Abstract
Coevolved mutualisms often exhibit high levels of partner specificity. Obligate pollination mutualisms, such as the fig–fig wasp and yucca–yucca moth systems, represent remarkable examples of such highly species-specific associations; however, the evolutionary processes underlying these patterns are poorly understood. The prevailing hypothesis suggests that the high degree of specificity in pollinating seed parasites is the fortuitous result of specialization in their ancestors because these insects are derived from endophytic herbivores that are themselves highly host-specific. Conversely, we show that in the Glochidion–Epicephala obligate pollination mutualism, pollinators are more host-specific than are closely related endophytic leaf-feeding taxa, which co-occur with Epicephala on the same Glochidion hosts. This difference is probably not because of shifts in larval diet (i.e. from leaf- to seed-feeding), because seed-eating lepidopterans other than Epicephala do not show the same degree of host specificity as Epicephala. Species of a tentative sister group of Epicephala each attack several distantly related plants, suggesting that the evolution of strict host specificity is tied to the evolution of pollinator habit. These results suggest that mutualists can attain higher host specificity than that of their parasitic ancestors and that coevolutionary selection can be a strong promoter of extreme reciprocal specialization in mutualisms.
Keywords: Caloptilia, Cuphodes, Diphtheroptila, Gracillariidae, Phyllanthaceae
1. Introduction
Parasitic lifestyles usually favour extreme specialization to one or few host species because they require complex adaptations to circumvent host defences and sustain life on a single host (Ehrlich & Raven 1964; Price 1980; Thompson 1994; Strauss & Zangerl 2002). Although specific mechanisms underlying host specialization may vary among taxa, the broad general understanding is that host–parasite coevolution promotes specialization in parasitic organisms (Thompson 1994, 2005). Strict host specificity of parasites is often linked to high species diversity because specialization to different hosts can result in host-associated speciation (Mitter et al. 1988; Farrell 1998; Schluter 2000; Coyne & Orr 2004).
In contrast, the evolutionary processes that determine the level of specialization in mutualisms are far less understood. Although many mutualisms do not evolve to exhibit high degrees of specificity (e.g. most plant–pollinator and plant–seed disperser interactions), reciprocal partner specialization is often found in intimate mutualisms, such as those between myrmecophytic plants and their resident ants (Davidson & McKey 1993; Heil & McKey 2003; Guimarães et al. 2007), ants/termites and their cultivated fungi (Mueller et al. 1998; Aanen et al. 2002; Currie et al. 2003) or various invertebrates and their endosymbiotic micro-organisms (Moran & Telang 1998; Hosokawa et al. 2006). Both ultimate and proximate causes of specialization have been proposed, including selection for elimination of less-cooperative partners (Heil et al. 2005; Poulsen & Boomsma 2005) and chemical or physical mechanisms of partner discrimination (Federle et al. 1997; Brouat et al. 2001; Edwards et al. 2006; Grangier et al. 2009). However, the general understanding of the evolutionary conditions favouring specialization in mutualisms is still very limited (Thompson 1994, 2005), and modern molecular approaches continue to refine our view of how mutualists are associated with one another on both local and broad geographical scales (Molbo et al. 2003; Mikheyev et al. 2006; Quek et al. 2007; Visser et al. 2009).
Perhaps, the most remarkable cases of reciprocal specialization between mutualists are found in obligate pollination mutualisms (Janzen 1979; Pellmyr 2003; Herre et al. 2008). The fig–fig wasp and yucca–yucca moth mutualisms are well-known examples of such highly species-specific associations, in which the plants are pollinated by one or, rarely, two insect species, which in turn are highly host-specific seed parasites of the plants they pollinate. Figs and yuccas have diversified into more than 700 and 40 species, respectively, and a corresponding high diversity of pollinator species have evolved, each of which is obligately mutualistic with one or few fig/yucca hosts (Weiblen 2002; Pellmyr 2003; Herre et al. 2008). This level of specificity is unusual among pollination mutualisms because, although selection may favour plants depending on specialized visitors for effective conspecific pollination, pollinators are generally expected to maximize the range of plants they visit to optimize resource use (Pellmyr 2002; Gómez & Zamora 2006). The high specificity of pollinating seed parasites is therefore considered to be the result of their inherently parasitic lifestyle (Thompson 1994, 2005) because seed-feeding insects commonly specialize to a narrow range of host plants. Indeed, detailed ecological and phylogenetic studies of the yucca moth family Prodoxidae have found that close relatives of the pollinators are also highly host-specific herbivores (Pellmyr & Thompson 1992; Pellmyr 1999; Pellmyr et al. 2006), suggesting that the high degree of pollinator-specificity is driven by the parasitic part of the interaction and cannot be attributed to mutualistic selection (Thompson 1994, 2005).
However, a growing body of evidence suggests that the current view of host specificity in pollinating seed parasites may require revision. Within the yucca moth lineage, two cheater species have independently lost their pollinating behaviour and oviposit in young fruits to exploit the seeds that other yucca moth species have pollinated (Pellmyr et al. 1996; Pellmyr 1999). In contrast to their pollinating relatives, each of these cheater species has evolved to use four to six yucca hosts (Pellmyr 1999; Segraves & Pellmyr 2004), suggesting that host specificity in the pollinators may not be determined solely by the herbivorous habit of the moths (Pellmyr 2003). In the fig system, non-pollinating agaonid wasps that are closely related to and co-occur with pollinating fig wasps tend to be less host specific than are the pollinators (Weiblen & Bush 2002; Marussich & Machado 2007; but see Lopez-Vaamonde et al. 2001; Jousselin et al. 2006, 2008). In addition, fig herbivores in general are dominated by insects that feed on several locally available fig hosts (Novotny et al. 2002, 2006). Given that shared pollinators can result in hybridization among closely related, co-occurring figs (Machado et al. 2005), selection may favour figs relying on specialist pollinators to achieve effective conspecific pollination. Thus, these observations indicate that pollinating seed parasites may in fact attain a higher degree of host specificity than that of their parasitic ancestors owing to coevolutionary selection arising after the evolution of pollination mutualism.
We tested whether host specificity is greater in pollinating seed parasites than in their herbivorous ancestors in a recently discovered mutualism between Phyllantheae plants (Phyllanthaceae) and Epicephala moths (Gracillariidae; Kato et al. 2003; Kawakita & Kato 2004a,b, 2009). Currently, an estimated 500 species of Phyllantheae plants exist that are pollinated by the ovipositing females of Epicephala moths (Kawakita & Kato 2009; Kawakita 2010). Among them are plants of the genus Glochidion, which is the largest radiation and comprises more than 300 species distributed throughout the Asian–Australian tropics (Govaerts et al. 2000). Previous detailed assessment of pollinator specificity in the Japanese species of Glochidion has found that, although some Epicephala species are associated with two closely related Glochidion hosts in different parts of their ranges, each Epicephala species is specialized to only one of the several co-occurring hosts in all populations studied, thus showing very strict local host specificity (Kawakita & Kato 2006). The Glochidion–Epicephala system is ideal for studying the evolution of high host-specificity in pollinating seed parasites because Glochidion plants are host to two other genera of herbivores (Diphtheroptila and Caloptilia) that belong to the subfamily Gracillariinae together with Epicephala (Kumata 1982; Kuroko 1982); this situation allows for comparison of host specificity among phylogenetically related genera that share the same host plants. Diphtheroptila are leaf miners that use young Glochidion leaves, whereas Caloptilia are leaf miners as early instar larvae and, as they develop into late instars, construct leaf rolls or induce leaf galls, depending on species (Kumata 1982). Furthermore, Glochidion plants are attacked by lepidopteran seed parasites that belong to the families Carposinidae, Tortricidae and Pyralidae, whose host specificity may be determined by a common mechanism with that of Epicephala owing to their shared larval diet.
In this study, we first conducted a molecular phylogenetic analysis of Gracillariinae to determine the relative phylogenetic positions of Epicephala, Diphtheroptila and Caloptilia within the subfamily. We then analysed host specificity of the abovementioned Glochidion-associated herbivores (Diphtheroptila, Caloptilia and non-gracillariid seed-feeding lepidopterans) to test whether the level of host specialization is indeed higher in Epicephala. We also investigated the degree of host specificity in a candidate sister genus of Epicephala to determine whether the high pollinator specificity is an ancestral condition predating the evolution of the pollination mutualism in Epicephala.
2. Material and methods
(a). Sampling
To determine the phylogenetic positions of Epicephala, Diphtheroptila and Caloptilia within the subfamily Gracillariinae, we first conducted a molecular phylogenetic analysis of the subfamily based on the mitochondrial cytochrome oxidase subunit I (COI) and the nuclear elongation factor-1 alpha (EF-1α), arginine kinase (ArgK) and 18S rRNA genes. Within Gracillariinae, Epicephala and Diphtheroptila belong to the Parectopa group as proposed by Kumata (1988), which is characterized by a highly distinct morphological synapomorphy (i.e. the female ostium bursae opens on the sternite of the seventh abdominal segment). Caloptilia belongs to the Gracillaria group (Kumata 1982) and thus is probably distantly related to Epicephala. We sampled a total of 45 non-Epicephala gracillariine species for phylogenetic analysis with a particular emphasis on the Parectopa group, including putative new taxa that have morphological affinities to Epicephala (full list of species are provided in the electronic supplementary material, table S1). An effort was made to sample moths from a broad range of angiosperm hosts to avoid sampling bias in our inference of generic relationships. Representatives of the subfamilies Oecophyllembiinae (Eumetriochroa hederae) and Lithocolletinae (Cameraria niphonica) were also sampled, and species of Bucculatricidae (Bucculatrix spp.) were used to root the entire gracillariid tree. Because we were unable to include many gracillariine genera in this analysis, our phylogenetic results remain inconclusive with regard to the sister group of Epicephala. However, firmly establishing the closest extant relative of Epicephala is not straightforward because a large number of unnamed lineages continue to be discovered (Vargas & Landry 2005; A. Kawakita & M. Kato 2009, personal observation) and phylogenetic relationships among gracillariine genera are difficult to resolve even by analyses of large molecular data sets (A. Kawahara, University of Maryland 2009, personal communication), probably owing to rapid radiation of extant genera. Rather, our purpose was to determine how closely Diphtheroptila and Caloptilia are related to Epicephala within the subfamily and to identify genera, if any, that are more closely related to Epicephala. Such genera share similar evolutionary background with Epicephala and thus provide an opportunity to test whether the level of host specificity in Epicephala is in fact exceptional in light of the overall evolutionary trend among its closest relatives.
To compare the degree of host specialization in Epicephala with those of Diphtheroptila, Caloptilia and the non-gracillariid seed feeders, we analysed the mitochondrial COI and nuclear EF-1α genes for these moths. Sampling was conducted within precisely the same geographical range as that used to assess pollinator specificity in our previous study (Kawakita & Kato 2006); this enabled a direct comparison of host specialization among herbivores using the same sets of Glochidion species. Diphtheroptila and Caloptilia were collected by sampling the leaves containing the larvae and were reared to adults in the laboratory. Seed-parasitic lepidopterans belonging to Carposinidae, Tortricidae and Pyralidae were sampled by collecting the fruits and were also reared to adults. To minimize the possibility of analysing multiple siblings, only one moth per tree per sampling event was used for the molecular analysis. Diphtheroptila sp. attacking Bridelia balansae and Caloptilia recitata infesting Rhus succendanea were used to root the Diphtheroptila and Caloptilia trees, respectively, based on the results of the phylogenetic analysis of Gracillariinae.
We also sampled moths of the leaf-miner genus Cuphodes because this genus was identified as the putative sister taxon of Epicephala in the Gracillariinae phylogeny (see §3). Cuphodes moths are known to occur on species of Diospyros (Ebenaceae) (Issiki 1957), Fabaceae (Kuroko 1982; Robinson et al. 2001) and Rhamnaceae (M. Kato 1991, personal observation), but their host ranges have not been investigated in detail. We therefore sampled Cuphodes from various species of Diospyros, Fabaceae and Rhamnaceae that occur in approximately the same geographical region as the above-sampled Glochidion herbivores to determine the degree of host specificity in a close relative of Epicephala. Analysis was done using COI, EF-1α and additionally, ArgK because we initially found one of the exemplars to fall into largely different clades in the COI and EF-1α phylogenies, which we suspected as the result of genetic introgression.
Full details of host association, sample size and locality information for the sampled gracillariid moths are provided in figure S1 and electronic supplementary material, tables S1 and S2.
(b). Molecular phylogenetic analysis
We extracted moth genomic DNA from thoracic muscle using a NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany). PCR amplification and direct sequencing of the COI, EF-1α, ArgK and 18S rRNA genes were conducted using the primers and protocols detailed in Kawakita et al. (2004) and Kawakita & Kato (2006, 2009). Heterozygous sites in EF-1α and ArgK were identified as double peaks of similar height in the chromatograms of both forward and reverse strands and accordingly coded using degenerate bases. Obtained sequences have been deposited in the GenBank database under accession numbers GU816251–GU816796.
Sequences of the protein-coding genes (COI, EF-1α and ArgK) contained no introns, and thus the alignment was straightforward. The alignment of 18S rRNA sequences was performed using ClustalX 2.0 (Larkin et al. 2007) software with default settings. Phylogenetic trees were constructed for each of the following four datasets: the four-gene dataset of Gracillariinae, the COI + EF-1α datasets of Diphtheroptila and Caloptilia, and the COI + EF-1α + ArgK dataset of Cuphodes. We focus on the analyses of combined datasets because initial analyses of individual genes suggested no strongly conflicting phylogenetic relationships among genes. However, there was a major incongruence in the placement of one Cuphodes specimen between the mitochondrial (COI) and nuclear gene datasets (EF-1α and ArgK; separate phylogenies are provided in the electronic supplementary material, figure S2), suggestive of mitochondrial introgression. We therefore performed the combined analysis excluding this anomalous Cuphodes individual. We did not construct phylogenetic trees for non-gracillariid seed parasites because each of the three taxonomic groups (Carposinidae, Tortricidae and Pyralidae) was represented by a single species having minimal sequence variation (see §3).
Phylogenetic trees were constructed using maximum likelihood (ML) and Bayesian methods. We performed ML analyses using the program Treefinder (Jobb 2008) with substitution model chosen and fitted separately for each gene. Nodal support was assessed using bootstrap analyses with 1000 replications. Bayesian analyses were performed using MrBayes 3.1.2 (Ronquist & Huelsenbeck 2003) with substitution parameters unlinked among gene partitions. Appropriate models of base substitution were selected for individual genes using MrModeltest 2.3 (Nylander 2004).
Because neither Diphtheroptila nor Caloptilia was recovered as sister to Epicephala on the Gracillariinae phylogeny, we tested the robustness of this reconstruction using the likelihood-based approximately unbiased (AU) test (Shimodaira 2002) as implemented in Treefinder. We also determined whether Epicephala, Diphtheroptila and Caloptilia each colonized Phyllanthaceae plants independently by reconstructing the ancestral host association using BayesTraits (Pagel et al. 2004). Each terminal taxon was coded as either Phyllanthaceae or non-Phyllanthaceae feeder, and ancestral states were reconstructed on the above-obtained ML phylogeny using the ML criterion. To account for phylogenetic uncertainty, we also used a Bayesian framework by integrating post-burn-in trees resulting from the Bayesian phylogenetic analysis. Likelihood ratio or Bayes factor of greater than 5 was considered significant evidence for the occurrence of either state at ancestral nodes (Pagel 1999; Pagel et al. 2004).
Analyses of Diphtheroptila and Caloptilia datasets recovered several well-defined clades that are each associated with two or more plant species (see §3). To detect any host-associated divergence within each of these putative species, we performed an analysis of molecular variance (AMOVA) on each of COI and EF-1α datasets using Arlequin 2.0 software (Schneider et al. 2000). EF-1α sequences with multiple heterozygous sites were analysed as genotypes with unknown gametic phase. Analyses were not performed for Cuphodes and non-gracillariid seed feeders owing to small sample sizes per clade/species.
Full details of the molecular analyses are provided in the electronic supplementary material.
3. Results and discussion
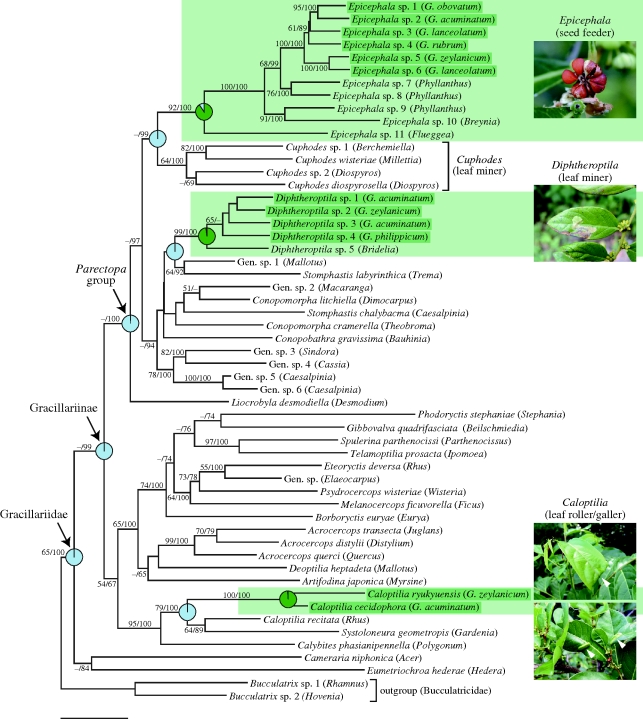
Phylogenetic analysis of Gracillariinae using the four-gene dataset suggested that Epicephala, Diphtheroptila and Caloptilia are not monophyletic and that they occupy separate positions in the phylogeny (figure 1). These relationships were recovered by both ML and Bayesian analyses, and an AU test rejected the hypothesis of either Diphtheroptila or Caloptilia forming a monophyletic group with Epicephala (p < 0.01 for both tests). Furthermore, Bayesian reconstruction of ancestral character states provided strong support for non-Phyllanthaceae plants as the ancestral host for Gracillariinae (figure 1). These results indicate that pollinating seed parasites (Epicephala) are not derived from leaf herbivores with which they share host plants, and that leaf-feeding Diphtheroptila and Caloptilia each colonized Glochidion plants independently. This contrasts with the situation in yucca moths, for which direct sisters of the pollinators are non-pollinating herbivores that feed on yuccas (Pellmyr & Leebens-Mack 1999; Pellmyr 2003). Nevertheless, the shared use of Glochidion by the three genera allows for a rigorous test of how different life histories affect patterns of host specificity in different herbivore clades by controlling for the effect of host plant species.
Figure 1.
Maximum-likelihood (ML) phylogeny of Gracillariinae based on 2548 bp of the combined mitochondrial COI and nuclear EF-1α, ArgK and 18S rRNA genes. Nodal numbers indicate ML bootstrap values followed by Bayesian posterior probabilities. Clades boxed in light green are those feeding on Phyllanthaceae plants, and species highlighted individually in dark green are those associated with Glochidion plants. Pie graphs show the relative likelihoods of alternative host associations at selected ancestral nodes: green, Phyllanthaceae host; blue, non-Phyllanthaceae host. Asterisks indicate significant differences in likelihoods (i.e. likelihood ratio or Bayes factor of greater than 5). Taxon names in parentheses indicate host plants (species name for Glochidion hosts and genus name for non-Glochidion hosts). Photographs show Glochidion fruit/leaves infested by Epicephala, Diphtheroptila and Caloptilia larvae; arrows indicate a leaf roll and leaf galls. Scale bar, 0.005 substitutions per site.
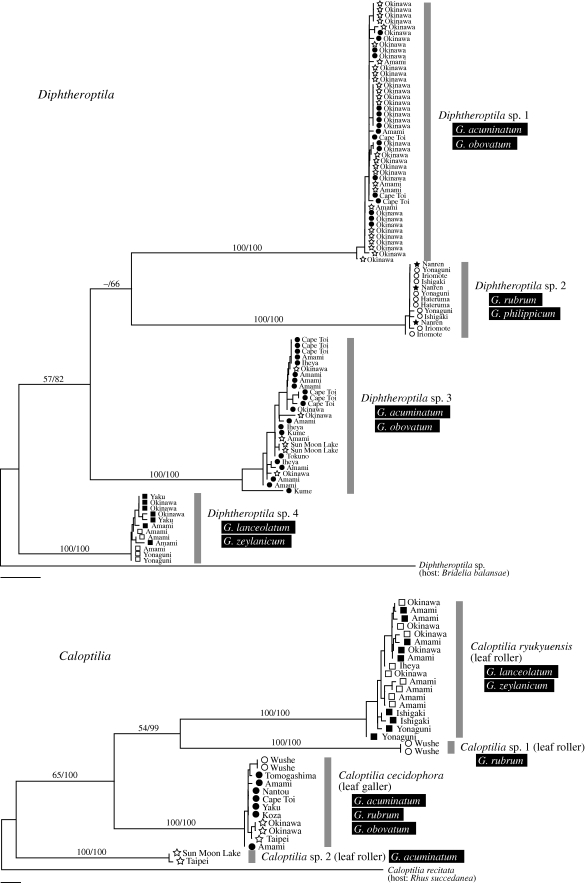
The analysis Diphtheroptila and Caloptilia COI + EF-1α datasets indicated that they each comprise four distinct clades throughout our sampling range (figure 2). Both COI and EF-1α recovered clades consisting of the same sets of individuals, suggesting that there is no gene flow among clades and thus they represent distinct species. Individuals belonging to different clades can also be distinguished by wing pattern, male genitalia morphology and larval feeding habit (A. Kawakita & M. Kato 2009, personal observation). The phylogenies of Diphtheroptila and Caloptilia further indicated that these moths commonly use more than one coexisting Glochidion species. We found no evidence for host race formation in these moths because the AMOVA analysis failed to detect host-associated genetic differentiation in either the COI or EF-1α gene (p > 0.1 for all tests). Although we cannot completely rule out the possibility of hidden divergence associated with Glochidion species, we consider it very unlikely that all the Diphtheroptila and Caloptilia species under consideration are at incipient stages of such host-associated divergence. Moreover, the level of host-associated differentiation, if any, is overwhelmingly lower than that found in Epicephala, in which individuals attacking different Glochidion hosts in any population are morphologically distinct and divergent by at least 4 per cent uncorrected pairwise sequence difference in the COI gene (Kawakita & Kato 2006). Therefore, these results provide strong evidence that Epicephala are more highly host specific than are their leaf-feeding relatives that use the same sets of Glochidion hosts.
Figure 2.
Maximum-likelihood (ML) phylogeny of Diphtheroptila and Caloptilia moths based on 1058 bp of the combined mitochondrial COI and EF-1α genes. Terminal symbols represent host Glochidion species followed by locality names. Numbers above branches indicate ML bootstrap values followed by Bayesian posterior probabilities. Individuals belonging to two of the Caloptilia clades were each identified morphologically as Caloptilia ryukyuensis and Caloptilia cecidophora. Symbols: filled circle, G. obovatum; open circle, G. rubrum; filled square, G. lanceolatum; open square, G. zeylanicum; filled star, G. philippicum; open star, G. acuminatum. Scale bars, 0.005 substitutions per site.
The observed increase in the level of host specialization in Epicephala, however, may simply be the result of a shift to seed feeding, rather than coevolutionary selection resulting from being a pollinator. We therefore determined the level of host specificity in seed-infesting lepidopterans that share the same larval food with Epicephala moths. Non-gracillariid moths that emerged from Glochidion fruits were morphologically identified as either Peragrarchis syncolleta (Carposinidae) or as undescribed species of Tritopterna (Tortricidae) or Cryptoblabes (Pyralidae). Analysis of the COI and EF-1α sequences in each of these moth taxa suggested that there is very little sequence variation among individuals sampled from four to five different Glochidion hosts and that none of the base substitutions found were diagnostic to host species (table 1). Although it is not straightforward to directly compare host specificity between Epicephala and non-gracillariid moths, the level of host specialization found in these groups is at the opposite extreme from the pattern expected if seed feeding is to promote higher host-specificity. Therefore, these results do not provide positive evidence that seed feeding favours a higher degree of host specialization and are consistent with the view that Epicephala host specificity is determined by factors other than larval diet.
Table 1.
Summary of genetic variations in non-gracillariid Glochidion seed parasites. Each moth species was sampled from four to five Glochidion hosts, and the number of base substitutions that were unique to individuals associated with a particular host (diagnostic sites) are given for each gene.
| bases sequenced |
diagnostic sites (variable sites) |
|||||
|---|---|---|---|---|---|---|
| family/species | host species(locality number) | moths sampled | COI | EF-1α | COI | EF-1α |
| Carposinidae | ||||||
| Peragrarchis syncolleta | G. acuminatum | 2 | 580 | 498 | 0 (1) | 0 (3) |
| G. lanceolatum | 1 | |||||
| G. obovatum | 3 | |||||
| G. rubrum | 1 | |||||
| Tortricidae | ||||||
| Tritopterna sp. | G. lanceolatum | 4 | 580 | 498 | 0 (2) | 0 (14) |
| G. obovatum | 1 | |||||
| G. philippicum | 1 | |||||
| G. rubrum | 3 | |||||
| G. zeylanicum | 4 | |||||
| Pyralidae | ||||||
| Cryptoblabes sp. | G. lanceolatum | 1 | 580 | 498 | 0 (0) | 0 (3) |
| G. obovatum | 2 | |||||
| G. rubrum | 2 | |||||
| G. zeylanicum | 2 | |||||
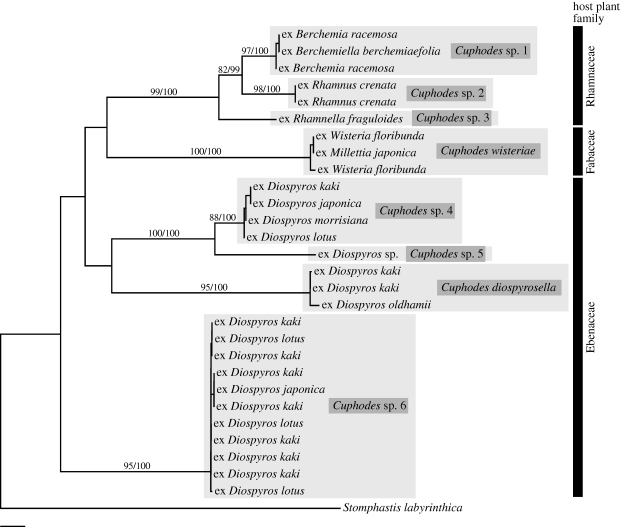
Another explanation for strict host specialization in Epicephala is that such high host specificity is a common feature among all the closest relatives of Epicephala, and that pollinator habit evolved against a background of high host specificity. To test this possibility, we examined host specificity in species of Cuphodes, which was tentatively suggested as the putative sister group of Epicephala in the Gracillariinae phylogeny (figure 1). Although an AU test did not reject the non-monophyly of the Epicephala + Cuphodes clade (p > 0.1), Epicephala and Cuphodes share distinct apomorphies not otherwise found in any genera of the Parectopa group (i.e. two pairs of bristles on the seventh and eighth abdominal segments in the males and anteriorly tilted posture of resting adults; A. Kawakita & M. Kato 2009, personal observation), suggesting that Cuphodes is probably one of the closest relatives of Epicephala.
Analysis of the COI + EF-1α + ArgK dataset in Cuphodes suggested that the sampled moths consist of eight putative species (figure 3; also see electronic supplementary material, figure S2), which can be distinguished by wing pattern, male genitalia morphology and larval mining pattern (A. Kawakita & M. Kato 2009, personal observation). The obtained phylogeny demonstrated that these species regularly use two to four closely related plants (figure 3), suggesting that the closest relatives of Epicephala do not show the same degree of host specificity as Epicephala. Although the use of different host plant families in Epicephala and Cuphodes may make direct comparison difficult, available evidence suggests that Cuphodes species exhibit much broader host ranges than do the species of Epicephala. For example, C. wisteriae uses Wisteria and Millettia, which are distantly related genera within Fabaceae, having diverged at least 50 Ma (Lavin et al. 2005), whereas the age of the Glochidion crown group is estimated to be less than 10 Ma (Kawakita & Kato 2009). Similarly, Cuphodes sp. 4 feeds on two genera (Berchemia and Berchemiella) of the Rhamnaceae family, although the antiquity of their divergence is unknown. Thus, these results indicate that the high degree of host specialization found in Epicephala is probably not an ancestral condition predating the evolution of pollinator habit.
Figure 3.
Maximum-likelihood (ML) phylogeny of Cuphodes based on 1601 bp of the combined mitochondrial COI and nuclear EF-1α and ArgK genes. Terminal taxon labels indicate host plant names. Host plant families are indicated using bars on the right. Numbers above branches indicate ML bootstrap values followed by Bayesian posterior probabilities. Scale bar, 0.01 substitutions per site.
Taken together, the present results on host specificity of Diphtheroptila, Caloptilia, Cuphodes and Glochidion-feeding, non-gracillariid lepidopterans all indicate that the level of host specialization in Epicephala is higher than would be expected if host specificity were determined solely by the herbivorous habit of the moths. Thus, our data are more consistent with the view that pollinator habit favours higher host specificity than the ancestral parasitic lifestyle. What, then, is the ultimate cause driving the strict host specificity of Epicephala? A previous analysis of floral scent in Glochidion (Okamoto et al. 2007) found clear differences in the chemical composition of floral volatiles among co-flowering Glochidion species. These differences are perceived by host-seeking Epicephala females (Okamoto et al. 2007) and probably facilitate the attraction of species-specific pollinators. Thus, selection may operate on Glochidion plants to produce distinct floral scents and attract specific pollinators and thereby to avoid incompatible hybridization. Although further experimentation is needed to determine whether interspecific crosses result in fruit production, any decrease in quantity and/or quality of hybrid fruits is likely to facilitate reciprocal specialization by Epicephala to species-specific floral volatiles. Thus, it is possible that the high plant–pollinator-specificity in obligate pollination mutualisms is driven by a plant's interest to avoid less advantageous hybridization.
Overall, our finding of strict host specificity in Epicephala is in marked contrast with previous findings in the yucca moth lineage (Pellmyr & Thompson 1992; Thompson 1994, 2005). The closest relatives of the pollinating yucca moths, Prodoxus, feeds on inflorescence stalks, fruit or, rarely, leaves of yucca plants and have very similar degrees of host specificity with the pollinating yucca moths (Pellmyr et al. 2006). We suggest that this difference is because of contrasting patterns of flowering phenology between yuccas and Glochidion. Because both pollinating (Tegeticula and Parategeticula) and non-pollinating (Prodoxus) yucca moths are short-lived (Powell 1984) and their life histories are strongly associated with yucca flowers (Pellmyr 1999, 2003; Pellmyr et al. 2006), the adult moths must emerge during a short period when host flowers are available. However, coexisting yucca species typically exhibit largely non-overlapping flowering periods (Pellmyr 2003); thus, there is little opportunity for both pollinating and non-pollinating yucca moths to select among multiple hosts within a single population. In contrast, most Glochidion species produce flowers and leaves continuously from spring to autumn, and as many as four species flower at the same time within our study area. Under such circumstances, both leaf-feeding and flower-infesting moths are provided with multiple available hosts, but the latter are more selective in their host choice owing to a broader range of coevolutionary traits with which they are constrained. The occurrence of multiple co-flowering host species is also the case in figs, for which preliminary analysis of host specificity in pollinating and non-pollinating fig wasps suggested that the former tends to be more host-specific (Weiblen & Bush 2002; Marussich & Machado 2007; but see Lopez-Vaamonde et al. 2001; Jousselin et al. 2006, 2008).
Although our data clearly indicate that pollinating seed parasites exhibit higher degrees of host specialization than those of their parasitic ancestors, a more direct test of host specificity would be to include non-pollinating Epicephala seed parasites in the analysis. The most basal lineage of Epicephala is a non-pollinator that attacks the seeds of Flueggea, a close relative of Glochidion within the tribe Phyllantheae. However, this species is currently known only from Flueggea suffruticosa in southwestern Japan (Kawakita & Kato 2009), where there are no other co-occurring Flueggea species. Also, a derived clade of Epicephala has secondarily lost the pollinating habit, and currently there are three species that are each specific to a single Phyllanthus host (Kawakita & Kato 2009). However, closely related Phyllanthus hosts are rarely available within the same population, which precludes a direct comparison of host specificity with pollinating Epicephala in this case as well. Within the yucca moth lineage, two derived species have independently lost their pollinating behaviour and oviposit in young fruits to exploit the seeds that other yucca moth species have pollinated (Pellmyr et al. 1996; Pellmyr 1999). These cheater species evolved to use four to six yucca hosts (Pellmyr 1999, 2003), which is consistent with our hypothesis that pollinator habit promotes host specificity in pollinating seed parasites. The cheater yucca moths are likely to have a broader phenological window for successful oviposition (Pellmyr 2003); thus, selection for host specialization may have been relaxed in these derived non-pollinators.
Although further research is required to identify coevolutionary forces driving pollinator specificity, our findings indicate that mutualistic selection probably favours strict host specificity of pollinating seed parasites in obligate pollination mutualisms. As shown in recent population-level analyses of gene flow in figs and yuccas (Machado et al. 2005; Smith et al. 2008, 2009), pollinator specificity is likely to strongly impact patterns of gene flow between coexisting plant species and play an important role in facilitating reproductive isolation between diverged populations. Thus, future studies of coevolution and codivergence in the Glochidion–Epicephala mutualism, as well as comparative analysis across systems, are likely to substantially improve our understanding of the role of coevolution in promoting speciation and diversification in obligate pollination mutualisms.
Acknowledgements
We thank D. Hembry for the identification of Tritopterna and critical reading of the manuscript; T. Kumata, I. Oshima and A. Kawahara for communication on gracillariid morphology and systematics; K. Tsuji and A. Kawahara for specimens; Y. Kameda and G. P. Svensson for field assistance; and two anonymous reviewers for comments that improved the paper. This work was supported by Japan Society for the Promotion of Science grants to A.K. and to M.K.
References
- Aanen D. K., Eggleton P., Lefèvre C. R., Frøslev T. G., Rosendahl S., Boomsma J. J.2002The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc. Natl Acad. Sci. USA 99, 14 887–14 892 (doi:10.1073/pnas.222313099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouat C., Garcia N., Andary C., McKey D.2001Plant lock and key: pairwise coevolution of an exclusion filter in an ant–plant mutualism. Proc. R. Soc. Lond. B 268, 2131–2141 (doi:10.1098/rspb.2001.1763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A.2004Speciation. Sunderland, MA: Sinauer Associates [Google Scholar]
- Currie C. R., Wong B., Stuart A. E., Schultz T. R., Rehner S. A., Mueller U. G., Sung G. H., Spatafora J. W., Straus N. A.2003Ancient tripartite coevolution in the attine ant–microbe symbiosis. Science 299, 386–388 (doi:10.1126/science.1078155) [DOI] [PubMed] [Google Scholar]
- Davidson D. W., McKey D.1993The evolutionary ecology of symbiotic ant–plant relationships. J. Hym. Res. 2, 13–83 [Google Scholar]
- Edwards D. P., Hassall M., Sutherland W. J., Yu D. W.2006Assembling a mutualism: ant symbionts locate their host plants by detecting volatile compounds. Insect. Soc. 53, 172–176 (doi:10.1007/s00040-006-0855-z) [Google Scholar]
- Ehrlich P. R., Raven P. H.1964Butterflies and plants: a study in coevolution. Evolution 18, 586–608 (doi:10.2307/2406212) [Google Scholar]
- Farrell B. D.1998‘Inordinate fondness’ explained: why are there so many beetles? Science 281, 555–559 (doi:10.1126/science.281.5376.555) [DOI] [PubMed] [Google Scholar]
- Federle W., Maschwitz U., Fiala B., Riederer M., Hölldobler B.1997Slippery ant-plants and skillful climbers: selection and protection of specific ant partners by epicuticular wax blooms in Macaranga (Euphorbiaceae). Oecologia 112, 217–224 (doi:10.1007/s004420050303) [DOI] [PubMed] [Google Scholar]
- Gómez J. M., Zamora R.2006Ecological factors that promote the evolution of generalization in pollination systems. In Plant–pollinator interactions: from generalization to specialization (eds Waser N., Ollerton J.), pp. 145–166 Chicago, IL: University of Chicago Press [Google Scholar]
- Govaerts R., Frodin D. G., Radcliffe-Smith A.2000World checklist and bibliography of Euphorbiaceae. Kew, UK: The Royal Botanic Gardens [Google Scholar]
- Grangier J., Dejean A., Malé P. J. G., Solano P. J., Orivel J.2009Mechanisms driving the specificity of a myrmecophyte–ant association. Biol. J. Linn. Soc. 97, 90–97 (doi:10.1111/j.1095-8312.2008.01188.x) [Google Scholar]
- Guimarães P. R., Jr, Rico-Gray V., Oliveira P. S., Izzo T. J., dos Reis S. F., Thompson J. N.2007Interaction intimacy affects structure and coevolutionary dynamics in mutualistic networks. Curr. Biol. 17, 1797–1803 [DOI] [PubMed] [Google Scholar]
- Heil M., McKey D.2003Protective ant–plant interactions as model systems in ecological and evolutionary research. Annu. Rev. Ecol. Evol. Syst. 34, 425–453 (doi:10.1146/annurev.ecolsys.34.011802.132410) [Google Scholar]
- Heil M., Rattke J., Boland W.2005Postsecretory hydrolysis of nectar sucrose and specialization in ant/plant mutualism. Science 308, 560–563 (doi:10.1126/science.1107536) [DOI] [PubMed] [Google Scholar]
- Herre E. A., Jandér K. C., Machado C. A.2008Evolutionary ecology of figs and their associates: recent progress and outstanding puzzles. Annu. Rev. Ecol. Evol. Syst. 39, 439–458 (doi:10.1146/annurev.ecolsys.37.091305.110232) [Google Scholar]
- Hosokawa T., Kikuchi Y., Nikoh N., Shimada M., Fukatsu T.2006Strict host–symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4, 1841–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issiki S.1957Lepidoptera. In Iconographia Insectorum Japonicorum in Coloribus Naturalibus, Part 1 (eds Esaki T., Issiki S., Inoue H.), pp. 1–318 Osaka, Japan: Hoikusha. [In Japanese.] [Google Scholar]
- Janzen D. H.1979How to be a fig. Annu. Rev. Ecol. Syst. 10, 13–51 (doi:10.1146/annurev.es.10.110179.000305) [Google Scholar]
- Jobb G.2008Treefinder version of October 2008. Program distributed by the author. Munich, Germany: See http://www.treefinder.de/ [Google Scholar]
- Jousselin E., van Noort S., Rasplus J. Y., Greeff J. M.2006Patterns of diversification of Afrotropical Otiteselline fig wasps: phylogenetic study reveals a double radiation across host figs and conservatism of host associations. J. Evol. Biol. 19, 253–266 (doi:10.1111/j.1420-9101.2005.00968.x) [DOI] [PubMed] [Google Scholar]
- Jousselin E., van Noort S., Berry V., Rasplus J. Y., Rønsted N., Erasmus J. C., Greeff J. M.2008One fig to bind them all: host conservatism in a fig wasp community unraveled by cospeciation analyses among pollinating and nonpollinating fig wasps. Evolution 62, 1777–1797 (doi:10.1111/j.1558-5646.2008.00406.x) [DOI] [PubMed] [Google Scholar]
- Kato M., Takimura A., Kawakita A.2003An obligate pollination mutualism and reciprocal diversification in the tree genus Glochidion. Proc. Natl Acad. Sci. USA 100, 5264–5267 (doi:10.1073/pnas.0837153100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakita A.2010Evolution of obligate pollination mutualism in the tribe Phyllantheae (Phyllanthaceae). Plant Species Biol. 25, 3–19 (doi:10.1111/j.1442-1984.2009.00266.x) [Google Scholar]
- Kawakita A., Kato M.2004aEvolution of obligate pollination mutualism in New Caledonian Phyllanthus (Euphorbiaceae). Am. J. Bot. 91, 410–415 (doi:10.3732/ajb.91.3.410) [DOI] [PubMed] [Google Scholar]
- Kawakita A., Kato M.2004bObligate pollination mutualism in Breynia (Phyllanthaceae): further documentation of pollination mutualism involving Epicephala moths (Gracillariidae). Am. J. Bot. 91, 1319–1325 (doi:10.3732/ajb.91.9.1319) [DOI] [PubMed] [Google Scholar]
- Kawakita A., Kato M.2006Assessment of the diversity and species-specificity of the mutualistic association between Epicephala moths and Glochidion trees. Mol. Ecol. 15, 3567–3581 (doi:10.1111/j.1365-294X.2006.03037.x) [DOI] [PubMed] [Google Scholar]
- Kawakita A., Kato M.2009Repeated independent evolution of obligate pollination mutualism in the Phyllantheae–Epicephala association. Proc. R. Soc. B 276, 417–426 (doi:10.1098/rspb.2008.1226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakita A., Takimura A., Terachi T., Sota T., Kato M.2004Cospeciation analysis of an obligate pollination mutualism: have Glochidion trees (Euphorbiaceae) and pollinating Epicephala moths (Gracillariidae) diversified in parallel? Evolution 58, 2201–2214 [DOI] [PubMed] [Google Scholar]
- Kumata T.1982A taxonomic revision of the Gracillaria group occurring in Japan. Insecta Matsumurana 26, 1–186 [Google Scholar]
- Kumata T.1988Japanese species of the Acrocercops-group (Lepidoptera: Gracillariidae) part I. Insecta Matsumurana 38, 1–111 [Google Scholar]
- Kuroko H.1982Gracillariidae. In Moths of Japan, vol. 1 (eds Inoue H., Sugi S., Kuroko H., Moriuchi S., Kawabe A., Owada M.), pp. 176–202 Tokyo, Japan: Kodansha. [In Japanese.] [Google Scholar]
- Larkin M. A., et al. 2007ClustalW and ClustalX version 2.0. Bioinformatics 23, 2947–2948 (doi:10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- Lavin M., Herendeen P. S., Wojciechowski M. F.2005Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst. Biol. 54, 575–594 (doi:10.1080/10635150590947131) [DOI] [PubMed] [Google Scholar]
- Lopez-Vaamonde C., Rasplus J. Y., Weiblen G. D., Cook J. M.2001Molecular phylogenies of fig wasps: partial cocladogenesis of pollinators and parasites. Mol. Phylogenet. Evol. 21, 55–71 (doi:10.1006/mpev.2001.0993) [DOI] [PubMed] [Google Scholar]
- Machado C. A., Robbins N., Gilbert M. T. P., Herre E. A.2005Critical review of host specificity and its coevolutionary implications in the fig/fig-wasp mutualism. Proc. Natl Acad. Sci. USA 102, 6558–6565 (doi:10.1073/pnas.0501840102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marussich W. A., Machado C. A.2007Host-specificity and coevoluion among pollinating and nonpollinating New World fig wasps. Mol. Ecol. 16, 1925–1946 (doi:10.1111/j.1365-294X.2007.03278.x) [DOI] [PubMed] [Google Scholar]
- Mikheyev A. S., Mueller U. G., Abbot P.2006Cryptic sex and many-to-one coevolution in the fungus-growing ant symbiosis. Proc. Natl Acad. Sci. USA 103, 10 702–10 706 (doi:10.1073/pnas.0601441103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter C., Farrell B., Wiegmann B.1988The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification? Am. Nat. 132, 107–128 (doi:10.1086/284840) [Google Scholar]
- Molbo D., Machado C. A., Sevenster J. G., Keller L., Herre E. A.2003Cryptic species of fig-pollinating wasps: Implications for the evolution of the fig–wasp mutualism, sex allocation, and precision of adaptation. Proc. Natl Acad. Sci. USA 100, 5867–5872 (doi:10.1073/pnas.0930903100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., Telang A.1998The evolution of bacteriocyte-associated endosymbionts in insects. Bioscience 48, 295–304 (doi:10.2307/1313356) [Google Scholar]
- Mueller U. G., Rehner S. A., Schultz T. R.1998The evolution of agriculture in ants. Science 281, 2034–2038 (doi:10.1126/science.281.5385.2034) [DOI] [PubMed] [Google Scholar]
- Novotny V., Basset Y., Miller S. E., Weiblen G. D., Bremer B., Cizek L., Drozd P.2002Low host specificity of herbivorous insects in a tropical forest. Nature 416, 841–844 (doi:10.1038/416841a) [DOI] [PubMed] [Google Scholar]
- Novotny V., Drozd P., Miller S. E., Kulfan M., Janda M., Basset Y., Weiblen G. D.2006Why are there so many species of herbivorous insects in tropical forests? Science 313, 1115–1118 (doi:10.1126/science.1129237) [DOI] [PubMed] [Google Scholar]
- Nylander J. A. A.2004MrModeltest v2. Program distributed by the author. Sweden: Evolutionary Biology Centre, Uppsala University; See http://www.abc.se/~nylander/ [Google Scholar]
- Okamoto T., Kawakita A., Kato M.2007Interspecific variation of floral scent composition in Glochidion (Phyllanthaceae) and its association with host-specific pollinating seed parasite (Epicephala; Gracillariidae). J. Chem. Ecol. 33, 1065–1081 (doi:10.1007/s10886-007-9287-0) [DOI] [PubMed] [Google Scholar]
- Pagel M.1999The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 48, 612–622 [Google Scholar]
- Pagel M., Meade A., Barker D.2004Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673–684 (doi:10.1080/10635150490522232) [DOI] [PubMed] [Google Scholar]
- Pellmyr O.1999A systematic revision of the yucca moths in the Tegeticula yuccasella complex north of Mexico. Syst. Entomol. 24, 243–271 (doi:10.1046/j.1365-3113.1999.00079.x) [Google Scholar]
- Pellmyr O.2002Pollination by animals. Plant–animal interactions (eds Herrera C. M., Pellmyr O.), pp. 157–184 Oxford, UK: Blackwell Publishing [Google Scholar]
- Pellmyr O.2003Yuccas, yucca moths, and coevolution: a review. Ann. Missouri Bot. Gard. 90, 35–55 (doi:10.2307/3298524) [Google Scholar]
- Pellmyr O., Leebens-Mack J.1999Forty million years of mutualism: evidence for Eocene origin of the yucca–yucca moth association. Proc. Natl Acad. Sci. USA 96, 9178–9183 (doi:10.1073/pnas.96.16.9178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellmyr O., Thompson J. N.1992Multiple occurrences of mutualism in the yucca moth lineage. Proc. Natl Acad. Sci. USA 89, 2927–2929 (doi:10.1073/pnas.89.7.2927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellmyr O., Leebens-Mack J., Huth C. J.1996Non-mutualistic yucca moths and their evolutionary consequences. Nature 380, 155–156 (doi:10.1038/380155a0) [DOI] [PubMed] [Google Scholar]
- Pellmyr O., Balcázar-Lara M., Althoff D. M., Segraves K., Leebens-Mack J.2006Phylogeny and life history evolution of Prodoxus yucca moths (Lepidoptera: Prodoxidae). Syst. Entomol. 31, 1–20 (doi:10.1111/j.1365-3113.2005.00301.x) [Google Scholar]
- Poulsen M., Boomsma J. J.2005Mutualistic fungi control crop diversity in fungus-growing ants. Science 307, 741–744 (doi:10.1126/science.1106688) [DOI] [PubMed] [Google Scholar]
- Powell J. A.1984Biological interrelationships of moths and Yucca schottii. Univ. Calif. Publ. Entomol. 100, 1–93 [Google Scholar]
- Price P.1980Evolutionary biology of parasites. Princeton, NJ: Princeton University Press [Google Scholar]
- Quek S. P., Davies S. J., Ashton P. S., Itino T., Pierce N. E.2007The geography of diversification in mutualistic ants: a gene's-eye view into the Neogene history of Sundaland rain forests. Mol. Ecol. 16, 2045–2062 (doi:10.1111/j.1365-294X.2007.03294.x) [DOI] [PubMed] [Google Scholar]
- Robinson G. S., Ackery P. R., Kitching I. J., Beccaloni G. W., Hernández L. M.2001Host plants of the moth and butterfly caterpillars of the Oriental Region. London, UK: The National History Museum [Google Scholar]
- Ronquist F., Huelsenbeck J. P.2003MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- Schluter D.2000The ecology of adaptive radiation. Oxford, UK: Oxford University Press [Google Scholar]
- Schneider S., Roessli D., Excoffier L.2000Arlequin: a software for population genetics data analysis, v. 2.0. Geneva: Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva [Google Scholar]
- Segraves K. A., Pellmyr O.2004Testing the out-of-Florida hypothesis on the origin of cheating in the yucca–yucca moth mutualism. Evolution 58, 2266–2279 [DOI] [PubMed] [Google Scholar]
- Shimodaira H.2002An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (doi:10.1080/10635150290069913) [DOI] [PubMed] [Google Scholar]
- Smith C. I., Godsoe W. K. W., Tank S., Yoder J. B., Pellmyr O.2008Distinguishing coevolution of covicariance in an obligate pollination mutualism: asynchronous divergence in Joshua tree and its pollinators. Evolution 62, 2676–2687 (doi:10.1111/j.1558-5646.2008.00500.x) [DOI] [PubMed] [Google Scholar]
- Smith C. I., Drummond C. S., Godsoe W., Yoder J. B., Pellmyr O.2009Host specificity and reproductive success of yucca moths (Tegeticula spp. Lepidoptera: Prodoxidae) mirror patterns of gene flow between host plant varieties of the Joshua tree (Yucca brevifolia: Agavaceae). Mol. Ecol. 18, 5218–5229 (doi:10.1111/j.1365-294X.2009.04428.x) [DOI] [PubMed] [Google Scholar]
- Strauss S. Y., Zangerl A. R.2002Plant–insect interactions in terrestrial ecosystems. In Plant–animal interactions (eds Herrera C. M., Pellmyr O.), pp. 77–106 Oxford, UK: Blackwell Publishing [Google Scholar]
- Thompson J. N.1994The coevolutionary process. Chicago, IL: The University of Chicago Press [Google Scholar]
- Thompson J. N.2005The geographic mosaic of coevolution. Chicago, IL: The University of Chicago Press [Google Scholar]
- Vargas H. A., Landry B.2005New genus and species of Gracillariidae (Lepidoptera) feeding on flowers of Acacia macracantha Wild. (Mimosaceae) in Chile. Acta Ent. Chilena 29, 47–57 [Google Scholar]
- Visser A. A., Ros V. I. D., de Beer Z. W., Debets A. J. M., Hartog E., Kuyper T. W., Laessøe T., Slippers B., Aanen D. K.2009Levels of specificity of Xylaria species associated with fungus-growing termites: a phylogenetic approach. Mol. Ecol. 18, 553–567 (doi:10.1111/j.1365-294X.2008.04036.x) [DOI] [PubMed] [Google Scholar]
- Weiblen G. D.2002How to be a fig wasp. Annu. Rev. Entomol. 47, 299–330 (doi:10.1146/annurev.ento.47.091201.145213) [DOI] [PubMed] [Google Scholar]
- Weiblen G. D., Bush G. L.2002Speciation in fig pollinators and parasites. Mol. Ecol. 11, 1573–1578 (doi:10.1046/j.1365-294X.2002.01529.x) [DOI] [PubMed] [Google Scholar]