A.2005The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034 (doi:10.1126/science.1117619) [DOI] [PubMed] [Google Scholar]
- UK National Weather Service. 2009. See http://www.metoffice.gov.uk/climate/uk/ . (Accessed 10 January 2009)
- van Buskirk J., Mulvihill R. S., Leberman R. C.2009Variable shifts in spring and autumn migration phenology in North American songbirds associated with climate change. Global Change Biol. 15, 760–771 (doi:10.1111/j.1365-2486.2008.01751.x) [Google Scholar]
- van der Jeugd H. P., Eichhorn G., Litvin K. E., Stahl J., Larsson K., van der Graaf A. J., Drent R. H.2009Keeping up with early springs: rapid range expansion in an avian herbivore incurs a mismatch between reproductive timing and food supply. Global Change Biol. 15, 1057–1071 (doi:10.1111/j.1365-2486.2008.01804.x) [Google Scholar]
- Walther G. R., Post E., Convey P., Menzel A., Parmesan C., Beebee T. J. C., Fromentin J. M., Hoegh-Guldberg O., Bairlein F.2002Ecological responses to recent climate change. Nature 416, 389–395 (doi:10.1038/416389a) [DOI] [PubMed] [Google Scholar]
- Wang R. H., Farrona S., Vincent C., Joecker A., Schoof H., Turck F., Alonso-Blanco C., Coupland G., Albani M. C.2009PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427 (doi:10.1038/nature07988) [DOI] [PubMed] [Google Scholar]
- Weinig C., Ungerer M. C., Dorn L. A., Kane N. C., Toyonaga Y., Halldorsdottir S. S., Mackay T. F. C., Purugganan M. D., Schmitt J.2002Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162, 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch S. M., Roe J. L., Dong Z. S.2003A genetic neural network model of flowering time control in Arabidopsis thaliana. Agron. J. 95, 71–81 [Google Scholar]
- Welch S. M., Dong Z. S., Roe J. L., Das S.2005Flowering time control: gene network modelling and the link to quantitative genetics. Aust. J. Agric. Res. 56, 919–936 (doi:10.1071/AR05155) [Google Scholar]
- Wilczek A. M., et al. 2009Effects of genetic perturbation on seasonal life-history plasticity. Science 323, 930–934 (doi:10.1126/science.1165826) [DOI] [PubMed] [Google Scholar]
- Wilkie J. D., Sedgley M., Olesen T.2008Regulation of floral initiation in horticultural trees. J. Exp. Bot. 59, 3215–3228 (doi:10.1093/jxb/ern188) [DOI] [PubMed] [Google Scholar]
- Williams J. W., Jackson S. T.2007Novel climates, no-analog communities, and ecological surprises. Front. Ecol. Environ. 5, 475–482 (doi:10.1890/070037) [Google Scholar]
- Williams J. W., Jackson S. T., Kutzbacht J. E.2007Projected distributions of novel and disappearing climates by 2100 AD. Proc. Natl Acad. Sci. USA 104, 5738–5742 (doi:10.1073/pnas.0606292104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis C. G., Ruhfel B., Primack R. B., Miller-Rushing A. J., Davis C. C.2008Phylogenetic patterns of species loss in Thoreau's woods are driven by climate change. Proc. Natl Acad. Sci. USA 105, 17 029–17 033 (doi:10.1073/pnas.0806446105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland A. J., Borner A., Korzun V., Li W. M., Petrovic S., Sayers E. J.1998The influence of photoperiod genes on the adaptability of European winter wheats (Reprinted from Wheat: Prospects for global improvement, 1998). Euphytica 100, 385–394 (doi:10.1023/A:1018327700985) [Google Scholar]
- Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T.2005TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46, 1175–1189 (doi:10.1093/pcp/pci151) [DOI] [PubMed] [Google Scholar]
- Zeilinger M. N., Farre E. M., Taylor S. R., Kay S. A., Doyle F. J.2006A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol. Syst. Biol. 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Friedl M. A., Schaaf C. B.2006Global vegetation phenology from moderate resolution imaging spectroradiometer (MODIS): evaluation of global patterns and comparison with in situ measurements. J. Geophys. Res. Biogeosci. 111, G04017 [Google Scholar]
- Zhang Q. Z., et al. 2008Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl Acad. Sci. USA 105, 21 028–21 033 (doi:10.1073/pnas.0810585105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Z., Li Z. M., Mei L., Yao J. L., Hu C. G.2009PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuation in expression level. Planta 229, 847–859 (doi:10.1007/s00425-008-0885-z) [DOI] [PubMed] [Google Scholar]
- Zobell O., Coupland G., Reiss B.2005The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol. 7, 266–275 (doi:10.1055/s-2005-865621) [DOI] [PubMed] [Google Scholar]