Abstract
Lepidoptera (butterflies and moths) represent one of the most diverse animals groups. Yet, the phylogeny of advanced ditrysian Lepidoptera, accounting for about 99 per cent of lepidopteran species, has remained largely unresolved. We report a rigorous and comprehensive analysis of lepidopteran affinities. We performed phylogenetic analyses of 350 taxa representing nearly 90 per cent of lepidopteran families. We found Ditrysia to be a monophyletic taxon with the clade Tischerioidea + Palaephatoidea being the sister group of it. No support for the monophyly of the proposed major internested ditrysian clades, Apoditrysia, Obtectomera and Macrolepidoptera, was found as currently defined, but each of these is supported with some modification. The monophyly or near-monophyly of most previously identified lepidopteran superfamilies is reinforced, but several species-rich superfamilies were found to be para- or polyphyletic. Butterflies were found to be more closely related to ‘microlepidopteran’ groups of moths rather than the clade Macrolepidoptera, where they have traditionally been placed. There is support for the monophyly of Macrolepidoptera when butterflies and Calliduloidea are excluded. The data suggest that the generally short diverging nodes between major groupings in basal non-tineoid Ditrysia are owing to their rapid radiation, presumably in correlation with the radiation of flowering plants.
Keywords: Lepidoptera, molecular systematics, phylogeny, rapid radiation
1. Introduction
Lepidoptera (moths and butterflies) with over 160 000 described and 500 000 estimated species are among the most diverse animal groups (Kristensen et al. 2007). Together with Coleoptera, Hymenoptera and Diptera, they cover well beyond one half of all described organism species (Hunt et al. 2007; Sharkey 2007; Yeates et al. 2007). Their fascinating appearance renders them the most adored group of insects. They have been bred for sericultural purposes for at least 5000 years, and the detailed life histories of countless numbers of species have been investigated since then. Lepidoptera include many serious pest species and they have been popular organisms in various model systems (Roe et al. 2010). The phylogenetics of some major lepidopteran groups have been intensively studied on the basis of their morphology, and more recently also based on DNA data. While the interrelationships of the earliest lineages of Lepidoptera have been intensively studied for decades (Kristensen & Skalski 1998; Kristensen et al. 2007), the largest radiation of Lepidoptera, the Ditrysia has eluded analytical studies until recently (Regier et al. 2009). However, even this analysis was based on a limited set of taxa and taxon sampling heavily biased towards ‘advanced’ Lepidoptera.
Hence, the order itself lacks a rigorous evolutionary framework. In addition to a lack of phylogenetic analyses with comprehensive taxon sampling, we can see two additional reasons for the existing situation. First, groups of ditrysian Lepidoptera are morphologically homogeneous, and their phylogenetic affinities are therefore especially difficult to unravel (Kristensen & Skalski 1998), and second, there has been a shortage of phylogenetically informative genetic markers suitable for routine phylogenomic analyses. Owing to recent pioneering work in designing appropriate nuclear genomic markers (Wahlberg & Wheat 2008), there is now a good set of suitable genetic markers for use in lepidopteran phylogenomics.
The aim of this research was to clarify broad patterns of lepidopteran affinities using a wide array of molecular markers and a comprehensive taxon sampling. We performed phylogenetic analyses of 350 taxa that represent 43 out of 45 recognized lepidopteran superfamilies and nearly 90 per cent of lepidopteran families. These taxa altogether represent over 99 per cent of described moth and butterfly species. The analyses are based on eight gene regions, of which seven are from the nuclear genome and one from the mitochondrial genome. Our results show several unexpected patterns as well as corroborate many previous groupings.
2. Material and methods
(a). Material acquisition and taxon sampling
The taxon sampling was planned largely based on the tentative lepidopteran phylogeny of Minet (1991) and Kristensen et al. (2007). Basically, all taxa at the subfamily level for which suitable material was available were included, supplemented with many taxa for which phylogenetic affinities have remained ambiguous, a total of 350 taxa (see the electronic supplementary material, table S1). The study covers all the lepidopteran superfamilies except Simaethistoidea and Whalleyanioidea, for which recent material is not known. Of the 124 lepidopteran families listed in Kristensen et al. (2007), 111 (89.5%) are included in this study (see the electronic supplementary material, table S1). They are divided into 252 named subfamilies, accounting for 75.2 per cent of the subfamilies listed in Kristensen et al. (2007). The classification and nomenclature follow Kristensen et al. (2007), except in Lypusidae and Cimelioidea, where subsequent classifications were followed (Yen & Minet 2007; Heikkilä & Kaila 2010). The bulk of the DNA material was gathered from the DNA collections of the authors. Notable additions were received from the ATOLep DNA collection at the University of Maryland (http://www.leptree.net), the Australian National Collection (ANIC) at the Commonwealth Scientific and Industrial Research Organization, and several private collections.
(b). Molecular techniques
Usually, legs preserved in 100 per cent alcohol were used for DNA extraction, but sometimes also other body parts or larvae were used. In several cases, air-dried specimens up to 10 years old yielded DNA of good quality, indicating that also nuclear DNA of unrelaxed material kept in dry and cool conditions can successfully be amplified using standard extraction and sequencing protocols even after a relatively long time of preservation (Wahlberg & Wheat 2008). Notably, material of the ANIC and some private collections were found to be very useful, emphasizing the importance of proper preservation conditions in museum collections, and at the same time warning against water relaxation of specimens, as such an operation almost certainly fragments the nuclear DNA. Remaining parts of specimens were stored to serve as vouchers. Total genomic DNA was extracted and purified using Qiagen's DNeasy extraction kit. DNA amplification and sequencing were carried out following protocol explained in detail elsewhere (Wahlberg & Wheat 2008). Sequencing was performed mainly with an ABI 3730 capillary sequencer (Oulu), and a smaller part with an ABI PRISMR 3130×l capillary sequencer (Turku). One mitochondrial (COI) and seven protein-coding nuclear gene regions (EF-1α, Wingless, RpS5, MDH, GAPDH, CAD and IDH) were sequenced, accounting for a total of 6303 bp with gaps. For details on the sequencing success and GenBank accession numbers of each species and gene, see the electronic supplementary material, table S2.
(c). Phylogenetic analyses
The sequence alignments were carried out manually using BioEdit 7.0.4.1. (Hall 1999). Overall, alignment of the gene regions was straightforward, however, there are short regions in Wingless and RpS5 in which unambiguous alignment was not possible across all sequences. Consequently, we excluded these regions from the data, leaving 6157 bp for analysis. To minimize the risk of any kind of confusion during the sequencing protocol and errors in alignments, we constructed neighbour-joining and maximum likelihood trees separately for each gene and checked them carefully for identical sequences and other doubtful patterns. Furthermore, to minimize the risk of wrong identification, all the specimens were cross-checked with their DNA barcodes in BOLD (Barcode of Life Data Systems, http://www.barcodinglife.org/views/login.php) (Ratnasingham & Hebert 2007), where reference specimens were available for many of the species used in this study (see the electronic supplementary material, table S1).
In a few cases where material of excellent quality was not available, sequence data were constructed by combining sequences from two individuals. This was done only if sequences successful in both individuals showed perfect or very close identity, suggesting no doubt about their conspecificity.
Since we were particularly interested in basal splitting events of ditrysian Lepidoptera, we evaluated the information value of third positions of codons in each gene separately. Changes in third positions represent mostly synonymous substitutions, and are hence unlikely to provide useful information at the deeper level of lepidopteran phylogeny, but may instead considerably increase the amount of uninformative ‘noise’ (homoplasy). After examining gene trees and a number of trials with various gene and taxon sets, we excluded third positions from the data in all genes except EF-1α, which evolves more slowly than any of the other genes (Wahlberg & Wheat 2008) and which, based on our evaluation, showed better resolution with third positions than without them. The trials with third positions included showed overall lower support values, and a few deviating groupings were regularly poorly supported and considered unlikely to be true. The effect of third positions was however not crucial as differences between the trials were not remarkable. With third positions excluded from all genes, except EF-1a, the data consisted of 4451 bp.
The phylogenetic analyses were carried out with both the model-based (maximum likelihood and Bayesian inference) and parsimony methods. We rooted our trees with Micropterigidae, arguably the sister group to other Lepidoptera (Kristensen & Skalski 1998; Wiegmann et al. 2002; Kristensen et al. 2007). The maximum likelihood analyses were carried out under the GTR + G model, chosen by Modeltest v. 3.7, and the data were partitioned into eight gene regions. The maximum likelihood analysis was implemented using the online version of RAxML (http://phylobench.vital-it.ch/raxml-bb/index.php) (Stamatakis et al. 2008). Supports for nodes were evaluated with 1000 bootstrap replicates of the data. We considered groups supported by over 50 per cent and strongly supported by over 90 per cent bootstrap support values.
The Bayesian analyses were carried out on a subset of the data using BEAST v. 1.4.8. (Drummond & Rambaut 2007). This analysis specifically aimed at clarifying basal branching events in Ditrysia and was performed with a subset of 118 species, because independent runs of the full set of taxa failed to converge in 20 million generations. The data were partitioned into the mitochondrial gene region (COI), the full EF-1α gene region and the combined nuclear genes with third codon positions removed (CAD, GAPDH, IDH, MDH, RpS5 and Wingless). The tree prior was set to the birth–death process, the independent models for the three partitions were all set to the GTR + G model, while all other priors were left to defaults. The branch lengths were allowed to vary under a relaxed clock model with an uncorrelated lognormal distribution. The analyses were run three times independently for 20 million generations, with every 1000th generation sampled. Using Tracer software (part of the BEAST package), we confirmed that the three runs had converged to a stationary distribution after the burn-in stage, which left a total of 36 000 samples describing the posterior distribution. Note that the age of the root was arbitrarily set to 100. Since Bayesian posterior probability values have a tendency toward ‘overcredibility’ (Suzuki et al. 2002; Kolaczkowski & Thornton 2009), we interpreted the Bayesian posterior probabilities conservatively and considered groups supported only if over 0.9, or preferably full 1.0, posterior probability was achieved. However, we did not consider posterior probability estimates entirely uninformative because groups strongly supported in posterior probabilities were usually found in maximum likelihood analysis as well.
The parsimony analysis was carried out using TNT (Goloboff et al. 2008). We used the ‘modern technology search’, including sectorial search, ratchet, drift and tree fusing; data compressed; gap = 5th state; ‘set initial level’—more than 60, random seed 100. The search was interrupted at 169 h 8 min (2 392 708 427 536 rearrangements tried by then). The best score (40 007 steps) was hit six times, and with 19 trees retained. The trees were further swapped using the tree bisection and reconnection in a ‘traditional search’. A total of 11 808 equally parsimonious trees were found. Bremer supports were calculated with a script for TNT that uses anticonstraints (Peña et al. 2006).
3. Results and discussion
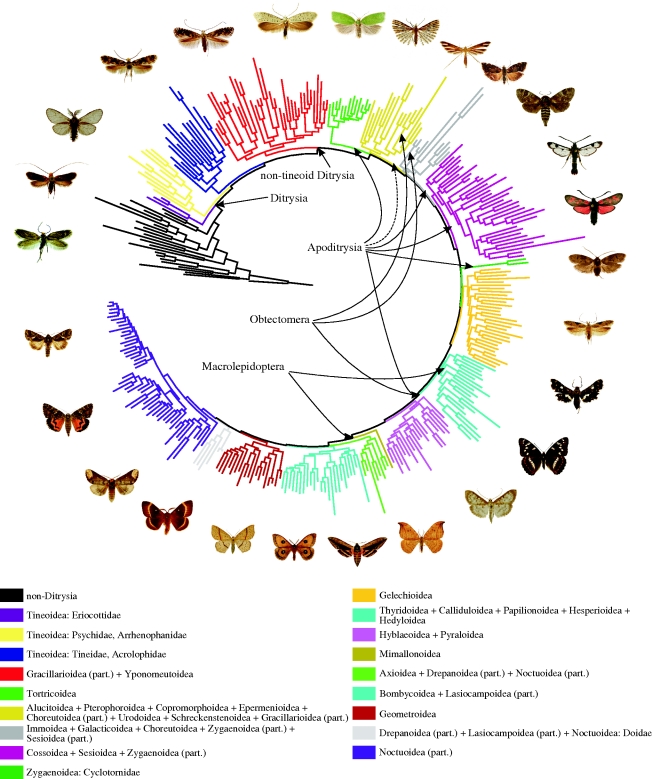
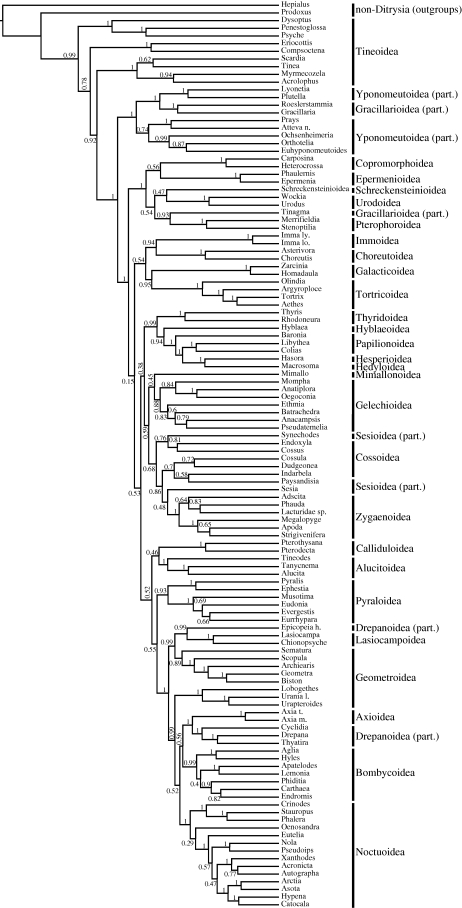
All the methods agreed on the broad patterns of relationships recovered (figures 1 and 2; see the electronic supplementary material, figures S1 and S2). The best obtained maximum likelihood tree is presented in figure 1 and in more detail in the electronic supplementary material, figure S1. We used this tree as a basis for most of our conclusions. A Bayesian tree of a ditrysian subset of taxa is presented in figure 2. Results of this tree were used to draw conclusions only when very high posterior probability was achieved. The result of the unconstrained parsimony analysis is in general agreement with those obtained using model-based analyses (see the electronic supplementary material, figure S2). The main discrepancies are lower general resolution with many polytomies, and a general pattern of clustering of the most divergent taxa together. This implies the vulnerability of the parsimony analysis to long-branch attraction (Felsenstein 1978). This appears particularly strong in the pattern of long-branched basal non-tineoid ditrysians (bucculatricid complex, Lithocolletiinae + Phyllocnistinae and most divergent putative zygaenoids, Heterogyniidae and Epipyropidae), among which even non-ditrysians Tischeriidae and Nepticulidae, with very long branches, grouped (see the electronic supplementary material, figure S2). Another probable reason for the underperformance of parsimony analysis is its vulnerability to large amounts of homoplasy (Whitfield & Kjer 2008); a feature almost inherently present in nucleotide data. In such cases, likelihood or Bayesian methods applying complex evolutionary models have reported to generally outperform parsimony methods (Gadagkar & Kumar 2005; Gaucher & Miyamoto 2005).
Figure 1.
Overview of the 350-taxon RAxML maximum likelihood analysis. The tree was rooted on Micropteryx, a taxon likely to be a sister group to all other Lepidoptera. Non-ditrysian clades are all shown in black. Major ditrysian branches are coloured and their content indicated at the superfamily level. Putative ditrysian clades are shown by arrows in the middle of the circle. A solid line indicates complete inclusion and a dashed line partial inclusion in the named clade.
Figure 2.
Cladogram of the Bayesian tree of the 118-taxon subset of ditrysian taxa. Posterior probabilities estimated under the GTR + G model (three BEAST runs of 20 million generations each) are shown above the branches. Ditrysian superfamilies are shown on the right.
(a). Major phylogenetic patterns
Our results consistently support the sister group relationship between Agathiphagidae and Heterobathmiidae. The monophyly of Glossata, including all Lepidoptera except Micropterigoidea, Agathiphagoidea and Heterobathmioidea, is supported. Of the other named clades below Ditrysia, we found support for the monophyly of Heteroneura, while our results do not support the monophyly of Coelolepida, Myoglossata and Neolepidoptera. Eulepidoptera become monophyletic after the inclusion of Andesianidae, which always group with Incurvarioidea, not with Tischerioidea as suggested by Simonsen (2009). We found support for the sister group relationship between Tischerioidea and Palaephatoidea, as suggested in Davis (1986) and Nielsen (1989). They together form the sister group to Ditrysia.
Morphological evidence of the monophyly of Ditrysia is convincing (Nielsen 1989; Kristensen & Skalski 1998), and this huge assemblage is supported by our molecular evidence, as well (but see note about parsimony analysis above). Tineoidea may not be monophyletic, as they are often divided into three separate and always strongly supported lineages, Eriocottidae, Psychidae, including Arrhenophanidae, and Tineidae, including Acrolophidae. The shift of Lypusidae from Tineoidea to Gelechioidea as in Heikkilä & Kaila (2010) is supported. The monophyly of non-tineoid Ditrysia is well supported, although morphological evidence of this assemblage has been scanty (Minet 1991; Kristensen & Skalski 1998).
While non-tineoid Ditrysia and many superfamilies therein appear monophyletic, the relationships within the clade remain largely questionable. Despite low support at many basal nodes, we do not consider the observed patterns uninformative for two reasons. First, the position of non-tineoid ditrysian superfamilies remained constant in various analyses we carried out. Second, there seems to be remarkable congruence between our result and that of Regier et al. (2009), even though the two studies are based largely on non-overlapping data. Hence, we consider it justified to keep the tentative branches uncollapsed. Regier et al. (2009) concluded that the low ‘backbone’ supports are most likely owing to the short internode branch lengths along the ‘backbone’, reflecting rapid radiation of major ditrysian lineages in the past. This feature, common in many other insect groups as well (Whitfield & Kjer 2008), renders resolving the basal internode relationships challenging.
The supposedly more advanced Ditrysia, i.e. superfamilies other than Tineoidea, Gracillarioidea, Yponomeutoidea and Gelechioidea, have been thought to form three internested clades: Apoditrysia, Obtectomera and Macrolepidoptera (Minet 1986, 1991; Nielsen 1989). None of them appeared monophyletic. This result is consistent with that of Regier et al. (2009) as a broad pattern, but there are several differences in details. Apoditrysia become monophyletic if Douglasiidae and Gelechioidea as in Kaila (2004) are included. The pattern is largely the same as the one observed by Regier et al. (2009), but with a significant conflict in the position of Choreutoidea, Alucitoidea and Urodoidea, which Regier et al. (2009) often found among the most basally branching ditrysian lineages after Tineoidea. A reason behind this discrepancy might be the rather unrepresentative taxon sampling in lower Ditrysia in their study. Obtectomera are monophyletic if Copromorphoidea and Immoidea are excluded. Immoidea were not included in Regier et al. (2009) and Copromorphoidea was similarly found to often fall outside Obtectomera. Overall, we found Obtectomera to be a more coherent assemblage than did Regier et al. (2009). Macrolepidoptera sensu Minet (1991) get support only if butterflies (including Hedyloidea and Hesperioidea) and Calliduloidea are excluded; a result consistent with that of Regier et al. (2009).
Gracillarioidea never come out as a monophyletic entity, with Douglasiidae consistently coming out as an apoditrysian taxon. Lithocolletiinae and the remaining Gracillarioidea are not always associated with each other. The status of the bucculatricid complex as a gracillarioid taxon is ambiguous. Bucculatricidae are linked with Ogmograptis and Tritymba, both Australian taxa, the first tentatively associated with Bucculatricidae, and the second formerly placed in Plutellidae (Yponomeutoidea). This complex may alternatively be linked with Zygaenoidea. The remaining Gracillarioidea are found to be either a sister group to, or embedded in, Yponomeutoidea. While almost consistently occurring, this connection is strongly supported only in a Bayesian analysis. Yponomeutoidea (with Tritymba excluded) are usually found to be monophyletic (although not in parsimony analysis or Bayesian analysis of a subset of taxa), but similarly consist of only loosely connected taxa. Lyonetiidae appear polyphyletic and Yponomeutidae paraphyletic, with Plutellidae and Lyonetiinae embedded therein. Glyphipterigidae are paraphyletic, with Acrolepiidae and the New Zealand, Tasmanian ‘megaplutellids’ included.
While being found within the core Macrolepidoptera, the other members of the ‘butterfly assemblage’ of Minet (1991), including Cimelioidea, Geometroidea and Drepanoidea, do not form a clade. Regier et al. (2009) came to the same conclusion. Within Macrolepidoptera, the status of Mimallonoidea as a sister taxon to all the others is often found, which fits their peculiarity of having both microlepidopteran and macrolepidopteran morphological features (Minet 1991). Like Regier et al. (2009), we found Mimallonoidea to be an unstable taxon, which in various trials associated with ‘microlepidoptera’, Bombycoidea or other groups, or even formed its own lineage, leaving its affinities unclear.
Of the most species-rich ditrysian superfamilies, the monophyly of Tortricoidea, Pyraloidea and Noctuoidea are supported, with the exception that Doidae associate with Drepanoidea instead of Noctuoidea. This finding is identical with that observed in Regier et al. (2009). Gelechioidea also appear monophyletic, although with low statistical support. A particularly problematic assemblage of ditrysian Lepidoptera concentrates around Cossoidea, Sesioidea and Zygaenoidea. None of these morphologically heterogeneous superfamilies appear monophyletic, but together they form a near-monophyletic assemblage, with the enigmatic and unstable zygaenoid families Epipyropidae and Cyclotornidae and, curiously, Tinthiinae of Sesiidae falling outside. This pattern is similar to that first suggested by Scott (1986), and observed also by Regier et al. (2009), albeit with less comprehensive taxon sampling. Even though Tinthiinae cannot be firmly associated with any other taxa, they never group with Sesiidae, and hence the wasp-like appearance of Sesiidae appears to have evolved twice independently. The unassigned Australian Heliocosma group is found within this loose assemblage of taxa.
Several small apoditrysian superfamilies form loose coalitions in the basal region of ditrysian Lepidoptera. Even though each of these superfamilies is found to be monophyletic, their relationships to other groups remain without good support. An assemblage formed of Alucitoidea, Pterophoroidea, Epermenioidea, Schreckensteinioidea, Copromorphoidea and Urodoidea is often found, supplemented with Douglasiidae (Gracillarioidea) and Millieriinae (Choreutoidea). Near that lies another loose concentration of small superfamilies, including Immoidea, Galacticoidea and the core Choreutoidea. These three superfamilies alternatively form a monophylum with Tortricoidea. Millieriinae never associate with Choreutidae, and Choreutoidea as currently delimited are thus polyphyletic.
True butterflies (Papilionoidea) appear paraphyletic with good support, both Hesperioidea and Hedyloidea nesting within them. A similar result was reported earlier based on molecular data, but this result was obscured by morphological evidence (Wahlberg et al. 2005), and later again by Regier et al. (2009). Also supporting the finding of Regier et al. (2009), Papilionidae seems to be the sister lineage of the other butterflies and skippers. Thyridoidea and Calliduloidea are found to be closely related to butterflies, with the association of Thyridoidea and butterflies being strongly supported in the Bayesian analysis.
The position of Hyblaeoidea, with only one species sampled, is unstable, and we cannot make firm statements about their affinities. They associate sometimes with Pyraloidea, but more often with Thyridoidea and butterflies. With Hyblaeoidea excluded, the clade including Pyraloidea and core Macrolepidoptera becomes well supported, placing most lepidopterans with tympanal organs together. In Macrolepidoptera, Bombycoidea form a monophyletic group, with Anthelidae included, as recently suggested (Regier et al. 2008; Zwick 2008). Lasiocampoidea are not found as a sister group to it, contradicting the findings of Regier et al. (2008). The core groups of Drepanoidea are monophyletic, with Doidae and Cimelioidea associating with them and Epicopeiidae falling outside, forming a sister group to Lasiocampoidea. This unexpected finding may also be supported by morphology (J. Holloway 2009, personal communication). The association of Lasiocampoidea with Epicopeiidae or the placement of Lasiocampoidea as distinct from Bombycoidea are, however, not well supported findings, and firm statements of their affinities cannot be drawn. Drepanoidea were not found to be a sister group to Geometroidea in any trials.
The monophyly of Geometroidea remains uncertain. Uraniidae, often with Sematuridae, usually form a monophylum with Geometridae, but this is not supported in the Bayesian analysis of the limited subset of taxa. The position of Sematuridae is similarly unstable. It may be the sister group to Uraniidae or Geometridae, may be nested within Geometridae, or alternatively, may form its own lineage within Macrolepidoptera. With the exception of the affinities of Epicopeiidae, these observations are in general agreement with those reported by Regier et al. (2009). Noctuoidea without Doidae are a well-supported monophyletic entity. They are divided into six well-supported clades, of which the isolation of the Euteliinae + Stictopterinae clade from ‘quadrifine’ Noctuidae is a novel finding.
(b). Lower-level interrelationships
An overview of lower-level lepidopteran taxa that were not found monophyletic compared with the classification of Kristensen et al. (2007) is presented in the electronic supplementary material, table S3. The putative monophyly of Tineoidea cannot be ruled out with certainty. The superfamily is divided into three well-supported clades, which follows the traditional division (Robinson 1988; Davis & Robinson 1998) otherwise, but with Arrhenophanidae and Acrolophidae embedded in Psychidae and Tineidae, respectively. Within Psychidae, many interrelationships are well-resolved. Typhoniinae get support as being a sister to the other taxa. The next splitting event is found between Placodominae and the remaining Psychidae. Naryciinae are found paraphyletic with respect to Taleporiinae. In Tineidae, interrelationships remain mostly weakly supported. Harmacloninae often form a sister group to the remaining Tineidae. The two representatives of Myrmecozelinae do not form a monophyletic group, as postulated by Robinson (2009).
In Yponomeutoidea, our results strongly support the monophyly of Ypsolophidae as consisting of Ypsolophinae and Ochsenheimeriinae (Kyrki 1990). In Yponomeutidae, Praydinae and Attevinae are often found as sisters, but in the presented maximum likelihood tree this is obscured by the inclusion of the unstable Cemiostominae (Lyonetiidae) within it. The two subfamilies of Lyonetiidae never formed a monophylum, but both groups are somewhat unstable and branch leading to Cemiostominae is long. Bedelliidae are also unstable, though always found within Yponomeutoidea. The New Zealand and Tasmanian ‘megaplutellids’, here represented by Proditrix and Doxophyrtis, are found close to Orthoteliinae of Glyphipterigidae, as suggested by Heppner (2003). Overall, although we tentatively consider Yponomeutoidea monophyletic, it is heterogeneous, and many interrelationships remain to be clarified.
Our findings provide support for the position of Chlidanotinae as a sister group to the other two subfamilies of Tortricidae. Olethreutinae and Tortricinae are both found to be monophyletic and sister groups to each other. Isonomeutis, considered an unusual member of Copromorphoidea (Dugdale et al. 1998), shows affinities with Alucitoidea. This association is supported by morphology (L. Kaila 2009, personal observation). Tineodidae are paraphyletic, with Alucitidae nested within it.
Neither Cossoidea nor Sesioidea form monophyletic assemblages, but are embedded within each other; a result generally consistent to that reported in Regier et al. (2009). Zygaenoidea are found subordinate to this assemblage. Sesioidea are found to be polyphyletic, with Tinthiinae falling outside the Sesioidea–Cossoidea assemblage and Castniidae, Brachodidae and Sesiidae each associating with various taxa of Cossoidea or the unassigned Heliocosma group of species. Regier et al. (2009) also found Castniidae always associating with Cossoidea rather than Sesiidae. Affinities and composition of the Australian Heliocosma group remain to be examined in more detail. Piestoceros has been placed under Psychidae in collections and lists (Nielsen et al. 1996), although this group has never really been studied. We find it related to Heliocosma. Cossidae are found to be polyphyletic, with Dudgeonidae, Metarbelinae and Cossulinae associated with groups other than ‘core Cossidae’ i.e. Cossinae + Zeuzerinae. Alternatively, Cossidae can be considered paraphyletic as Sesioidea and Zygaenoidea fall within it in this study.
Zygaenoidea are a heterogeneous group of moths defined by hardly any shared characters (Epstein et al. 1998). Zygaenoidea are consistently found subordinate to the Cossoidea–Sesioidea complex, as reported in Regier et al. (2009). With the exception of the parasitic Epipyropidae and Cyclotornidae, which have very long branches and cannot safely be placed anywhere, the superfamily is usually found to be monophyletic, and most interrelationships within the superfamily remain without strong support. The ‘core Zygaenoidea’ are divided into two lineages, roughly following the patterns found by Regier et al. (2009), but with some notable exceptions. Dalceridae are not found as a part of the ‘limacodid group’, but closer to Zygaenidae and a sister group to Phaudinae, which in turn are supported as falling outside of Zygaenidae, as recently suggested (Fänger et al. 1998; Niehuis et al. 2006). Lacturidae, Heterogynidae, Dalceridae and Phaudinae together form a sister group to ‘core Zygaenidae’. The ‘limacodid group’ is consistently found, but with Dalceridae excluded and with inclusion of Himantopteridae and Anomoeotidae, which were not included in Regier et al.'s (2009) study. These two closely related families group with Aididae, together forming a strongly supported clade. It is therefore probable that Himantopteridae, Anomoeotidae, Somabrachyidae and Aididae form a monophyletic clade within the Limacodidae, rendering this family paraphyletic. Even though there is no molecular evidence supporting the placement of Epipyropidae and Cyclotornidae within Zygaenoidea, the morphology of their immature stages is in favour of this position (Epstein et al. 1998).
Within Gelechioidea our results support some of the groupings of Hodges (1998) and Kaila (2004), but most affinities remain without support. Xyloryctinae get support as being a sister to Coleophorinae rather than Scythridinae. Autostichidae become monophyletic only with Glyphidoceridae and Deoclonidae included. Stathmopoda is not closely related to Oecophora, and it is therefore unlikely that Oecophoridae (sensu Hodges 1998) are monophyletic, a result also observed by Kaila (2004). Elachistidae (sensu Hodges 1998 or Kaila 2004) never form a monophyletic entity. The division of Pyraloidea into Pyralidae and Crambidae (Solis & Mitter 1992) is supported with high confidence. In Pyralidae, the result is consistent with that presented in Regier et al. (2009). In Crambidae, our results are not consistent with those reported earlier (Solis & Maes 2002), with their main division between ‘spilomeline’ and ‘pyraustine’ groups not supported. Our results broadly agree with the results of Regier et al. (2009).
In Lasiocampoidea, Chionopsychinae were found to be the first diverging lineage and Lasiocampinae were a sister to Poecilocampinae, the latter finding being contradictory to that observed by Regier et al. (2008). Within Geometridae, our findings are in moderate agreement with those of Young (2006) and agree almost perfectly with those observed by other recent studies (Yamamoto & Sota 2007; Regier et al. 2009; Wahlberg et al. in press). Larentiinae + Sterrhinae are found to form the sister group to all other Geometridae. Archiearinae, formerly considered the first diverging lineage, are supported as subordinate to Larentiinae + Sterrhinae and sister to the lineage comprising Geometrinae, Oenochrominae and Ennominae.
While all maximum likelihood analyses supported Oenosandridae as being the sister group to all other Noctuoidea, this result was not unambiguously achieved in the Bayesian analysis. The monophyly of Notodontidae, Nolidae, Erebidae and Noctuidae is supported. Euteliinae + Stictopterinae form a separate, strongly supported lineage, which is found as a sister group to Noctuinae. Formerly, the group has been associated with ‘quadrifine Noctuidae’ or Erebiidae (Lafontaine & Fibiger 2006; Mitchell et al. 2006). As first observed by Weller et al. (1994) and later further supported by Mitchell et al. (2006), the arctiids and lymantriids, formerly considered valid families within Noctuoidea, are deeply embedded in Noctuidae. We found them both within Erebidae, which is consistent with the findings of Mitchell et al. (2006).
(c). Lepidoptera-flowering plant co-radiation
Lepidoptera are the largest group of insects that are almost exclusively dependent on angiosperm plants (Powell et al. 1998) and are clearly a difficult group to resolve phylogenetically (Regier et al. 2009). The other megadiverse clades of insects have phytophagous groups, but large components of those clades feed on other resources. Lepidoptera are likely to have diversified in concert with angiosperm plants, which have an evolutionary history reaching back about 150 Myr (Magallón & Castillo 2009). Even though angiosperms prevail as the hosts of most monotrysian Lepidoptera as well, it is possible that the advent of Ditrysia coincided with the timing of the great diversification of flowering plants, thus facilitating fast adaptive radiation of these Lepidoptera (Whitfield & Kjer 2008). Our results with short basal branches in the ditrysian clade suggest that the initial expansion onto angiosperm plants may have happened rapidly and led to a rapid radiation of lineages that we now recognize as superfamilies. However, a formal analysis of this hypothesis is beyond the scope of this study.
4. Conclusions
This study, to our knowledge the most comprehensive analytical study on ditrysian lepidopteran phylogeny so far, suggests that the ditrysian Lepidoptera are monophyletic, with Tischerioidea + Palaephatoidea being their sister group. The superfamilies and most families are connected to each other with short nodes, but the findings are, in spite of the weak statistical support for many groupings, often in close agreement with another recent study (Regier et al. 2009). The non-tineoid Ditrysia is well supported. Even though none of the proposed major clades of more advanced Ditrysia, i.e. Apoditrysia, Obtectomera and Macrolepidoptera was supported as currently delineated, they get support from the present analysis after some adjustment. Many recognized superfamilies and families were found to be either para- or polyphyletic, though generally with weak support. The generally short nodes supporting the monophyly of most superfamilies imply a rapid radiation in the past, presumably in concert with the diversification of flowering plants. We anticipate that a more comprehensive taxon and gene sampling, supported by a rigorous analysis of comprehensive morphological data, will, in the future, provide a more robust backbone for the phylogeny of Lepidoptera.
Acknowledgements
A notable amount of material for this study was provided by the Leptree project, headed by C. Mitter (US NSF award no. 0531769). The following people also helped in obtaining DNA material for the study: K. Ambil, E. D. Edwards, S. Haapala, P. Hirvonen, R. Hoare, M. Horak, J. Itämies, D. Janzen (US NSF awards nos DEB0072730 and DEB0515699), J. Junnilainen, U. Jürivete, J. Kaitila, A. Kallies, A. Karhu, O. Karsholt, T. Klemetti, C. Kokx, H. Kronholm, J. Kullberg, J.-F. Landry, J. Lehto, P. Malinen, V. S. Monteys, T. Mutanen, E. Nieukerken, K. Nupponen, O. Pellmyr, T. Roslin, P. Välimäki, B. Wikström and J. Zwier. We are grateful to M. Heikkilä for commenting and editing the text, K. Kosola for improving language, and L. Kvist, P. Kärkkäinen and R. Zahiri for their technical help. We also thank R. Hoare, M. Horak and C. Young for hosting our visits to Australia and New Zealand, C. Peña for providing efficient facilities for sequence handling, two anonymous referees for useful comments, as well as M. Horak, J. Holloway, C. Mitter and many other colleagues for fruitful discussions. This work was funded by the Academy of Finland through grants awarded to L.K. (project 1110906) and N.W. (projects 118369 and 129811).
References
- Davis D. R.1986A new family of monotrysian moths from austral South America (Lepidoptera: Palaephatidae), with a phylogenetic review of the Monotrysia. Smith. Contr. Zool. 434, 1–202 [Google Scholar]
- Davis D., Robinson G. S.1998The Tineoidea and Gracillarioidea. In Handbook of zoology, vol. IV, Arthropoda: Insecta, part 35, Lepidoptera, moths and butterflies, vol. 1 (ed. Kristensen N. P.), pp. 91–117 Berlin, Germany: Walter de Gruyter [Google Scholar]
- Drummond A. J., Rambaut A.2007BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugdale J. S., Kristensen N. P., Robinson G. S., Scoble M. J.1998The smaller Microlepidoptera-grade superfamilies. In Handbook of zoology, vol. IV, Arthropoda: Insecta, part 35, Lepidoptera, moths and butterflies, vol. 1 (ed. Kristensen N. P.), pp. 217–232 Berlin, Germany: Walter de Gruyter [Google Scholar]
- Epstein M. E., Geertsema H., Naumann C. M., Tarmann G. M.1998The Zygaenoidea. In Handbook of zoology, vol. IV, Arthropoda: Insecta, part 35, Lepidoptera, moths and butterflies, vol. 1 (ed. Kristensen N. P.), pp. 159–180 Berlin, Germany: Walter de Gruyter [Google Scholar]
- Fänger H., Yen S.-H., Naumann C. M.1998External morphology of the last instar larva of Phauda mimica Strand, 1915 (Lepidoptera: Zygaenoidea). Entomol. Scand. 29, 429–450 [Google Scholar]
- Felsenstein J.1978Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27, 401–410 (doi:10.2307/2412923) [Google Scholar]
- Gadagkar S. R., Kumar S.2005Maximum likelihood outperforms maximum parsimony even when evolutionary rates are heterotachous. Mol. Biol. Evol. 22, 2139–2141 (doi:10.1093/molbev/msi212) [DOI] [PubMed] [Google Scholar]
- Gaucher E. A., Miyamoto M. M.2005A call for likelihood phylogenetics even when the process of sequence evolution is heterogeneous. Mol. Phyl. Evol. 37, 928–931 (doi:10.1016/j.ympev.2005.03.027) [DOI] [PubMed] [Google Scholar]
- Goloboff P. A., Farris J. S., Nixon K. C.2008TNT, a free program for phylogenetic analysis. Cladistics 24, 774–786 (doi:10.1111/j.1096-0031.2008.00217.x) [Google Scholar]
- Hall T. A.1999BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleid Acids Symp. Ser. 41, 95–98 [Google Scholar]
- Heikkilä M., Kaila L.2010Reassessment of the enigmatic Lepidopteran family Lypusidae (Lepidoptera: Tineoidea; Gelechioidea). Syst. Entomol. 35, 71–89 (doi:10.1111/j.1365-3113.2009.00483.x) [Google Scholar]
- Heppner J. B.2003Primitive sedge moths from New Zealand and Tasmania: transfer of Proditrix and relatives to Orthoteliinae (Lepidoptera: Glyphipterigidae). Lepid. News 1–2, 31–42 [Google Scholar]
- Hodges R. W.1998The Gelechioidea. In Handbook of zoology, vol. IV, Arthropoda: Insecta, part 35, Lepidoptera, moths and butterflies, vol. 1 (ed. Kristensen N. P.), pp. 131–158 Berlin, Germany: Walter de Gruyter [Google Scholar]
- Hunt T., et al. 2007A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318, 1913–1916 (doi:10.1126/science.1146954) [DOI] [PubMed] [Google Scholar]
- Kaila L.2004Phylogeny of the superfamily Gelechioidea (Lepidoptera: Ditrysia): an exemplar approach. Cladistics 20, 303–340 (doi:10.1111/j.1096-0031.2004.00027.x) [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B., Thornton J. W.2009Long-branch attraction bias and inconsistency in Bayesian phylogenetics. PLoS One 4, e7891 (doi:10.1371/journal.pone.0007891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen N. P., Skalski A. W.1998Phylogeny and palaeontology. In Handbook of zoology, vol. IV, Arthropoda: Insecta, part 35, Lepidoptera, moths and butterflies, vol. 1 (ed. Kristensen N. P.), pp. 7–25 Berlin, Germany: Walter de Gruyter [Google Scholar]
- Kristensen N. P., Scoble M., Karsholt O.2007Lepidoptera phylogeny and systematics: the state of inventorying moth and butterfly diversity. Zootaxa 1668, 699–747 [Google Scholar]
- Kyrki J.1990Tentative reclassification of holarctic Yponomeutoidea (Lepidoptera). Nota Lepid. 13, 28–42 [Google Scholar]
- Lafontaine J. D., Fibiger M.2006Revised higher classification of the Noctuidae (Lepidoptera). Can. Entomol. 138, 610–635 (doi:10.4039/N06-012) [Google Scholar]
- Magallón S., Castillo A.2009Angiosperm diversification through time. Am. J. Bot. 96, 349–365 (doi:10.3732/ajb.0800060) [DOI] [PubMed] [Google Scholar]
- Minet J.1986Ebauche d'une classification moderne de l'ordred des Lépidoptères. Alexanor 14, 291–313 [Google Scholar]
- Minet J.1991Tentative reconstruction of the ditrysian phylogeny (Lepidoptera: Glossata). Entomol. Scand. 22, 69–957 [Google Scholar]
- Mitchell A., Mitter C., Regier J. C.2006Systematics and evolution of the cutworm moths (Lepidoptera: Noctuidae Noctuoidea): evidence from two protein-coding nuclear genes. Syst. Entomol. 31, 21–46 (doi:10.1111/j.1365-3113.2005.00306.x) [Google Scholar]
- Niehuis O., Yen S.-H., Naumann C. M., Misof B.2006Higher phylogeny of zygaenid moths (Insecta: Lepidoptera) inferred from nuclear and mitochondrial sequence data and the evolution of larval cuticular cavities for chemical defence. Mol. Phylogenet. Evol. 39, 812–829 (doi:10.1016/j.ympev.2006.01.007) [DOI] [PubMed] [Google Scholar]
- Nielsen E. S.1989Phylogeny of major lepidopteran groups. In The Hierarchy of Life (eds Fernholm B., Bremer K., Jörnvall H.), pp. 281–294 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- Nielsen E. S., Edwards E. D., Rangsi T. V.1996Checklist of the Lepidoptera of Australia. Monographs on Australian Lepidoptera, vol. 4 Canberra, Australia: CSIRO publishing [Google Scholar]
- Peña C., Wahlberg N., Weingartner E., Kodandaramaiah U., Nylin S., Freitas A. V. L., Brower A. V. Z.2006Higher level phylogeny of Satyrinae butterflies (Lepidoptera: Nymphalidae) based on DNA sequence data. Mol. Phylogenet. Evol. 40, 29–49 (doi:10.1016/j.ympev.2006.02.007) [DOI] [PubMed] [Google Scholar]
- Powell J. A., Mitter C., Farrell B.1998Evolution of larval food preferences in Lepidoptera. In Handbook of zoology, vol. IV, Arthropoda: Insecta, part 35, Lepidoptera, moths and butterflies, vol. 1 (ed. Kristensen N. P.), pp. 403–422 Berlin, Germany: Walter de Gruyter [Google Scholar]
- Ratnasingham S., Hebert P. D.2007BOLD: the barcode of life data systems. Mol. Ecol. Notes 7, 355–364 (doi:10.1111/j.1471-8286.2007.01678.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier J. C., Cook C. P., Mitter C., Hussey A.2008A phylogenetic study of the ‘bombycoid complex’ (Lepidoptera) using five protein-coding nuclear genes, with comments on the problem of macrolepidopteran phylogeny. Syst. Entomol. 33, 175–189 [Google Scholar]
- Regier J. C., et al. 2009Toward reconstructing the evolution of advanced moths and butterflies (Lepidoptera: Ditrysia): an initial molecular study. BMC Evol. Biol. 9, 280 (doi:10.1186/1471-2148-9-280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. S.1988A phylogeny for the Tineoidea (Lepidoptera). Entomol. Scand. 19, 117–129 [Google Scholar]
- Robinson G.2009Biology, distribution and diversity of tineid moths. Kuala Lumpur, Malaysia: Natural History Museum [Google Scholar]
- Roe A. D., et al. 2010Evolutionary framework for Lepidoptera model systems. In Genetics and molecular biology of lepidoptera (eds Goldsmith M., Marec F.), pp. 1–24 Boca Raton, FL: CRC Press [Google Scholar]
- Scott J. A.1986On the monophyly of the Macrolepidoptera, including a reassessment of their relationship to Cossoidea and Cstnioidea, and a reassignment of Mimallonidae to Pyraloidea. J. Res. Lepid. 25, 30–38 [Google Scholar]
- Sharkey M. J.2007Phylogeny and classification of Hymenoptera. Zootaxa 1668, 521–548 [Google Scholar]
- Simonsen T.2009Wing vestiture of the newly described monotrysian Lepidoptera family Andesianidae Davis and Gentili suggests affinity with the putative Tischerioidea–Ditrysia clade (Insecta: Lepidoptera). Stud. Neotrop. Fauna Environ. 44, 109–114 (doi:10.1080/01650520902956042) [Google Scholar]
- Solis M. A., Maes K. V. N.2002Preliminary phylogenetic analysis of the subfamilies of Crambidae (Pyraloidea: Lepidoptera). Belgian J. Entomol. 4, 53–95 [Google Scholar]
- Solis M. A., Mitter C.1992Review and phylogenetic analysis of the Pyralidae (sensu stricto) (Lepidoptera: Pyralidae) and placement of the Epipaschiinae. Syst. Entomol. 17, 79–90 (doi:10.1111/j.1365-3113.1992.tb00323.x) [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A.2008Rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 75, 758–771 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Glazko G. V., Nei M.2002Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc. Natl Acad. Sci. USA 99, 16 138–16 143 (doi:10.1073/pnas.212646199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg N., Wheat C. H.2008Genomic outposts serve the phylogenetic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Syst. Biol. 57, 231–242 (doi:10.1080/10635150802033006) [DOI] [PubMed] [Google Scholar]
- Wahlberg N., et al. 2005Synergistic effects of combining morphological and molecular data in resolving the phylogeny of butterflies and skippers. Proc. R. Soc. B 272, 1577–1586 (doi:10.1098/rspb.2005.3124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg N., Snäll N., Viidalepp J., Ruohomäki K., Tammaru T.In pressThe evolution of female flightlessness among Ennominae of the Holarctic forest zone (Lepidoptera, Geometridae). Mol. Phylogenet. Evol. (doi:10.1016/j.ympev.2010.01.025) [DOI] [PubMed] [Google Scholar]
- Weller S. J., Martin J. A., Constable J. L.1994Phylogeny of Noctuoid moths and the utility of combining independent nuclear and mitochondrial genes. Syst. Biol. 43, 194–211 [Google Scholar]
- Whitfield J. B., Kjer K. M.2008Ancient rapid radiations of insects: challenges for phylogenetic analysis. Annu. Rev. Entomol. 53, 449–472 (doi:10.1146/annurev.ento.53.103106.093304) [DOI] [PubMed] [Google Scholar]
- Wiegmann B. M., Regier J. C., Mitter C.2002Combined molecular and morphological evidence of the phylogeny of the earliest lepidopteran lineages. Zoologica Scripta 31, 67–81 (doi:10.1046/j.0300-3256.2001.00091.x) [Google Scholar]
- Yamamoto S., Sota T.2007Phylogeny of the Geometridae and the evolution of winter moths inferred from a simultaneous analysis of mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 44, 711–723 (doi:10.1016/j.ympev.2006.12.027) [DOI] [PubMed] [Google Scholar]
- Yeates D. K., Wiegmann B. M., Courtney G. W., Meier R., Lambkin C., Pape T.2007Phylogeny and systematics of Diptera: two decades of progress and prospects. Zootaxa 1668, 565–590 [Google Scholar]
- Yen S.-H., Minet J.2007Cimelioidea: a new superfamily name for the gold moths (Lepidoptera: Glossata). Zool. Stud. 46, 262–271 [Google Scholar]
- Young C. J.2006Molecular relationships of the Australian Ennominae (Lepidoptera: Geometridae) and implications for the phylogeny of the Geometridae from molecular and morphological data. Zootaxa 1264, 1–147 [Google Scholar]
- Zwick A.2008Molecular phylogenetics of Anthelidae and other bombycoid taxa (Lepidoptera: Bombycoidea). Syst. Entomol. 33, 190–209 [Google Scholar]