Abstract
Recent climate change has triggered profound reorganization in northeast Atlantic ecosystems, with substantial impact on the distribution of marine assemblages from plankton to fishes. However, assessing the repercussions on apex marine predators remains a challenging issue, especially for pelagic species. In this study, we use Bayesian coalescent modelling of microsatellite variation to track the population demographic history of one of the smallest temperate cetaceans, the harbour porpoise (Phocoena phocoena) in European waters. Combining genetic inferences with palaeo-oceanographic and historical records provides strong evidence that populations of harbour porpoises have responded markedly to the recent climate-driven reorganization in the eastern North Atlantic food web. This response includes the isolation of porpoises in Iberian waters from those further north only approximately 300 years ago with a predominant northward migration, contemporaneous with the warming trend underway since the ‘Little Ice Age’ period and with the ongoing retreat of cold-water fishes from the Bay of Biscay. The extinction or exodus of harbour porpoises from the Mediterranean Sea (leaving an isolated relict population in the Black Sea) has lacked a coherent explanation. The present results suggest that the fragmentation of harbour distribution range in the Mediterranean Sea was triggered during the warm ‘Mid-Holocene Optimum’ period (approx. 5000 years ago), by the end of the post-glacial nutrient-rich ‘Sapropel’ conditions that prevailed before that time.
Keywords: cetacean, climate change, habitat fragmentation, population genetics, coalescence
1. Introduction
Changes in the environment can affect the behaviour and fitness of individual organisms and the viability of populations. Populations can respond to changing habitats by adapting (through natural selection or phenotypic plasticity), moving (to avoid habitat of reduced suitability, or take advantage of emerging habitat), by adjusting population size or some combination of the above. Both natural selection and genetic drift can shape populations as they evolve in this context.
In the North Atlantic, studies have increasingly reported strong shifts in plankton and fish assemblages with the contemporaneous climate and ocean warming (Beaugrand et al. 2002, 2008; Richardson & Schoeman 2004; Perry et al. 2005). These shifts in marine habitat and community structure are expected to drive major changes in the distribution, density and dispersal of apex predators such as marine mammals, with reduction in density in marginal parts of the habitat leading to fragmentation, local displacement or extinction if the habitat becomes unsuitable (Ryther 1969; Learmonth et al. 2006; O'Corry-Crowe 2008; de Bruyn et al. 2009; Marx & Uhen 2010). Such evidence is, however, hard to capture for highly mobile marine predators (Kintisch 2006) such as cetacean species (Learmonth et al. 2006; O'Corry-Crowe 2008). Key to answering how these apex predators will deal with current and future change in their environment lies in (i) understanding how cetaceans dealt with past climate changes and (ii) determining the impact of changing climate on cetaceans over contemporary time scales (O'Corry-Crowe 2008).
Past shifts in the Earth's climate have led to the alteration or loss of existing environments and the creation of new ones. These changes have been associated with speciation events, adaptive radiations and extinctions, population expansions and contractions, and changes in individual dispersal and breeding behaviour (Hewitt 2000, 2004; Hofreiter & Stewart 2009). Recent studies supported such processes in cetacean species by providing evidence that past changes in the environment, and in particular global change in temperature and diatoms richness that are dominant marine primary producers at the basis of marine food web, were significantly linked to the species diversity of mysticete and odontocete whales (Pastene et al. 2007; O'Corry-Crowe 2008; Marx & Uhen 2010). The contemporaneous climate change is however unique with respect to its speed and to the fact that it embeds in global change where both natural and anthropogenic influences act in synergy on ecosystems (Brook et al. 2008; Pimm 2008, 2009). Understanding how these factors affect marine systems is therefore a crucial issue.
In this study, we investigated the demographic history of a small coastal cetacean widely distributed in the North Atlantic, the harbour porpoise Phocoena phocoena, with regards to recent variation in its habitat. At around 1.5 m in length and 50 kg in weight, it is the smallest cetacean in the North Atlantic (Read 1999), and often considered as a species living in the ‘fast lane’ (Read & Hohn 1995; Lockyer 2007). Reproductive costs of harbour porpoises are high (Lockyer 2007), as females are often gestating and lactating at the same time and parturition occurs shortly before mating (Lockyer 2003). Given their limited capacity to store energy, it is assumed that harbour porpoises must feed frequently without prolonged periods of fasting (Koopman et al. 1996, 2002). Some authors suggested that fasting periods exceeding as short as 3 days could affect body condition (Kastelein et al. 1997; Lockyer 2007). Thus, temporary shortages in prey availability can negatively impact on the animals and are likely to be responsible for changes in their distribution (Santos & Pierce 2003; Santos et al. 2004; Johnston et al. 2005; Herr et al. 2009). Relatively continuous accessibility to adequate prey is therefore critical. Any changes in prey availability may affect energy stores, and ultimately survival (MacLeod et al. 2007). Therefore, we expect the distribution of this species to be strongly tied to variation in the primary and secondary productivity that provides the basis for apex consumers (Lockyer 2007).
The harbour porpoise is currently distributed fairly continuously throughout cold to temperate coastal waters of the North Pacific, the North Atlantic and in the Black Sea, but it is absent from the Mediterranean (Read 1999). The southeastern part of the distribution range in the North Atlantic is of particular interest in regard to recent climate change, as it covers the biogeographic transition between arctic/boreal species and subtropical species (Southward et al. 1995). Changes in distribution patterns and abundance in this region are thus more evident than elsewhere in Europe for many marine species (Southward et al. 1995; Beaugrand et al. 2008). In this southeastern part of the North Atlantic, the harbour porpoise displays a disjunct distribution range. The absence of porpoises in the Mediterranean splits the Black Sea range from the Atlantic range. Black Sea porpoises are therefore recognized as a subspecies P. p. relicta distinct from the Atlantic one P. p. phocoena (Rosel et al. 1995), and from an evolutionary perspective, as an evolutionary significant unit (ESU; Moritz 1994, 2002) independent from the Atlantic one (Tolley & Rosel 2006; Viaud-Martinez et al. 2007). More recently, in a study covering the entire eastern Atlantic range, Fontaine et al. (2007) discovered a second disjunction, a population restricted to the cold-water upwelling zone along the Atlantic coasts of Iberia (IB) (Fiùza 1983) and distinct from porpoises in the northern Bay of Biscay (NBB), which are part of the main North Atlantic range, extending from French waters fairly continuously to the Arctic (figure 1). Fontaine et al. (2007) argued that oceanographic conditions, specifically those conditioning food availability, impose strong constraints on the dispersal, and thus the genetic structure of the species. If this hypothesis is correct, the existing genetic structure across the North Atlantic range will have a recent origin, as the oceanographic conditions in the eastern North Atlantic, in the Mediterranean Sea and in the Black Sea have changed dramatically over the course of the Holocene (Alley et al. 2003; Mayewski et al. 2004; Osborn & Briffa 2006; Springer et al. 2008; Rohling et al. 2009).
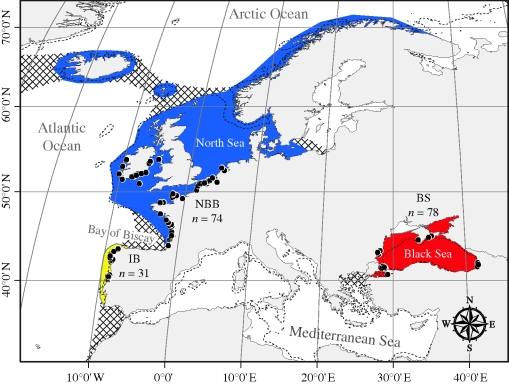
Figure 1.
Sampling and distribution range of harbour porpoises in European waters. The filled and hatched surfaces indicate the known and the possible distribution range, respectively. The coloured surfaces show the genetically distinct populations identified in Fontaine et al. (2007): the Black Sea porpoises (red, n = 78), the Iberian porpoises (yellow, n = 31) and the porpoises in northern Bay of Biscay waters (blue, n = 78). The dotted line shows the margin of the continental shelf (isobath −200 m) and the black circles the sampling locations.
As a cold temperate species, porpoises are unlikely to favour the current conditions of the southern Bay of Biscay and the Mediterranean Sea, with deep, warm and oligotrophic (nutrient-poor) waters (Fontaine et al. 2007). However, in the recent past, these waters have at times been cold and nutrient-rich, and it seems likely that porpoise ranges have closely tracked this nutrient availability. This would suggest the Iberian porpoise range could have been continuous with that of North Atlantic waters during recent cold periods that affected European climate (Grove 2004). There have been a number of notable cold periods between the Last Glacial Maximum (LGM, 18 000 years before present (yr BP)) and the most recent cold period, known as the ‘Little Ice Age’ (LIA). The period of continuity between the Black Sea and Atlantic ranges is a much more long standing question (Frantzis et al. 2001). The straits of Bosphorus and Dardanelle have been periodically closed during the Quaternary, but have been open continuously since approximately 8400 years ago (Major et al. 2002; Rohling et al. 2009). Analyses of genetic polymorphism at the non-coding region of the mitochondrial genome (the control region, mtDNA CR) (Tolley & Rosel 2006; Viaud-Martinez et al. 2007) suggested, however, a much older splitting time between Atlantic and Black Sea mitochondrial clades (at least 175 000 years ago).
In order to investigate the splitting process among the three porpoise populations identified in the Northeast Atlantic, we extracted historical information from genetic polymorphism data using model-based Bayesian coalescent approaches, and correlated this genetic inference with palaeo-oceanographic and historical records. Specifically, we tested whether the genetic divergence observed among harbour porpoise populations in the Northeast Atlantic is compatible with the recent changes reported in the seascape and marine assemblages, or is more likely owing to ancient vicariance processes.
2. Material and methods
(a). Microsatellite genotypes
Individual genotypes at 10 microsatellite loci were from Fontaine et al. (2007). To avoid computation time becoming intractable for the coalescent approach described below, we restricted the analyses to 183 harbour porpoises at the borders of the genetic barriers evidenced in Fontaine et al. (2007): 74 animals from the northern Bay of Biscay, 31 collected along the Iberian coast and 78 from the Black Sea (figure 1). Details regarding descriptive statistics on the microsatellite dataset can be found in Fontaine et al. (2007).
(b). Data analysis
We analysed the datasets using the ‘isolation with migration’ (IM) model of population divergence (Hey & Nielsen 2004). The model uses coalescent simulations within a Bayesian inference framework to estimate marginal probability distributions for six demographic parameters scaled by the mutation rate μ: the time since population-splitting (T = t μ), measures of neutral population genetic diversity of the two current and one ancestral population (θ1, θ2, θA) proportional to their effective population sizes Ne (θ = 4Ne μ) and bidirectional migration rates (M1 = m1/μ, M2 = m2/μ) (Hey & Nielsen 2004).
We analysed population-splitting first comparing the IB with the NBB population. Then, we compared the two ESUs, i.e. harbour porpoises from the Black Sea (BS) with those in the Atlantic (AT, including IB and NBB). This procedure was adopted because previous works suggested that the Black Sea harbour porpoises separated from Atlantic populations long before IB porpoises separated from the NBB populations. This is supported by complete lineage sorting of mtDNA haplotypes between porpoises from the Black Sea and the North Atlantic (Tolley & Rosel 2006; Viaud-Martinez et al. 2007), in contrast with the lack of private mitochondrial and microsatellite alleles between the Iberian and northern Bay of Biscay (Tolley & Rosel 2006; Fontaine et al. 2007; Viaud-Martinez et al. 2007). Pooling Atlantic populations may induce some bias in our inference regarding the Black Sea–Atlantic comparison, because the model assumes panmixia within each population. However, a recent simulation study showed that the model estimates were robust to such departure (Strasburg & Rieseberg 2010). On the other hand, comparing each pair of populations separately (IB–BS and NBB–BS), thus ignoring a population in the Atlantic (IB or NATL) that is more closely related to the other, profoundly violates the basic assumptions of the model (Hey & Nielsen 2004) and could have a more profound impact on the model estimates (Strasburg & Rieseberg 2010).
We conducted Markov Chain Monte Carlo (MCMC) simulations using the IM program (Hey & Nielsen 2004), assuming a stepwise mutation model of microsatellite evolution and uniform prior distributions over parameter ranges. Prior boundaries were empirically determined to ensure that the posterior distributions fell completely within the prior range. The modes of the posterior probability distributions were thus taken as the maximum-likelihood estimate (MLE) of the parameters. We estimated the credibility intervals as the 90 per cent highest probability density intervals (HPDIs) (i.e. the shortest span that includes 90% of the probability density of a parameter).
Convergence of such a kind of analysis and dataset is a tricky issue. To ensure reliable convergence towards the stationary distribution, we monitored multiple independent runs for the two datasets, each with 50–100 independent heated chains under Metropolis coupling to improve mixing (Hey & Nielsen 2004). Each run was initiated with a burn-in period of at least 300 000 updates, with each multi-chain simulation lasting several months (the longest runs were up to 3.1 × 107 iterations, see the electronic supplementary material, table S1, for details regarding runs). Mixing properties of the MCMC were assessed by visual inspection of the parameter trend plots and by examining the level of autocorrelation between initial and final parameter values (using the effective sample size, ESS). Analyses were considered to have converged upon the stationary distribution if independent runs generated similar posterior distributions, with each having a lowest ESS of 50 for each estimated parameter as recommended in Hey & Nielsen (2004).
To convert posterior estimates of the model scaled by the mutation rate μ (i.e. T and θ) into demographic units (i.e. t and Ne), we used a value of μ = 5.0 × 10−4 mutation per generation, which is considered as the average mutation rate over many species (Brinkmann et al. 1998; Estoup & Angers 1998; Ellegren 2000; Estoup et al. 2002; Sun et al. 2009). Assuming a generation time (G) for harbour porpoise ranging between 5 (GL, the average age of females at first parturition) (Lockyer 2003) and 7 years (GH, as suggested by Tolley & Rosel 2006), population-splitting time (T) can be converted to calendar years (t) and estimates of population mutation rates (θ1, θ2 and θA) can be converted into estimates of effective population size parameters (N1, N2 and NA, respectively, in a number of individuals) as described in Hey & Nielsen (2004). Migration parameters in the model (M1 and M2) can be used to obtain population migration rates (2N1m1 and 2N2m2, i.e. the effective number of migrants per generation into population 1 and 2, respectively) using the MLE estimates of current θ and of M (2N1m1 = θ1M1/2 and 2N2m2= θ2M2/2) (Hey & Nielsen 2004).
3. Results and discussion
(a). Convergence of IM runs
After several steps of trimming, the replicated IM simulations displayed good mixing properties of the coupled MCMCs as suggested by the parameter trend line plots (data not shown) with an ESS of at least 71 for the IB–NBB analyses and 53 for AT–BS analyses (details on these runs are provided in the electronic supplementary material, table S1). IM replicates provided consistent similar posterior probability densities for all parameters (electronic supplementary material, figures S1 and S2). All these indications suggested that we have sufficiently explored the parameter space. We selected runs that best converged (i.e. those displaying the highest ESS estimates) to convert them into demographic units (figure 2 and table 1) and to interpret the outputs.
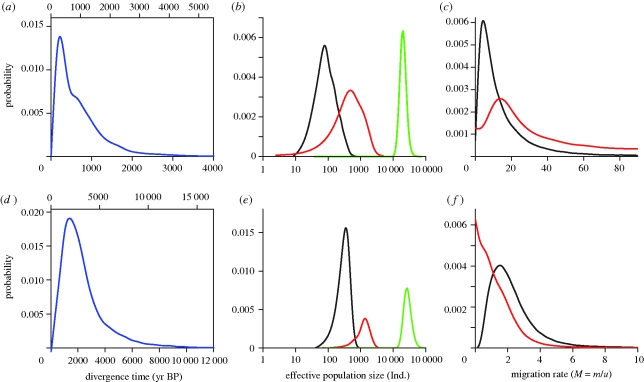
Figure 2.
Marginal posterior probability densities for each parameter of the IM model. Curves are shown for the analyses comparing harbour porpoise populations from (a–c) Iberian (IB) versus northern Bay of Biscay (NBB), and (d–f) the Black Sea (BS) versus Atlantic (AT). The bottom and top x-axes of plots (a,d) show the time scales converted into calendar years before present (yr BP) using two generation times 5 and 7 years (bottom and top x-axes, respectively). (b) Black line, IB; red line, NBB; green line, ancestral. (c) Migration rate into IB, black line; migration rate into NBB, red line. (e) Black line, BS; red line, AT; green line, ancestral. (f) Migration rate into BS, black line; migration rate into AT, red line.
Table 1.
Maximum-likelihood estimates and 90% HPDIs (between brackets) of parameter estimates under the IM model. (2N1m1 and 2N2m2 are the effective number of migrants per generation, respectively, into population 1 and 2. 1, population 1; 2, population 2; A, ancestral population.)
| parameter | Iberian (1) × northern (2) Bay of Biscay | Black Sea (1) × Atlantic (2) |
|---|---|---|
| t (years)a | 175–245 (55–2079) | 1325–1855 (125–6,335) |
| N1 | 79 (22–328) | 362 (148–573) |
| N2 | 353 (148–2243) | 1393 (543–2453) |
| NA | 19 688 (13 313–30 113) | 26 925 (16 875–45 225) |
| 2N1m1 = θ1m1/2 | 0.34 | 0.54 |
| 2N2m2 = θ2m2/2 | 4.65 | 0 |
aSplitting time estimates (T = t μ) were converted into calendar years using a per-generation mutation rate (μ) of 5.0 × 10−4 and two generation times (G) 5 and 7 years.
(b). Iberian versus northern Bay of Biscay
Results from the IM model for the isolation of IB porpoises from those of the NBB are shown in figure 2a–c and summarized in table 1. Estimated posterior probability distributions of effective population sizes (figure 2b and table 1) show that a large ancestral population, with a modal effective size (NA) of ca 20 000 individuals (HPDI: 13 313–30 113), split into two smaller populations: a very small population off the Iberian Atlantic coasts with an effective size of ca 80 individuals (HPDI: 22–328), and a larger population in the northern Bay of Biscay with an effective size of ca 350 individuals (HPDI: 148–2243) (table 1). The splitting time was clearly resolved, with a sharply peaked posterior distribution (figure 2a) and a MLE between 175 and 245 yr BP (HPDI: 55–2079), depending on the generation time used (5 or 7 years). Migration rate distributions (figure 2c) show a clear signal of migration in both directions with, however, 14 times higher gene flow from Iberia towards the northern Bay of Biscay after population-splitting (table 1).
The genetic inference thus indicates the barrier to gene flow located in the southern Bay of Biscay (Fontaine et al. 2007) rose during the past millennium and most probably during the few last centuries. It is now widely recognized that worldwide anomalies in climatic and oceanic conditions occurred during the past 2000 years (Osborn & Briffa 2006). The most significant according to Osborn & Briffa (2006) include the ‘Medieval Warm Period’ (1200–800 yr BP), the ‘LIA’ (600–150 yr BP) and the modern warm period. During the LIA that culminated 300 years ago, dramatic cooling in oceanographic and climatic conditions in the North Atlantic, and over Europe in particular, had a substantial impact on human activities, for which there are historical records, but also more generally on terrestrial and marine ecosystems (Grove 2004). Fishery records in Europe, which date back as far as the tenth century for some species (Alheit & Hagen 1997), report both significant increases in capture of cold-water fishes (e.g. herring Clupea harengus) as far south as in the Bay of Biscay and the southward retreat of warm-water species (e.g. sardine) during the LIA (Alheit & Hagen 1997). Given that cold-water fishes, like herring or sandeel (Ammoditidae sp.), are a major component of porpoise diet throughout its current range (Read 1999; Santos & Pierce 2003), it is highly probable that the harbour porpoise's past range was likewise extended southward into the Bay of Biscay. The IM demographic estimates in terms of timing, geneflow and the relative size of the subranges are consistent with such a scenario where harbour porpoises north and south of the Bay of Biscay formed a single range during the LIA cold conditions, and began to fragment with the onset of the current warm period 200–300 yr BP (Osborn & Briffa 2006), together with substantial shifts in the marine community in the eastern North Atlantic and especially in the Bay of Biscay (Poulard & Blanchard 2005). The relatively cold nutrient-enriched conditions generated by coastal upwelling along the Iberian Atlantic coasts (Fiùza 1983) have allowed a small porpoise population and other cold temperate species to persist in these waters up to now (OSPAR Commission 2000). The predominant northward migration signal detected in the genetic data supports a northward exodus of Iberian porpoises during their isolation, as expected for a Northern Hemisphere temperate species in response to ocean warming.
(c). Atlantic versus Black Sea
Results from the IM model for the isolation of Black Sea porpoises from those in the Atlantic are shown in figures 2d–f and summarized in table 1. The estimates suggest a very large ancestral effective population size (NA) of ca 30 000 individuals (HPDI: 16 875–45 225). According to the model, this ancestral population split into much smaller populations (ca 360 (HPDI: 148–573) individuals in the Black Sea and ca 1400 (HPDI: 543–2453) individuals in the Atlantic). The population split is estimated to have occurred 1325–1855 years ago (HPDI: 125–6335, figure 2d), depending on the generation time used. Migration rate posterior distributions (figure 2f) show a clear signal of migration, though very low, from the Atlantic to the Black Sea, but no evidence of gene flow in the reverse direction (table 1 and figure 2f).
Previous studies suggested a much older separation time of Black Sea from Atlantic porpoises based on the analysis of genetic polymorphism at the non-coding region of the mtDNA CR (Tolley & Rosel 2006; Viaud-Martinez et al. 2007). We ran further IM simulations using published and new mtDNA sequences (see electronic supplementary material, methods, table S1, S2 and figure S3). Combined microsatellite and mtDNA data did not change inference compared with the results described above, but results from the mtDNA locus alone showed a strong discrepancy with the nuclear loci (electronic supplementary material, figure S3; Viaud-Martinez et al. 2007). The Atlantic–Black Sea splitting time estimate based only on the mtDNA CR and converted into calendar years using the formerly admitted range of mutation rates (between 3.3 and 4.3 × 10−8 substitutions per site and per year (s.s.yr−1) (Tolley & Rosel 2006; Viaud-Martinez et al. 2007) was up to three orders of magnitude greater than at nuclear loci. However, any estimate of divergence time is conditioned on the uncertainty regarding the mutation rate estimates, which is very difficult to assess and highly variable among species (Nabholz et al. 2008a). The IM algorithm accounts for variation in the mutation rate among loci by estimating the relative locus-specific mutation rate as a mutation rate scalar (Hey & Nielsen 2004). Provided we have a sound mutation rate estimate for at least one locus allows us to deduce the mutation rate of the others. Assuming that the average mutation rate for microsatellite loci is 1 × 10−4 mutations per year (considering a generation time of 5 years), the mtDNA CR substitution rate can be estimated from our combined analysis at 1.3 × 10−6 s.s.yr−1 (HPDI: 8.4 × 10−7–2.3 × 10−6). This is at least two orders of magnitude faster than ‘traditional’ substitution rate estimates derived from interspecific phylogenetic datasets or the fossil record (Hoelzel et al. 1991; Henn et al. 2009) but, within the intervals of those found with the mean human mtDNA CR pedigree rate estimate derived from a meta-analysis (9.5 × 10−7 s.s.yr−1) (Howell et al. 2003), with ancient DNA datasets in Adélie penguin Pygoscelis adeliae (9.6 × 10−7 s.s.yr−1) (Lambert et al. 2002), in Southern elephant seal Mirounga leonina 9.8 × 10−7 s.s.yr−1 HPDI: 1.7 × 10−9–2.1 × 10−6) (de Bruyn et al. 2009) and in Steller sea lions Eumetopias jubatus 2.7 10−7 s.s.yr−1 (Phillips et al. 2009). Such a high intraspecific estimate is reminiscent to the time dependency of mtDNA mutation rates recently recognized as a general phenomenon applying to many species (e.g. primates, marine mammals, birds and fishes) (Lambert et al. 2002; Howell et al. 2003; Ho et al. 2005, 2007; Bandelt 2008; de Bruyn et al. 2009; Endicott et al. 2009; Henn et al. 2009). Higher mutation rate estimates are obtained from events calibrated with more recent dates, such as pedigrees, and lower estimates result when events are calibrated with older dates.
The anomalous behaviour of the mitochondrion did not only concern the substitution rate, but also estimates of ancestral population genetic diversity (θ) from mtDNA CR alone. mtDNA θ estimates showed a diametrically opposed pattern to that of nuclear microsatellite loci, with estimates of ancestral population genetic diversity being much smaller than the current population diversity (electronic supplementary material, figure S3 and fig. 4 in Viaud-Martinez et al. (2007)). Under neutral evolution of the genetic markers used, historical variation in population size should affect genetic polymorphism at both nuclear and mitochondrial loci in a similar manner (albeit with larger drift effects on the mitochondrion). The opposing pattern we observed here could be the outcome of the large stochastic variance in the coalescence process affecting a single locus (Hey & Machado 2003), but it is worthwhile considering other potential causal factors. First, selective processes may have acted on the mtDNA genome (e.g. selective sweeps). Given the central role of mitochondria in heat and energy production, and the strong energetic constraints faced by porpoises which are small, warm-blooded marine carnivores with a demanding reproductive schedule, detecting a signal of selection on the mitochondrion would seem hardly surprising. Second, heterogeneous mutation rates along the mtDNA CR are poorly captured by the substitution model. Mutational hotspots resulting in high levels of homoplasy could provide a false signal of population expansion (Pakendorf & Stoneking 2005; Galtier et al. 2006; Strasburg & Rieseberg 2010). Finally, the discrepancy may be the result of complex and multiple determinants of genetic diversity on the mtDNA genome. A recent study examining mtDNA evolution in mammals showed that mitochondrial rates of evolution generally do not depart from neutral expectations, although they are highly stochastic among species, and essentially unpredictable from knowledge of species biology (Nabholz et al. 2008b; Galtier et al. 2009).
As harbour porpoises currently inhabit productive coastal waters of the North Atlantic and Black Sea (Read 1999), we expected that porpoises were present in the Mediterranean at a time when palaeoceanographic conditions were more productive than the present-day oligotrophic conditions. Geological records indicate that such conditions prevailed in the Mediterranean Sea throughout the deglaciation period following the LGM (approx. 18 000 yr BP) until the Mid-Holocene warm period (approx. 6000 yr BP) (Kerwin et al. 1999; Jimenez-Espejo et al. 2007; Rohling et al. 2009). The strongest contrast with current conditions was in the eastern Mediterranean basin, where organic-rich sediments, termed ‘Sapropel’, accumulated under strong hydrological changes generated by intense fresh water discharges and substantially enhanced primary productivity (Calvert et al. 1992; Rohling et al. 2009). At the end of this Sapropel episode, approximately 5500 years ago, primary productivity abruptly decreased toward present-day conditions. Demographic estimates from the IM model based on microsatellite data or combined microsatellite and mtDNA data are consistent with porpoise range fragmentation in the Mediterranean starting around 5500 years ago at the end of this Sapropel event, while productive conditions persisted in the northern Aegean Sea and the Black Sea. The posterior estimates of Atlantic–Black Sea time of separation based on microsatellite data firmly place this isolation event in the post-LGM period and point to the post-Sapropel period. The signal of migration from the Atlantic to the Black Sea captured in the IM analysis (figure 2f and table 1) probably reflects migration events during the early stage of the divergence process and is consistent with the proposal that the last remnants of Mediterranean porpoises (the ‘admixed’ link between Atlantic and Black Sea) ended up retreating to the Black Sea.
Post-glacial and very recent climate-driven environmental changes have triggered deep reorganization in Northeast Atlantic ecosystems, with changes detected in plankton and fish communities. Our study provides strong evidences of such a response for a small coastal cetacean that has to meet a particularly demanding energetic budget to survive. Harbour porpoise populations in the Northeast Atlantic waters have undergone recent dramatic changes in connectivity and distribution probably related to climate-driven variation in habitat suitability. Our inferences naturally depend upon a specific model of population evolution (IM model), which necessarily simplifies the population demographic history, and on a specific mutation rate. Departures from the model, such as departure from panmixia within population or change in the effective size of diverging populations, may induce some bias in our estimates. However, the model appeared quite robust to these departures (Strasburg & Rieseberg 2010). The biological congruence between our genetic inferences and historical and palaeoceanographic records suggests the model captures the essential aspects of this fragmentation process.
It seems inevitable that marine predators will need to adapt to a changing spatial distribution of primary and secondary production within pelagic marine ecosystems as they did in the past (Marx & Uhen 2010). This reaction to climate change could however be exacerbated on a much shorter time scale by the depletion of major commercial fish stocks induced by intense commercial fisheries (Worm et al. 2006, 2009) and/or by detrimental interactions with human activities such as incidental catches, which probably constitute a much more serious short-term threat for small coastal cetaceans such as harbour porpoises (Stenson 2003; Hovelsrud et al. 2008). The combination of these effects raises serious concerns regarding the ability of top predators to adapt to such rapidly changing habitat conditions. Addressing these concerns will be a particular challenge for future research and management policy.
Acknowledgements
We are grateful to all fishermen, stranding networks and volunteers that contributed in the collection of samples used in the present study. We thank A. Jacomme, A. Estoup, C. Moritz, R. Cheddadi, E. J. Rohling, L. Crespin, R. Streiff, T. Malausa, S. Piry, S. J. Goodman, G. Roderick, J. Hey and N. Galtier for their valuable help and/or comments during the development of this research. This work was funded by the Belgian Science Policy (Contract SSTC EV/12/46A). M.C.F. was supported by research fellowships from the Belgian National Fund for Scientific Research (FRS-FNRS, mandate ‘Aspirant’), and by the AGAPE Marie-Curie Fellowship programme, and K.A.T. by the Norwegian Research Council. This paper is an MARE publication no. 187.
References
- Alheit J., Hagen E.1997Long-term climate forcing of European herring and sardine populations. Fish Oceanogr. 6, 130–139 (doi:10.1046/j.1365-2419.1997.00035.x) [Google Scholar]
- Alley R. B., et al. 2003Abrupt climate change. Science 299, 2005–2010 (doi:10.1126/science.1081056) [DOI] [PubMed] [Google Scholar]
- Bandelt H. J.2008Clock debate: when times are changing: time dependency of molecular rate estimates: tempest in a teacup. Heredity 100, 1–2 (doi:10.1038/sj.hdy.6801054) [DOI] [PubMed] [Google Scholar]
- Beaugrand G., Reid P. C., Ibanez F., Lindley J. A., Edwards M.2002Reorganization of North Atlantic marine copepod biodiversity and climate. Science 296, 1692–1694 (doi:10.1126/science.1071329) [DOI] [PubMed] [Google Scholar]
- Beaugrand G., Edwards M., Brander K., Luczak C., Ibanez F.2008Causes and projections of abrupt climate-driven ecosystem shifts in the North Atlantic. Ecol. Lett. 11, 1157–1168 (doi:10.1111/j.1461-0248.2008.01218.x) [DOI] [PubMed] [Google Scholar]
- Brinkmann B., Klintschar M., Neuhuber F., Huhne J., Rolf B.1998Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am. J. Hum. Genet. 62, 1408–1415 (doi:10.1086/301869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook B. W., Sodhi N. S., Bradshaw C. J. A.2008Synergies among extinction drivers under global change. Trends Ecol. Evol. 23, 453–460 (doi:10.1016/j.tree.2008.03.011) [DOI] [PubMed] [Google Scholar]
- Calvert S. E., Nielsen B., Fontugne M. R.1992Evidence from nitrogen isotope ratios for enhanced productivity during formation of eastern Mediterranean sapropels. Nature 359, 223–225 (doi:10.1038/359223a0) [Google Scholar]
- de Bruyn M., Hall B. L., Chauke L. F., Baroni C., Koch P. L., Hoelzel A. R.2009Rapid response of a marine mammal species to Holocene climate and habitat change. PLoS Genet. 5, e1000554 (doi:10.1371/journal.pgen.1000554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H.2000Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet. 16, 551–558 (doi:10.1016/S0168-9525(00)02139-9) [DOI] [PubMed] [Google Scholar]
- Endicott P., Ho S. Y. W., Metspalu M., Stringer C.2009Evaluating the mitochondrial timescale of human evolution. Trends Ecol. Evol. 24, 515–521 (doi:10.1016/j.tree.2009.04.006) [DOI] [PubMed] [Google Scholar]
- Estoup A., Angers B.1998Microsatellites and minisatellites for molecular ecology: theoretical and empirical considerations. In Advances in molecular ecology (ed. Carvalho G.), pp. 55–86 Amsterdam, The Netherlands: IOS Press [Google Scholar]
- Estoup A., Jarne P., Cornuet J. M.2002Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 11, 1591–1604 (doi:10.1046/j.1365-294X.2002.01576.x) [DOI] [PubMed] [Google Scholar]
- Fiùza A. F. G.1983Upwelling patterns of Portugal. In Coastal upwelling, its sedimentary record. Part A. Responses of the sedimentary regime to present coast upwelling (eds Suess E., Thiede J.), pp. 85–98 New York, NY: Plenum Press [Google Scholar]
- Fontaine M. C., et al. 2007Rise of oceanographic barriers in continuous populations of a cetacean: the genetic structure of harbour porpoises in Old World waters. BMC Biol 5, 30 (doi:10.1186/1741-7007-5-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantzis A., Gordon J., Hassidis G., Komenou A.2001The enigma of harbour porpoise presence in the Mediterranean Sea. Mar. Mamm. Sci. 17, 937–943 (doi:10.1111/j.1748-7692.2001.tb01307.x) [Google Scholar]
- Galtier N., Enard D., Radondy Y., Bazin E., Belkhir K.2006Mutation hot spots in mammalian mitochondrial DNA. Genome Res. 16, 215–222 (doi:10.1101/gr.4305906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Nabholz B., Glemin S., Hurst G. D. D.2009Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550 (doi:10.1111/j.1365-294X.2009.04380.x) [DOI] [PubMed] [Google Scholar]
- Grove J. M.2004Little ice ages: ancient and modern. Routledge Studies in Physical Geography and Environment. London, UK: Routledge [Google Scholar]
- Henn B. M., Gignoux C. R., Feldman M. W., Mountain J. L.2009Characterizing the time dependency of human mitochondrial DNA mutation rate estimates. Mol. Biol. Evol. 26, 217–230 (doi:10.1093/molbev/msn244) [DOI] [PubMed] [Google Scholar]
- Herr H., Fock H. O., Siebert U.2009Spatio-temporal associations between harbour porpoise Phocoena phocoena and specific fisheries in the German Bight. Biol. Conserv. 142, 2962–2972 (doi:10.1016/j.biocon.2009.07.025) [Google Scholar]
- Hewitt G. M.2000The genetic legacy of quaternary ice ages. Nature 405, 907–913 (doi:10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- Hewitt G. M.2004Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. Lond. B 359, 183–195 Discussion 195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J., Machado C. A.2003The study of structured populations: new hope for a difficult and divided science. Nat. Rev. Genet. 4, 535–543 (doi:10.1038/nrg1112) [DOI] [PubMed] [Google Scholar]
- Hey J., Nielsen R.2004Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167, 747–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. Y., Phillips M. J., Cooper A., Drummond A. J.2005Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22, 1561–1568 (doi:10.1093/molbev/msi145) [DOI] [PubMed] [Google Scholar]
- Ho S. Y., Kolokotronis S. O., Allaby R. G.2007Elevated substitution rates estimated from ancient DNA sequences. Biol. Lett. 3, 702–705 (doi:10.1098/rsbl.2007.0377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzel A. R., Hancock J. M., Dover G. A.1991Evolution of the cetacean mitochondrial d-loop region. Mol. Biol. Evol. 8, 475–493 [DOI] [PubMed] [Google Scholar]
- Hofreiter M., Stewart J.2009Ecological change, range fluctuations and population dynamics during the Pleistocene. Curr. Biol. 19, R584–R594 (doi:10.1016/j.cub.2009.06.030) [DOI] [PubMed] [Google Scholar]
- Hovelsrud G. K., McKenna M., Huntington H. P.2008Marine mammal harvests and other interactions with humans. Ecol. Appl. 18, S135–S147 (doi:10.1890/06-0843.1) [DOI] [PubMed] [Google Scholar]
- Howell N., Smejkal C. B., Mackey D. A., Chinnery P. F., Turnbull D. M., Herrnstadt C.2003The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates. Am. J. Hum. Genet. 72, 659–670 (doi:10.1086/368264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Espejo F. J., Martinez-Ruiz F., Sakamoto T., Iijima K., Gallego-Torres D., Harada N.2007Paleoenvironmental changes in the western Mediterranean since the last glacial maximum: high resolution multiproxy record from the Algero-Balearic basin. Palaeogeogr. Palaeoclimatol. Palaeoecol. 246, 292–306 (doi:10.1016/j.palaeo.2006.10.005) [Google Scholar]
- Johnston D. W., Westgate A. J., Read A. J.2005Effects of fine-scale oceanographic features on the distribution and movements of harbour porpoises Phocoena phocoena in Bay of Fundy. Mar. Ecol. Prog. Ser. 295, 279–293 (doi:10.3354/meps295279) [Google Scholar]
- Kastelein R. A., Van der Sijs S. J., Staal C., Nieuwstraten S. H.1997Blubber thickness in harbour porpoises (Phocoena phocoena). In The biology of harbour porpoise (eds Read A. J., Wiepkema P. R., Nachtigall P. E.), pp. 179–199 Woerden, The Netherlands: De Spil Publishers [Google Scholar]
- Kerwin M., Overpeck J., Webb R. S., DeVernal A., Rind D. H., Healy R. J.1999The role of oceanic forcing in Mid-Holocene northern hemisphere climatic change. Paleoceanography 14, 200–210 (doi:10.1029/1998PA900011) [Google Scholar]
- Kintisch E.2006As the seas warm. Science 313, 776–779 (doi:10.1126/science.313.5788.776) [DOI] [PubMed] [Google Scholar]
- Koopman H. N., Iverson S. J., Gaskin D. E.1996Stratification and age-related differences in blubber fatty acids of the male harbour porpoise (Phocoena phocoena). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 165, 628–639 [DOI] [PubMed] [Google Scholar]
- Koopman H. N., Pabst D. A., McLellan W. A., Dillaman R. M., Read A. J.2002Changes in blubber distribution and morphology associated with starvation in harbour porpoise (Phocoena phocoena): evidence for regional variation in blubber structure and function. Physiol. Biochem. Zool. 75, 498–512 (doi:10.1086/342799) [DOI] [PubMed] [Google Scholar]
- Lambert D. M., Ritchie P. A., Millar C. D., Holland B., Drummond A. J., Baroni C.2002Rates of evolution in ancient DNA from Adelie penguins. Science 295, 2270–2273 (doi:10.1126/science.1068105) [DOI] [PubMed] [Google Scholar]
- Learmonth J. A., MacLeod C. D., Santos M. B., Pierce G. J., Crick H. Q. P., Robinson R. A.2006Potential effects of climate change on marine mammals. Oceanogr. Mar. Biol. 44, 431–464 [Google Scholar]
- Lockyer C.2003Harbour porpoises (Phocoena phocoena) in the North Atlantic: biological parameters. In Harbour porpoises in the North Atlantic, vol. 5 (eds Haug T., Desportes G., Vikingsson G. A., Witting L.), pp. 71–90 Tromso, Norway: NAMMCO Scientific Publications [Google Scholar]
- Lockyer C.2007All creatures great and smaller: a study in cetacean life history energetics. J. Mar. Biol. Assoc. UK 87, 1035–1045 (doi:10.1017/S0025315407054720) [Google Scholar]
- MacLeod C. D., Santos M. B., Reid R. J., Scott B. E., Pierce G. J.2007Linking sandeel consumption and the likelihood of starvation in harbour porpoises in the Scottish North Sea: could climate change mean more starving porpoises? Biol. Lett. 3, 185–188 (doi:10.1098/rsbl.2006.0588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major C., Ryan W., Lericolais G., Hajdas I.2002Constraints on Black Sea outflow to the Sea of Marmara during the last glacial-interglacial transition. Mar. Geol. 190, 19–34 (doi:10.1016/S0025-3227(02)00340-7) [Google Scholar]
- Marx F. G., Uhen M. D.2010Climate, critters, and cetaceans: cenozoic drivers of the evolution of modern whales. Science 327, 993–996 (doi:10.1126/science.1185581) [DOI] [PubMed] [Google Scholar]
- Mayewski P. A., et al. 2004Holocene climate variability. Quat. Res. 62, 243–255 (doi:10.1016/j.yqres.2004.07.001) [Google Scholar]
- Moritz C.1994Defining ‘evolutionary significant unit’ for conservation. Trends Ecol. Evol. 9, 373–375 (doi:10.1016/0169-5347(94)90057-4) [DOI] [PubMed] [Google Scholar]
- Moritz C.2002Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 51, 238–254 (doi:10.1080/10635150252899752) [DOI] [PubMed] [Google Scholar]
- Nabholz B., Glemin S., Galtier N.2008aStrong variations of mitochondrial mutation rate across mammals: the longevity hypothesis. Mol. Biol. Evol. 25, 120–130 (doi:10.1093/molbev/msm248) [DOI] [PubMed] [Google Scholar]
- Nabholz B., Mauffrey J.-F., Bazin E., Galtier N., Glemin S.2008bDetermination of mitochondrial genetic diversity in mammals. Genetics 178, 351–361 (doi:10.1534/genetics.107.073346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Corry-Crowe G.2008Climate change and the molecular ecology of Arctic marine mammals. Ecol. Appl. 18, S56–S76 (doi:10.1890/06-0795.1) [DOI] [PubMed] [Google Scholar]
- Osborn T. J., Briffa K. R.2006The spatial extent of 20th-century warmth in the context of the past 1200 years. Science 311, 841–844 (doi:10.1126/science.1120514) [DOI] [PubMed] [Google Scholar]
- OSPAR Commission. Quality Status Report 2000: Region IV—Bay of Biscay and Iberian Coast. OSPAR Commission, London, UK: 2000. [Google Scholar]
- Pakendorf B., Stoneking M.2005Mitochondrial DNA and human evolution. Annu. Rev. Genomics Hum. Genet. 6, 165–183 (doi:10.1146/annurev.genom.6.080604.162249) [DOI] [PubMed] [Google Scholar]
- Pastene L. A., et al. 2007Radiation and speciation of pelagic organisms during periods of global warming: the case of the common minke whale, Balaenoptera acutorostrata. Mol. Ecol. 16, 1481–1495 (doi:10.1111/j.1365-294X.2007.03244.x) [DOI] [PubMed] [Google Scholar]
- Perry A. L., Low P. L., Ellis J. R., Reynolds J. D.2005Climate change and distribution shifts in marine fishes. Science 308, 1912–1915 (doi:10.1126/science.1111322) [DOI] [PubMed] [Google Scholar]
- Phillips C. D., Trujillo R. G., Gelatt T. S., Smolen M. J., Matson C. W., Honeycutt R. L., Patton J. C., Bickham J. W.2009Assessing substitution patterns, rates and homoplasy at HVRI of Steller sea lions, Eumetopias jubatus. Mol. Ecol. 18, 3379–3393 (doi:10.1111/j.1365-294X.2009.04283.x) [DOI] [PubMed] [Google Scholar]
- Pimm S. L.2008Biodiversity: climate change or habitat loss—which will kill more species? Curr. Biol. 18, R117–R119 (doi:10.1016/j.cub.2007.11.055) [DOI] [PubMed] [Google Scholar]
- Pimm S. L.2009Climate disruption and biodiversity. Curr. Biol. 19, R595–R601 (doi:10.1016/j.cub.2009.05.055) [DOI] [PubMed] [Google Scholar]
- Poulard J.-C., Blanchard F.2005The impact of climate change on the fish community structure of the eastern continental shelf of the Bay of Biscay. ICES J. Mar. Sci. 62, 1436–1443 (doi:10.1016/j.icesjms.2005.04.017) [Google Scholar]
- Read A. J.1999Harbour porpoise (Phocoena phocoena). In Handbook of marine mammals (eds Ridgway S. H., Harrison R.), pp. 323–350 London, UK:Academic Press [Google Scholar]
- Read A. J., Hohn A. A.1995Life in the fast lane: the life history of harbour porpoises from the Gulf of Maine. Mar. Mamm. Sci. 11, 423–440 (doi:10.1111/j.1748-7692.1995.tb00667.x) [Google Scholar]
- Richardson A. J., Schoeman D. S.2004Climate impact on plankton ecosystems in the Northeast Atlantic. Science 305, 1609–1612 (doi:10.1126/science.1100958) [DOI] [PubMed] [Google Scholar]
- Rohling E. J., Abu-Zieb R., Casford J. L. S., Hayes A., Hoogakker B. A. A.,.2009The marine environment: present and past. In The physical geography of the Mediterranean (ed. Woodward J. C.), pp. 33–67 Oxford, UK: Oxford University Press [Google Scholar]
- Rosel P., Dizon A. E., Haygood M. G.1995Variability of the mitochondrial control region in populations of the harbour porpoise, Phocoena phocoena, on interoceanic and regional scales. Con. J. Fish. Aquat. Sci. 52, 1210–1219 [Google Scholar]
- Ryther J. H.1969Photosynthesis and fish production in the sea. Science 166, 72–76 (doi:10.1126/science.166.3901.72) [DOI] [PubMed] [Google Scholar]
- Santos M. B., Pierce G. J.2003The diet of harbour porpoise (Phocoena phocoena) in the northeast Atlantic. Oceanogr. Mar. Biol. 41, 355–390 [Google Scholar]
- Santos M. B., Pierce G. J., Learmonth J. A., Reid R. J., Ross H. M., Patterson I. A. P., Reid D. G., Beare D.2004Variability in the diet of harbor porpoises (Phocoena phocoena) in Scottish waters 1992–2003. Mar. Mamm. Sci. 20, 1–27 (doi:10.1111/j.1748-7692.2004.tb01138.x) [Google Scholar]
- Southward A. J., Hawkins S. J., Burrows M. T.1995Seventy years' observations of changes in distribution and abundance of zooplankton and intertidal organisms in the western English Channel in relation to rising sea temperature. J. Therm. Biol. 20, 127–155 (doi:10.1016/0306-4565(94)00043-I) [Google Scholar]
- Springer G. S., Rowe H. D., Hardt B., Edwards R. L., Cheng H.2008Solar forcing of Holocene droughts in a stalagmite record from West Virginia in east-central North America. Geophys. Res. Lett. 35, L17703 (doi:10.1029/2008GL034971) [Google Scholar]
- Stenson G. B.2003Harbour porpoise (Phocoena phocoena) in the North Atlantic: abundance, removals, and sustainability of removals. In Harbour porpoises in the North Atlantic, vol. 5 (eds Haug T., Desportes G., Vikingsson G. A., Witting L.), pp. 271–302 Tromso, Norway: NAMMCO Scientific Publications [Google Scholar]
- Strasburg J. L., Rieseberg L. H.2010How robust are ‘isolation with migration’ analyses to violations of the IM model? A simulation study. Mol. Biol. Evol. 27, 297–310 (doi:10.1093/molbev/msp233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. X., Mullikin J. C., Patterson N., Reich D. E.2009Microsatellites are molecular clocks that support accurate inferences about history. Mol. Biol. Evol. 26, 1017–1027 (doi:10.1093/molbev/msp025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley K. A., Rosel P. E.2006Population structure and historical demography of eastern North Atlantic harbour porpoises inferred through mtDNA sequences. Mar. Ecol. Prog. Ser. 327, 297–308 (doi:10.3354/meps327297) [Google Scholar]
- Viaud-Martinez K. A., et al. 2007Morphological and genetic differentiation of the Black Sea harbour porpoise Phocoena phocoena. Mar. Ecol. Prog. Ser. 338, 281–294 (doi:10.3354/meps338281) [Google Scholar]
- Worm B., et al. 2006Impacts of biodiversity loss on ocean ecosystem services. Science 314, 787–790 (doi:10.1126/science.1132294) [DOI] [PubMed] [Google Scholar]
- Worm B., et al. 2009Rebuilding global fisheries. Science 325, 578–585 (doi:10.1126/science.1173146) [DOI] [PubMed] [Google Scholar]