Abstract
The differences in the richness and prevalence of human pathogens among different geographical locations have ramifying consequences for societies and individuals. The relative contributions of different factors to these patterns, however, have not been fully resolved. We conduct a global analysis of the relative influence of climate, alternative host diversity and spending on disease prevention on modern patterns in the richness and prevalence of human pathogens. Pathogen richness (number of kinds) is largely explained by the number of birds and mammal species in a region. The most diverse countries with respect to birds and mammals are also the most diverse with respect to pathogens. Importantly, for human health, the prevalence of key human pathogens (number of cases) is strongly influenced by disease control efforts. As a consequence, even where disease richness is high, we might still control prevalence, particularly if we spend money in those regions where current spending is low, prevalence is high and populations are large.
Keywords: disease, diversity gradients, global climate, human health
1. Introduction
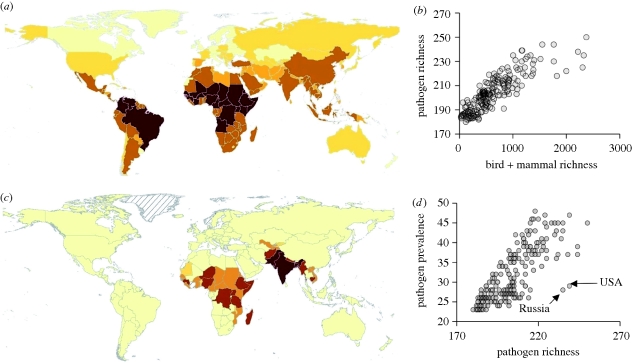
Pathogens cause roughly one in five human deaths (nearly 11 million per year), are responsible for 51 per cent of years of life lost globally and have long affected human demographics (Anderson & May 1991; WHO 2004, 2008). They have also been identified as drivers of human behaviour (Fincher & Thornhill 2008a; Nettle 2009; Park & Schaller 2009; Preston et al. 2009; Zhu 2009), the politics and political stability of countries (Thornhill et al. 2009), human fertility (Guegan et al. 2001), global economies (Thornhill et al. 2009) and more generally the course and dynamics of human history (Denevan 1992; Morens et al. 2004). Pathogen richness (the number of kinds), prevalence (number of cases) and their consequences vary dramatically among regions (figure 1a), as they have since humans first began to spread around the world (Wolfe et al. 2007). Yet the dominant causes of such variation remain incompletely understood.
Figure 1.
Map showing geographical variation in human pathogen richness (a: darker red indicates greater richness; maximum 249) and the regions in which additional spending on disease control might have the greatest effect on human pathogen prevalence (c: on a logged scale, darker red indicates more additional spending recommended; maximum $8.5 billion). (b) Scatter plot of the relationship between human pathogen richness and the richness of birds + mammals and (d) between human pathogen prevalence and richness.
Researchers have linked the presence and prevalence of some pathogens to climate, as has been highlighted in recent discussions of climate change and disease (Lafferty 2009b). The prevalence of malaria (Pascual et al. 2006; Chaves et al. 2008), plague (Snall et al. 2009) and dengue (Cazelles et al. 2005) are affected both by climate and changes in climate. Where it has been considered, the richness of pathogens also correlates with climate or its proxy (latitude), in humans (Guernier et al. 2004; Jones et al. 2008), non-human primates (Nunn et al. 2005) and some bird species (Calvete 2003).
Yet while the distribution, richness and prevalence of human pathogens covary with climate, other non-climate factors might also be important (Guernier et al. 2004). In particular, additional variation in human pathogen richness might be explained by differences among regions in the richness of alternative hosts or vectors (Hawkins & Pausas 2004; Cumming & Guegan 2006). Host richness could influence pathogen richness in humans in three non-exclusive mechanisms. A greater number of alternative host or vector species can decrease the probability of local extinction of a given pathogen (Hechinger & Lafferty 2005; Nunn et al. 2005). Alternatively, regions with greater host richness probably have a higher total richness of pathogens, such that the species pool of pathogens capable of jumping to humans may be higher (Jones et al. 2008; Pedersen & Davies in press). In this scenario, host species richness might influence the emergence of human pathogens rather than the probability of local extinction. The first two scenarios are cases in which diversity might beget more diversity. Finally, the patterns of alternative host species richness may simply capture the integrated effects of drivers of diversification (e.g. historical climate, isolation, topography and geographical area) more effectively than simple measures of modern climate or geography.
Another possibility—the premise on which public health spending is based—is that differences in disease control effort among regions affect the prevalence and richness of parasites and infectious diseases (Lafferty 2009a,b). For example, human efforts to control malaria over the last century have varied between regions, impacting malaria's distribution and prevalence (Hay et al. 2004). Finally, the age of particular habitats or biomes may influence the diversity of many taxa, with lineages having had less time to colonize and diversify in younger biomes (Rohde 1992; Rosenzweig 1992). The habitat constituted by human bodies has been present in some regions longer than others. Therefore, we might hypothesize that pathogen richness would increase, the longer humans are permanently settled in a region. Over time, pathogens may have an opportunity to overcome dispersal barriers (Araujo & Ferreira 2000; Araujo et al. 2008; Moodley et al. 2009) or even to evolve to colonize humans as a novel host (Jones et al. 2008). When humans first migrated into the Americas, they left behind many diseases present in Europe (Wolfe et al. 2007). On the other hand, many of those diseases eventually overcame this initial dispersal limitation and colonized the Americas with catastrophic consequences (Denevan 1992).
Here we use data from political regions of the world to construct models of human pathogen richness and prevalence that include: (i) climatic and geographical variables thought to be potentially important for the transmission, persistence or diversification rates of pathogens; (ii) two measures of the productivity available to human pathogens, human population density and total human population size; (iii) the richness of two major groups of alternative hosts for human parasites and diseases, birds and mammals; (iv) a measure of disease control effort (healthcare spending) and (v) a categorical measure of the time that humans have permanently occupied particular regions.
2. Material and methods
We modelled the relationship between spatial variation in human pathogen richness and prevalence, and various environmental variables, healthcare spending and human population.
(a). Data collection
(i). Pathogen richness
We compiled data on the number of human pathogens, including bacteria, viruses, helminthes and protists in each country or territory. Data were extracted from the Global Infectious Diseases and Epidemiology Network (GIDEON) database (http://www.cyinfo.com) on 15 May 2009. Separately, we extracted the estimates of the richness of pathogens with birds and/or mammals as their reservoirs. GIDEON is a medical database that provides continually updated data on the regional presence and the status of pathogens. The database has been used in a variety of recent studies of the consequences and patterns of pathogen richness and prevalence (Guernier et al. 2004; Smith et al. 2007; Fincher & Thornhill 2008a,b; Fincher et al. 2008). Globally, the GIDEON database includes 347 pathogens of humans. The average pathogen richness per political unit was 204 (n = 214, range = 180–250).
(ii). Pathogen prevalence
In addition to considering pathogen richness, we also considered one commonly used measure of prevalence. We used data from Fincher et al. (2008) in classifying prevalence for all types of each of seven classes of parasites: leishmaniasis, trypanosomes, malaria, schistosomes, filariae, spirochetes and leprosy, for a total of 22 pathogens (excluding three pathogens that occurred in every political region), each of which accounts for a significant proportion of global mortality. Each pathogen was coded as endemic (3), sporadic (2) or not endemic (1)—on the basis of the GIDEON reports. A single index of prevalence was derived across disease classes by summing their ranks such that if all disease classes were endemic in a region, that region would have a total prevalence of 22 × 3 = 66. In practice, the greatest prevalence observed for a region was 47, for Nigeria.
(iii). Climatic and geographical variables
Contemporary environmental variables were obtained from the WorldClim database (Hijmans et al. 2004) at a spatial resolution of 1° and extracted using ArcGIS (ESRI 2006). We integrated the 1° data to produce measures of climatic variables for the entire political regions. We did this in two ways. First, we calculated the average of each climatic variable among grid cells by political region. Second, we calculated the maximum, minimum and range for each climatic variable among grid cells for each political region. We selected three climatic variables for analyses that are clearly associated with diversity theory and/or directly linked to the biology of human pathogens: temperature, precipitation and actual evapotranspiration (AET). Unlike temperature and precipitation data, AET data were derived from MODIS 16 (Mu et al. 2007). Correlations between variations of these variables and pathogen richness have been suggested previously (Guernier et al. 2004). Temperature is implicated as a driver of diversity in metabolic theory (Brown et al. 2004) and temperature-dependent kinetics (Rohde 1992), wherein higher temperatures are predicted to be associated with faster generation times, mutation rates and ultimately rates of diversification. Species energy theory suggests that regions with more productivity, as measured by net primary productivity or AET, have lower rates of extinction and hence more rapid diversification rates (where diversification = speciation − extinction) (Rosenzweig 1968). Larger geographical areas may also allow the persistence of more species, across a wider range of conditions (e.g. greater topographic and soil heterogeneity). In addition, larger areas may have potential direct effects on diversification rates (Rosenzweig & Sandlin 1997). In turn, the geographical area of each political region (derived from the CIA World Factbook; http://www.cia.gov/cia/publications/factbook) was also included as a potential explanatory variable.
(iv). Human population data
In addition to the external environment, factors that may influence the pathogen richness include aspects of human population size and density. Countries or territories with greater numbers of individuals may be more likely to record a particular pathogen. Population density is also positively correlated with disease richness among primate species (Nunn et al. 2003) and carnivore species (Lindenfors et al. 2007), and epidemiological models suggest that disease prevalence and spread is heavily dependent on host densities (Anderson & May 1979). We include as potential explanatory variables both human population density (people per square kilometre) and size in a region. Data on human populations and the area of political units came from the WHO (2004) and, when the WHO database lacked data, from the CIA World Factbook (http://www.cia.gov/cia/publications/factbook).
(v). Disease control data
Disease control effort would ideally have a significant impact on pathogen richness and prevalence. Sufficient data existed from the vast majority of political units to include the following potential explanatory variables in our models (source for all: WHO 2004): per capita government spending on healthcare (measured both with average exchange rates in US dollars (USD) and via purchasing power parity (PPP)), per capita total spending on healthcare (includes non-government spending, measured in USD and PPP), general government spending on health as a percentage of total government spending, immunization rates (we tested four different proxies: per cent of 1-year olds immunized with three doses of Hib3 (Haemophilus influenzae type B) vaccine, per cent of 1-year olds immunized with three doses of Hepatitis B, per cent of 1-year olds immunized with MCV (Meningococcal conjugate vaccine) and per cent of neonates protected at birth against neonatal tetanus).
(vi). Alternative host data
Data on the diversity of birds in each political region were derived from the Avibase, World Bird Database (http://avibase.bsc-eoc.org/checklist.jsp?lang=EN), an online database of regional bird distribution. Mammal data for each political region are from the NatureServe mammal distribution maps (Patterson et al. 2007). The scale of our spatial analysis is restricted by the data with the lowest resolution (e.g. coarsest grain size). Although much finer resolution data are available for birds and mammals, these data could not be used because the distribution patterns of the several hundred kinds of human pathogens are more poorly understood than are the distribution of more than 12 000 bird species and 4000 mammal species. The fact that warbler species distributions are better understood than the distribution of human pathogens is a gap that clearly deserves research attention.
(vii). Human history data
The longer humans have permanently resided in a region, the more human pathogens may be present in that region. Because dates of first human settlement are contentious and reliable evidence is not available for many regions, we scored the time since first permanent human settlement in each region on a five-point scale: 1 = Africa (region of origin and first settlements); 2 = most of Europe, Asia, Australia, New Guinea (by approx. 40 000–60 000 BP); 3 = North America (by approx. 15 000–25 000 BP); 4 = South America (by approx. 1000–5000 years after North America (i.e. 10 000–24 000 BP). This timeframe also includes parts of Northern Europe that were covered in ice during the last glacial maximum and were repopulated during this time period); 5 = most islands (by approx. 1000–7000 BP).
(b). Statistical analysis
We identified the key predictors and pathways leading to pathogen richness and prevalence using a three-step approach. First, we evaluated correlation strengths among the large set of putative predictor variables to identify the best subset of variables that demonstrated the strongest correlations (r > 0.25) but captured different axes of variation (i.e. were not themselves strongly correlated, r < 0.50) (see electronic supplementary material, figure S1). Second, we constructed a generalized linear model (GLM) with Poisson errors for pathogen richness in R (http://www.r-project.org). To normalize residuals, human population size, geographical area and per capita healthcare spending (PPP) were log-transformed. The full model is provided as tables S1 and S2 in the electronic supplementary material). We then used a model simplification procedure, removing non-significant terms (α = 0.05) in a stepwise fashion (Crawley 2002), to explore the relative contributions of the various terms included in the start model. Next, we generated a GLM of pathogen prevalence using the same set of initial predictors. Because pathogen prevalence is influenced by richness, we also included log (pathogen richness) among the predictor variables. Standard model criticism was performed to check for non-constancy of variance and non-normality of errors (Crawley 2002). We saved model residuals and examined spatial autocorrelation using Moran's I correlograms. Since spatial data are only partially independent, regression approaches are not fully justified except where spatial autocorrelation in residuals is insignificant (Diniz Filho et al. 2003).
While stepwise model simplification procedures have been criticized (Whittingham et al. 2006), there remains no best method to select among multiple predictor variables without a priori information (Anderson 2008). Importantly, to avoid potential type I error (Mundry & Nunn 2009), our modelling procedure was not used in significance testing but rather to discriminate among potential causal pathways for the set of predictors that independently demonstrated strong and significant correlations with pathogen richness and/or prevalence (see §3).
Third, we used structural equation modelling to evaluate casual pathways inferred from our GLMs. We explored all significant terms retained with our GLM following model simplification, plus the key climate terms demonstrating strong univariate correlations with pathogen richness and prevalence, even when they were non-significant in the GLM, because previous work suggested climate to be an important predictor of pathogen richness (Guernier et al. 2004). Because of the difficulty of fitting nonlinear terms within structural equation models (SEMs) (Kline 1998), we additionally square-root-transformed richness and prevalence, and log-transformed AET and temperature, prior to model fitting. We compared three models: model one included direct and indirect effects of climate on pathogen richness and prevalence, as well as direct effects of human population size, disease control effort and mammal plus bird species richness. Model two excluded the non-significant pathways from within model one. Model three included only the strongest correlations inferred from the GLMs and SEM model two. These models were chosen to explore the relative importance of key predictor terms along a gradient of decreasing path complexity. Models were contrasted using goodness-of-fit statistics and Bayesian information criterion (BIC), which is related to, but penalizes additional parameters more strongly than, Akaike's information criterion (AIC). Model fitting was performed using the SEM R-library.
Last, because pathogen prevalence varies greatly from country to country, any effort to reduce total global prevalence should target particular regions. Deciding which regions to target will depend on the factors that determine the magnitude of pathogen prevalence in a particular location. Our GLM results identify these key factors that predict prevalence in a location. In turn, we explored the impact that a minimal increase in healthcare spending would have on global pathogen prevalence given the relationship between prevalence and the predictor variables in our final GLM (see §3). We used a ‘greedy’ algorithm to apportion $18 billion (US dollars, approx. 1% of annual private spending on healthcare (WHO 2008)) among regions in $100 000 increments, so that each additional increment maximized per capita reduction in global prevalence: 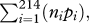 where ni is the human population size of country i and pi is the expected prevalence from the GLM.
where ni is the human population size of country i and pi is the expected prevalence from the GLM.
3. Results
Pathogen richness and prevalence are strongly correlated with mammal plus bird species richness, geographical area, human population size, disease control effort and various climate variables related to temperature, precipitation and productivity (electronic supplementary material, tables S1 and S2). The number of mammal plus bird species in a region is the strongest single correlate of pathogen richness (r = 0.85), whereas disease control effort is the strongest correlate of prevalence (r = 0.69) after pathogen richness.
Our full GLM, including all strongly correlating terms (§2), explains over 82 per cent of pathogen richness among geopolitical regions (estimated as per cent deviance explained; electronic supplementary material, table S1). However, by removing non-significant terms from the model, we show that just three variables; number of bird plus mammal species, disease control effort and total human population, are sufficient to explain approximately 82 per cent of the variation in pathogen richness (GLMs; table 1 and figure 1b). Despite evidently strong spatial autocorrelation in both response and predictor variables, we found no evidence of residual spatial autocorrelation (electronic supplementary material, figure S2), except at distances of less than 400 km, indicating that almost all variation in pathogen richness is explained by predictor variables included within our model.
Table 1.
GLM of pathogen richness: pseudo-r2 = 0.82, AIC = 1367.5.
| coefficient | estimate | z-value | p-value |
|---|---|---|---|
| intercept | 5.213e+00 | 173.633 | <2e−16 |
| bird + mammal richness | 8.501e−05 | 5.959 | 2.53e−09 |
| health spending (per capita) | −8.131e−03 | −2.484 | 0.012985 |
| total human population | 1.138e−02 | 3.884 | 0.000103 |
Pathogen prevalence is also well predicted by our GLMs. The reduced model, which includes disease control effort and two aspects of climate (maximum temperature and minimum AET), explains variation in prevalence among regions as well as the full model which includes additional terms (83% deviance explained for both models; compare table 2 and table S2 in the electronic supplementary material). Because parsimony (and information theory) favours models with fewer parameters, we refer to this reduced model as our ‘favoured’ model.
Table 2.
GLM of pathogen prevalence: pseudo-r2 = 0.83, AIC = 998.57.
| estimate | z-value | p-value | |
|---|---|---|---|
| intercept | −4.967e+00 | −2.966 | 0.003021 |
| health spending (per capita) | −3.452e−02 | −3.485 | 0.000493 |
| total human population | −2.734e−02 | −2.341 | 0.019236 |
| maximum temperature (°C) | 1.158e−03 | 4.345 | 1.39e−05 |
| minimum AET | −2.893e−05 | −2.946 | 0.003220 |
| pathogen richness | 1.623e+00 | 4.939 | 0.003220 |
Our results both for pathogen richness and prevalence were robust to the alternative proxies of disease control effort (electronic supplementary material, tables S3 and S4). Total human population size was a better predictor in our models than human population density (not shown).
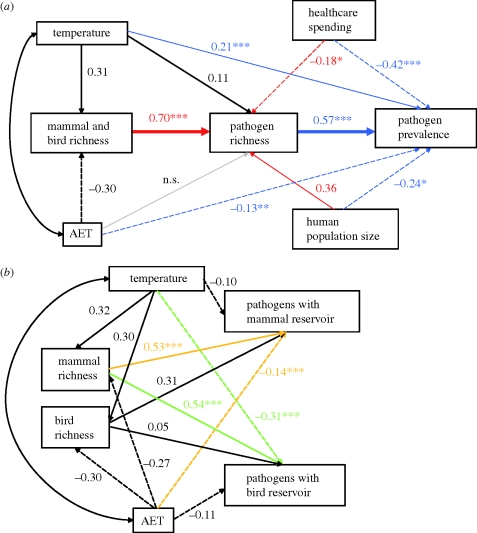
By fitting structural equation models, we show that bird plus mammal richness has the strongest direct effect on pathogen richness, whereas the direct effects of climate and healthcare spending are weak (figure 2a). By contrast, pathogen prevalence was most strongly affected by spending on disease control and pathogen richness, with additional effects owing to climate and human population size, as suggested by the GLMs. A simplified model, with a linear pathway linking mammal plus bird species richness to pathogen richness, which in turn drives pathogen prevalence, is favoured by goodness-of-fit statistics (adjusted goodness-of-fit index = 0.44 and 0.98, BIC = 156.15 and −4.62 for the model including all significant paths and the three-parameter model, respectively). However, it is not possible to explore all possible path combinations without inflating type I error rates.
Figure 2.
SEM illustrating standardized coefficients for pathways among variables retained within the GLMs. Arrows indicate the direction of causality. Double-headed arrows indicate modelled covariance structure. (a) Coloured arrows represent the significant pathways from the GLMs of disease richness (red) and disease prevalence (blue). The pathway from AET to disease richness is not supported in the SEM or GLMs. Pathways in bold represent the SEM favoured by goodness-of-fit statistics (adjusted goodness-of-fit index = 0.44 and 0.98, BIC = 156.15 and −4.62, for the full model (all significant paths shown) and the favoured three-parameter model, respectively). (b) Coloured arrows represent the pathways retained in the minimum adequate models for the GLMs of richness of pathogens with mammal (orange) and bird (green) reservoirs. Values adjacent to paths represent standardized coefficients, asterisks indicate p-values from GLMs: ***p < 0.001, **p < 0.01, *p < 0.05.
When pathogens with bird versus mammal reservoirs are considered separately, we find that the number of mammal species is the stronger predictor for both pathogen types (figure 2b). Notably, mammal species richness is better than bird species richness at predicting richness of pathogens with a bird reservoir (electronic supplementary material, tables S5 and S6).
The relationship between disease control effort and prevalence is linear on a semi-log plot, such that as effort increases returns on that effort diminish (electronic supplementary material, figure S1). Although it was not possible to differentiate clearly between our alternative proxies for disease control effort (electronic supplementary material, tables S3 and S4), our analyses suggest that the most efficient strategy for reducing per capita disease prevalence is to concentrate additional healthcare effort in a small subset of countries (figure 1c). With an increase in healthcare spending of just 1 per cent ($18 billion per year) targeted in the regions indicated in figure 1c, disease prevalence could be reduced by several per cent for over two billion people.
4. Discussion
We find that most of the variation in pathogen richness from one political region to the next is explained by mammal and bird richness, health spending and total human population. In itself, mammal plus bird richness explained most (72%) of the variation in pathogen richness. By contrast, pathogen prevalence is influenced by pathogen richness and disease control efforts, as well as the direct effects of climate and total human population.
Given the strong relationship between pathogen richness and bird and mammal richness, one can predict the number of kinds of pathogens that will be present simply by knowing the number of bird and mammal species present in a country. We can identify at least three mechanisms that may link pathogen richness and bird and mammal richness. The first possibility is that human pathogens, birds and mammals have all responded to similar drivers of diversification. Bird and mammal diversity tends to increase with primary productivity, whether as measured directly or in the form of AET (Hawkins et al. 2003). Productive environments are thought to have a reduced rate of extinction, whether of pathogens or hosts (Wright 1983), while hot, productive areas are also predicted to offer higher mutation rates, faster generation times and, as a consequence, accelerated speciation rates (Rohde 1992). Larger geographical areas typically offer, in addition to more hosts, a wider variety of habitats and climates (MacArthur & Wilson 1967). We included direct measures of primary productivity and area in our models. However, bird and mammal diversity may also be influenced by topographic complexity and isolation (Davies et al. 2007). Topographic complexity can increase habitat diversity and in turn increase species richness. Isolation reduces the chances of species arriving and/or new propagules rescuing the population if they go extinct (MacArthur & Wilson 1967). It is impossible to include in any model all of the environmental variables and their various transformations that potentially influence richness and so perhaps bird and mammal richness are, in a way, a proxy for the complex effects of many interacting environmental drivers.
A second possibility is that regions with high bird and mammal richness have higher total pathogen richness, and hence a higher probability of the origin or host switch of a pathogen that also or newly affects humans. In this model, much like the model recently offered to explain correlations between consumer and producer richness (Jetz et al. 2009), diversity begets diversity. The third possibility is that mammal and bird richness directly influences human pathogen richness by serving as alternative hosts. If either of the latter two links were the primary cause of the patterns we observed, we would expect to see pathogens with bird reservoirs best predicted by bird richness and pathogens with mammal reservoirs best predicted by mammal richness. Instead, mammal richness is the stronger predictor of the richness of pathogens both with non-human mammalian reservoirs and those with non-human bird reservoirs (electronic supplementary material, tables S5 and S6). Since no biological link is expected between these two variables, the correlation implies that mammal richness captures additional variables important for pathogen diversity rather than causing such patterns.
Last, we mention one intriguing alternative explanation for the correlation between pathogen richness and the number of mammal and bird species in a region: pathogen diversity might drive diversity of host clades. High pathogen richness might depress host population densities, thereby inhibiting competitive exclusion (Janzen 1970; Connell 1971; Fincher & Thornhill 2008b) or increase the pace of evolution and species diversification via co-evolutionary arms races (Dobzhansky 1950). Studies in primates support a link between number of pathogens and number of species in host clades (Nunn et al. 2004), suggesting a link between pathogens and host diversification. However, additional data on the distribution of non-human parasites are required to evaluate this hypothesis more fully.
Therefore, although there is a strong statistical relationship between bird and mammal diversity and pathogen richness, we cannot assume that the richness of alternative host species causes pathogen richness. More research is needed to uncover any causal links among these variables and the mechanisms that define the relationships. If we are right that bird and mammal richness indicate rather than cause human pathogen richness, there is no conflict between conservation and human health. In addition, given the incredible extent of ecosystem services humans receive from biological diversity (Costanza et al. 1997), reducing bird and mammal diversity in the hopes of reducing human pathogen richness would be unwarranted.
Compared with the influence of bird and mammal richness, the effort countries make on disease control is poorly correlated with human pathogen richness. The best models of richness do not include disease control measures. Although we cannot preclude that other measures of disease control effort would have explained more variation, environmental variables alone account for the vast majority of the variation in pathogen richness. In theory, regional or even global eradication of diseases can be cheaper in the long run than more local disease control (Ottesen et al. 2008), but in practice eradication successes are rare, with just one example of global eradication of a disease, that of smallpox (de Quadros 2002).
The effect of disease control effort was far greater on human pathogen prevalence (table 2) than on richness. In light of climate change, ecologists have tended to emphasize the links between climate and the prevalence of individual diseases (McMichael et al. 2006; Senior 2008; Semenza & Menne 2009), but recent reviews suggest the possible influence of other factors, including disease control measures (Reiter 2001; Gould & Higgs 2009; Lafferty 2009b; Ostfeld 2009). We found that climate had a direct effect on pathogen prevalence, but the impact was minor relative to the influence of disease control efforts. The idea that disease control can influence prevalence is the basis of billions of dollars of public health aid and medical care spent every year on contagious diseases. Critically, disease control effort can influence pathogen prevalence even if it does not affect pathogen richness. For example, while massive programmes to control malaria reduced malaria prevalence throughout the USA, it had no effect on disease richness; malaria is still present in the USA (Reiter 2001). In our analyses, the USA and Russia (with total annual healthcare spending of $2 trillion and $50 billion, respectively; WHO 2008) are interesting case examples. In both countries, pathogen richness is high and prevalence is relatively low (figure 1d), despite substantial differences in annual healthcare spending ($2 trillion and $50 billion, respectively). Such differences between pathogen richness and prevalence exemplify the enormous challenges disease eradication faces. This issue is simultaneously hopeful and worrisome for public health planning. For the vast majority of pathogens where eradication has failed, efforts at prevention can reduce prevalence in the short-term, but any reductions in healthcare efforts can lead to a resurgence of disease cases (e.g. dengue; Phillips 2008).
The time humans have lived in a region was not a significant predictor in either of our GLMs. However, Africa has marginally greater disease prevalence than other regions (including African versus non-African countries as a binary variable in the GLM explains an additional approximately 3 per cent of the variation in prevalence; ΔAIC = 5.6). One might expect such a pattern since as humans migrated out of Africa they shed some of their pathogens (Araujo et al. 2008). However, other explanations are also possible. Africa may suffer from the persistent effects of many years of low spending on disease control, rather than just the effects of current spending. Alternatively, disease prevalence may be elevated because of enhanced susceptibility owing to the high incidence of human immunodeficiency virus or the presence of a greater number of more closely related primates (particularly apes) (Pedersen & Davies in press). Regardless of its cause, the pathogen prevalence in Africa is greater than expected on the basis of its bird and mammal richness and climate alone.
Our analyses suggest that the most efficient strategy for reducing per capita global pathogen prevalence is to concentrate additional healthcare effort in particular countries based on the key predictors of pathogen prevalence, population size and current healthcare spending patterns. Assuming the relationship between pathogen prevalence and its key predictors (healthcare spending, pathogen richness, human population size, temperature and AET) modelled in table 2 and a modest 18 billion dollar additional global spending, representing just 1 per cent of the total private spending on healthcare per annum (WHO 2008), spending would be most effectively focused on regions with large populations (e.g. India and Pakistan) and also areas with almost non-existent spending on healthcare, such as Madagascar and much of eastern Africa (figure 1c). Our models predict that such a targeted effort would be able to reduce global pathogen prevalence by several per cent for over two billion people. However, these calculations assume monies are apportioned randomly among potential local strategies and programmes (which in the absence of global plans is not unrealistic). More focused efforts on easier to control pathogens, such as filariasis, which can be treated with a combination of two relatively cheap drugs (Ottesen et al. 2008), might yield even greater impacts on prevalence and ultimately human survival.
We may view ourselves as apart from nature, but our parasites and infectious diseases appear to have not noticed the shift in our worldview. Human pathogens remain subject to the same influences as the rest of biodiversity, despite our best efforts to influence them otherwise. Humans who live in regions with low biological diversity of pathogens can attribute that low diversity not to the success of public health, but rather to the migration of their ancestors to a climate that is relatively cool and dry, and where fewer alternative hosts and disease vectors exist. Except for cases in which we completely eradicate widespread diseases, we are unlikely to be able to change patterns of pathogen richness dramatically (except by changing patterns of climate). On the other hand, pathogen prevalence is much more sensitive to variation in health spending among regions. These two results together offer some hope in terms of controlling pathogens and their effects, both today and in a warmer future world. Pathogen richness, even when high, does not guarantee high prevalence, because of the potential impact of disease control effort. From the perspective of reducing global pathogen prevalence, we would argue that additional disease control effort would be best focused in those many regions where prevalence remains high, populations are large and healthcare spending per capita is relatively low. Because pathogens are not restricted by political boundaries and local epidemics can rapidly transform into global pandemics, reducing prevalence in one part of the world will also benefit other parts of the world. While it is clear that aspects of the distribution of pathogens have, in the past, affected all aspects of human life, the degree to which these pathogens will affect us in the future depends on how well we mediate their prevalence.
Acknowledgements
This work was funded by NASA award NNX09AK22G, DOE-PER and DOE-NICCR to R.R.D. We thank N. Haddad, K. Gross, A. Latimer, M. Sanchez, J. Solomon, B. Bartel and J. Morales for discussion. S. Altizer provided particularly insightful comments. L. Wolkovich helped with SEM. T.J.D. was supported in part as a postdoctoral associate at the National Center for Ecological Analysis and Synthesis, a center funded by a National Science Foundation grant and the University of California, Santa Barbara.
References
- Anderson D. R.2008Model based inference in the life sciences: a primer on evidence. CO, USA: Springer [Google Scholar]
- Anderson R. M., May R. M.1979Population biology of infectious diseases. I. Nature 280, 361–371 (doi:10.1038/280361a0) [DOI] [PubMed] [Google Scholar]
- Anderson R. M., May R. M.1991Infectious diseases of humans: dynamics and control. Oxford, UK: Oxford University Press [Google Scholar]
- Araujo A., Ferreira L. F.2000Paleoparasitology and the antiquity of human host-parasite relationships. Mem. Inst. Oswaldo Cruz, Rio de Janeiro 95, 89–93 [DOI] [PubMed] [Google Scholar]
- Araujo A., Reinhard K. J., Ferreira L. F., Gardner S. L.2008Parasites as probes for prehistoric human migrations? Trends Parasitol. 24, 112–115 [DOI] [PubMed] [Google Scholar]
- Brown J. H., Gillooly J. F., Allen A. P., Savage V. M., West G. B.2004Toward a metabolic theory of ecology. Ecology 85, 1771–1789 (doi:10.1890/03-9000) [Google Scholar]
- Calvete C.2003Correlates of helminth community in the red-legged partridge (Alectoris rufa L.) in Spain. J. Parasitol. 89, 445–451 (doi:10.1645/0022-3395(2003)089[0445:COHCIT]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Cazelles B., Chavez M., McMichael A. J., Hales S.2005Nonstationary influence of El Nino on the synchronous dengue epidemics in Thailand. PLoS Med. 2, 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves L. F., Kaneko A., Taleo G., Pascual M., Wilson M. L.2008Malaria transmission pattern resilience to climatic variability is mediated by insecticide-treated nets. Malar. J. 7, 100 (doi:10.1186/1475-2875-7-100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell J. H.1971On the role of natural enemies in preventing competitive exclusion in some marine animals and in rain forest trees. In Dynamics of populations (eds den Boer P. J., Gradwell G. R.), pp. 298–312 Wageningen, The Netherlands: Centre of Agricultural Publishing and Documentation [Google Scholar]
- Costanza R., et al. 1997The value of the world's ecosystems and natural capital. Nature 387, 253–260 (doi:10.1038/387253a0) [Google Scholar]
- Crawley M. J.2002Statistical computing: an introduction to data analysis using S-plus. Chichester, UK: John Wiley & Sons Ltd [Google Scholar]
- Cumming G. S., Guegan J. F.2006Food webs and disease: is pathogen diversity limited by vector diversity? Ecohealth 3, 163–170 (doi:10.1007/s10393-006-0028-6) [Google Scholar]
- Davies R. G., et al. 2007Topography, energy and the global distribution of bird species richness. Proc. R. Soc. B 274, 1189–1197 (doi:10.1098/rspb.2006.0061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Quadros C. A.2002History and prospects for viral disease eradication. Med. Microbiol. Immunol. 191, 75–81 (doi:10.1007/S00430-002-0120-7) [DOI] [PubMed] [Google Scholar]
- Denevan W.1992The pristine myth: the landscape of the Americas in 1492. Ann. Am. Assoc. Geogr. 82, 369–385 (doi:10.1111/j.1467-8306.1992.tb01965.x) [Google Scholar]
- Diniz Filho J. A. F., Bini L. M., Hawkins B. A.2003Spatial autocorrelation and red herrings in geographical ecology. Global Ecol. Biogeogr. 12, 53–64 (doi:10.1046/j.1466-822X.2003.00322.x) [Google Scholar]
- Dobzhansky T.1950Evolution in the tropics. Am. Sci. 38, 209 [Google Scholar]
- ESRI 2006ArcMap. Redlands, CA: ESRI [Google Scholar]
- Fincher C. L., Thornhill R.2008aAssortative sociality, limited dispersal, infectious disease and the genesis of the global pattern of religion diversity. Proc. R. Soc. B 275, 2587–2594 (doi:10.1098/rspb.2008.0688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher C. L., Thornhill R.2008bA parasite-driven wedge: infectious diseases may explain language and other biodiversity. Oikos 117, 1289–1297 (doi:10.1111/j.0030-1299.2008.16684.x) [Google Scholar]
- Fincher C. L., Thornhill R., Murray D. R., Schaller M.2008Pathogen prevalence predicts human cross-cultural variability in individualism/collectivism. Proc. R. Soc. B 275, 1279–1285 (doi:10.1098/rspb.2008.0094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould E. A., Higgs S.2009Impact of climate change and other factors on emerging arbovirus diseases. Trans. R. Soc. Trop. Med. Hyg. 103, 109–121 (doi:10.1016/j.trstmh.2008.07.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guegan J. F., Thomas F., Hochberg M. E., de Meeus T., Renaud F.2001Disease diversity and human fertility. Evolution 55, 1308–1314 [DOI] [PubMed] [Google Scholar]
- Guernier V., Hochberg M. E., Guegan J. F. O.2004Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2, 740–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins B. A., Pausas J. G.2004Does plant richness influence animal richness? The mammals of Catalonia (NE Spain). Divers. Distrib. 10, 247–252 (doi:10.1111/j.1366-9516.2004.00085.x) [Google Scholar]
- Hawkins B. A., et al. 2003Energy, water, and broad-scale geographic patterns of species richness. Ecology 84, 3105–3117 (doi:10.1890/03-8006) [Google Scholar]
- Hay S. I., Guerra C. A., Tatem A. J., Noor A. M., Snow R. W.2004The global distribution and population at risk of malaria: past, present, and future. Lancet Infect. Dis. 4, 327–336 (doi:10.1016/S1473-3099(04)01043-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechinger R. F., Lafferty K. D.2005Host diversity begets parasite diversity: bird final hosts and trematodes in snail intermediate hosts. Proc. R. Soc. B 272, 1059–1066 (doi:10.1098/rspb.2005.3070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans R. J., Cameron S., Parra J.2004WorldClim, version 1.2. A square kilometer resolution database of global terrestrial surface climate. Available at: http://biogeo.berkeley.edu
- Janzen D. H.1970Herbivores and the number of tree species in tropical forests. Am. Nat. 104, 501–528 [Google Scholar]
- Jetz W., Kreft H., Ceballos G., Mutke J.2009Global associations between terrestrial producer and vertebrate consumer diversity. Proc. R. Soc. B 276, 269–278 (doi:10.1098/rspb.2008.1005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. E., Patel N. G., Levy M. A., Storeygard A., Balk D., Gittleman J. L., Daszak P.2008Global trends in emerging infectious diseases. Nature 451, 990–994 (doi:10.1038/nature06536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline R. B.1998Principles and practice of structural equation modeling. New York, NY: Guilford Press [Google Scholar]
- Lafferty K. D.2009aCalling for an ecological approach to studying climate change and infectious diseases. Ecology 90, 932–933 (doi:10.1890/08-1767.1) [DOI] [PubMed] [Google Scholar]
- Lafferty K. D.2009bThe ecology of climate change and infectious diseases. Ecology 90, 888–900 (doi:10.1890/08-0079.1) [DOI] [PubMed] [Google Scholar]
- Lindenfors P., Nunn C. L., Jones K. E., Cunningham A. A., Sechrest W., Gittleman J. L.2007Parasite species richness in carnivores: effects of host body mass, latitude, geographical range and population density. Global Ecol. Biogeogr. 16, 496–509 (doi:10.1111/j.1466-8238.2006.00301.x) [Google Scholar]
- MacArthur R. H., Wilson E. O.1967The theory of island biogeography. Princeton, NJ: Princeton University Press [Google Scholar]
- McMichael A. J., Woodruff R. E., Hales S.2006Climate change and human health: present and future risks. Lancet 367, 859–869 (doi:10.1016/S0140-6736(06)68079-3) [DOI] [PubMed] [Google Scholar]
- Moodley Y., et al. 2009The peopling of the Pacific from a bacterial perspective. Science 323, 527–530 (doi:10.1126/science.1166083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D. M., Folkers G. K., Fauci A. S.2004The challenge of emerging and re-emerging infectious diseases. Nature 430, 242–249 (doi:10.1038/nature02759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Q., Heinsch F. A., Zhao M., Running S. W.2007Development of a global evapotranspiration algorithm based on MODIS and global meteorology data. Remote Sens. Environ. 111, 519–536 (doi:10.1016/j.rse.2007.04.015) [Google Scholar]
- Mundry R., Nunn C. L.2009Stepwise model fitting and statistical inference: turning noise into signal pollution. Am. Nat. 173, 119–123 [DOI] [PubMed] [Google Scholar]
- Nettle D.2009Ecological influences on human behavioural diversity: a review of recent findings. Trends Ecol. Evol. 24, 618–624 (doi:10.1016/j.tree.2009.05.013) [DOI] [PubMed] [Google Scholar]
- Nunn C. L., Altizer S., Jones K. E., Sechrest W.2003Comparative tests of parasite species richness in primates. Am. Nat. 162, 597–614 [DOI] [PubMed] [Google Scholar]
- Nunn C. L., Altizer S., Sechrest W., Jones K. E., Barton R. A., Gittleman J. L.2004Parasites and the evolutionary diversification of primate clades. Am. Nat. 164, S90–S103 [DOI] [PubMed] [Google Scholar]
- Nunn C. L., Altizer S. M., Sechrest W., Cunningham A. A.2005Latitudinal gradients of parasite species richness in primates. Divers. Distrib. 11, 249–256 (doi:10.1111/j.1366-9516.2005.00160.x) [Google Scholar]
- Ostfeld R. S.2009Climate change and the distribution and intensity of infectious diseases. Ecology 90, 903–905 (doi:10.1890/08-0659.1) [DOI] [PubMed] [Google Scholar]
- Ottesen E. A., Hooper P. J., Bradley M., Biswas G.2008The global programme to eliminate lymphatic filariasis: health impact after 8 years. PLoS Neglected Trop. Dis. 2, e317 (doi:10.1371/journal.pntd.0000317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Schaller M.2009Parasites, minds and cultures. Psychologist 22, 942–945 [Google Scholar]
- Pascual M., Ahumada J. A., Chaves L. F., Rodo X., Bouma M.2006Malaria resurgence in the East African highlands: temperature trends revisited. Proc. Natl Acad. Sci. USA 103, 5829–5834 (doi:10.1073/pnas.0508929103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B. D., Ceballos G., Sechrest W., Tognelli M. F., Brooks T., Luna L., Ortega P., Salazar I., Young B. E.2007Digital distribution maps of the mammals of the Western Hemisphere, version 3.0. Arlington, VI: NatureServe [Google Scholar]
- Pedersen A. B., Davies T. J.In press Cross species pathogen transmission and disease emergence in primates. EcoHealth. (doi:10.1007/s10393-010-0284-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. L.2008Dengue reborn: widespread resurgence of a resilient vector. Environ. Health Perspect. 116, A382–A388 (doi:10.1289/ehp.116-a382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston B. T., Capellini I., McNamara P., Barton R. A., Nunn C. L.2009Parasite resistance and the adaptive significance of sleep. BMC Evol. Biol. 9, 7 (doi:10.1186/1471-2148-9-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter P.2001Climate change and mosquito-borne disease. Environ. Health Perspect. 109, 141–161 (doi:10.2307/3434853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K.1992Latitudinal gradients in species diversity: the search for the primary cause. Oikos 65, 514–527 (doi:10.2307/3545569) [Google Scholar]
- Rosenzweig M. L.1968Net primary productivity of terrestrial environments: predictions from climatological data. Am. Nat. 102, 67–74 (doi:10.1086/282523) [Google Scholar]
- Rosenzweig M. L.1992Species diversity gradients: we know more and less than we thought. J. Mammal. 73, 715–730 (doi:10.2307/1382191) [Google Scholar]
- Rosenzweig M. L., Sandlin E. A.1997Species diversity and latitudes: listening to area's signal. Oikos 80, 172–176 (doi:10.2307/3546528) [Google Scholar]
- Semenza J. C., Menne B.2009Climate change and infectious diseases in Europe. Lancet Infect. Dis. 9, 365–375 (doi:10.1016/S1473-3099(09)70104-5) [DOI] [PubMed] [Google Scholar]
- Senior K.2008Climate change and infectious disease: a dangerous liaison? Lancet Infect. Dis. 8, 92–93 (doi:10.1016/S1473-3099(08)70008-2) [DOI] [PubMed] [Google Scholar]
- Smith K. F., Sax D. F., Gaines S. D., Guernier V., Guegan J. F.2007Globalization of human infectious disease. Ecology 88, 1903–1910 (doi:10.1890/06-1052.1) [DOI] [PubMed] [Google Scholar]
- Snall T., Benestad R. E., Stenseth N. C.2009Expected future plague levels in a wildlife host under different scenarios of climate change. Global Change Biol. 15, 500–507 (doi:10.1111/j.1365-2486.2008.01725.x) [Google Scholar]
- Thornhill R., Fincher C. L., Aran D.2009Parasites, democratization, and the liberalization of values across contemporary countries. Biol. Rev. 84, 113–131 (doi:10.1111/j.1469-185X.2008.00062.x) [DOI] [PubMed] [Google Scholar]
- Whittingham M. J., Stephens P. A., Bradbury R. B., Freckleton R. P.2006Why do we still use stepwise modelling in ecology and behaviour? J. Anim. Ecol. 75, 1182–1189 (doi:10.1111/j.1365-2656.2006.01141.x) [DOI] [PubMed] [Google Scholar]
- WHO 2004World health report 2004. Geneva, Switzerland: World Health Organization [Google Scholar]
- WHO 2008World health statistics 2008. Geneva, Switzerland: World Health Organization; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe N. D., Dunavan C. P., Diamond J.2007Origins of major human infectious diseases. Nature 447, 279–283 (doi:10.1038/nature05775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. H.1983Species-energy theory: an extension of species-area theory. Oikos 41, 496 (doi:10.2307/3544109) [Google Scholar]
- Zhu S.2009Psychosis may be associated with toxoplasmosis. Med. Hypotheses 73, 799–801 (doi:10.1016/j.mehy.2009.04.013) [DOI] [PubMed] [Google Scholar]