Abstract
Hatching failure is a surprisingly common phenomenon given that natural selection constantly works against it. In birds, an average of about 10 per cent of eggs across species fail to hatch, often owing to the death of embryos. While embryo mortality owing to inbreeding is both well-documented and evolutionarily plausible, this is not true for other sources of mortality. In fact, the basis for hatching failure in natural populations remains largely unexplained. Here, we demonstrate that embryo mortality in captive zebra finches (Taeniopygia guttata) follows from chromosomal aneuploidy or polyploidy. As part of microsatellite genotyping of a captive breeding population, we found 12 individuals (3.6%) with three alleles among 331 embryos that had died during development, while there were no such cases observed among 1210 adult birds. Subsequent genotyping of 1920 single nucleotide polymorphism markers distributed across the genome in birds with three alleles at microsatellite loci, and in greater than 1000 normal birds, revealed that the aberrant karyotypes involved cases of both trisomies and triploidy. Cases of both maternally and paternally inherited trisomies resulted from non-disjunction during meiosis. Maternally inherited cases of triploidy were attributable to failure of meiosis leading to diploid eggs, while paternally inherited triploidy could have arisen either from diploid sperm or from dispermy. Our initial microsatellite screening set only had the power to detect less than 10 per cent of trisomies and by extrapolation, our data therefore tentatively suggest that trisomy might be a major cause of embryo mortality in zebra finches.
Keywords: chromosomal anomalities, diploid sperm, meiotic error, non-disjunction, polyploidy, polyspermy
1. Introduction
Across a wide range of animal taxa, about 15 per cent of the eggs produced fail to hatch (Koenig 1982; Anderson 1990; Eberhard 1996; Morrow et al. 2002; Briskie & Mackintosh 2004; Spottiswoode & Møller 2004) either owing to eggs not being fertilized or owing to embryo mortality (Birkhead et al. 2008). While some of this mortality may be for environmental reasons (e.g. temperature, humidity, infection, pesticides), genetic factors may also play a major role. In humans, it is estimated that more than 20 per cent of all conceptions are aneuploid, i.e. they carry an abnormal number of some of the chromosomes (McFadden & Friedman 1997; Hassold & Hunt 2001), and it is only because of the selective abortion of aneuploid embryos that just a very small proportion of babies born alive suffer from trisomy (three copies of one chromosome; three copies of all chromosomes is referred to as triploidy). This high rate of aneuploidy in humans has not been found in any other animal species, though few studies have had the power to detect such aberrations with some accuracy. Cytological studies on commercial breeds of chicken have found modest rates of haploidy (1.5% of embryos), polyploidy (1.0%) and aneuploidy (0.4%) (Bloom 1972; Fechheimer 1981; Thorne et al. 1990), and have suggested that this may cause embryo mortality (Hailu et al. 1995). Visual inspection of chromosome spreads, however, might grossly underestimate the frequency of aneuploidy, since only the largest chromosomes can be identified with confidence.
To our knowledge, no systematic molecular genetic study of chromosomal aberrations has been carried out on birds. Here, we study the incidence and character of chromosomal abnormalities in a captive population of zebra finches (Taeniopygia guttata) using microsatellite and large-scale single nucleotide polymorphism (SNP) genotyping. In this breeding population held in aviaries, 8 per cent of eggs do not develop at all, and 27 per cent die during embryo development. This is somewhat higher than the 17 per cent of hatching failure found in a wild population of this species (Zann 1996) and higher than the average of 11.5 per cent found across 122 species of birds (median 9.2%, maximum 55%, data taken from Morrow et al. (2002) and Spottiswoode & Møller (2004)). To screen for potential chromosomal aberrations, we first performed microsatellite genotyping of 1742 zebra finches including adult birds, their offspring, as well as dead embryos. We then focused on those cases where we found individuals carrying three different alleles at one or more marker loci, using a genome-wide panel of nearly 2000 SNP markers providing high resolution. As a reference, more than 1000 normal birds were also genotyped with this large set of SNP markers.
2. Material and methods
(a). Subjects
Zebra finches were bred in aviaries at the Max Planck Institute for Ornithology, Seewiesen in Germany and originated from a captive population at Sheffield University (UK) (Forstmeier et al. 2007b). A total of 104 males and 108 females were bred for three to six months in 15 aviaries (volume: 6 m3) containing 12–15 birds (sex-ratio: 0.4–0.6). These produced 1194 eggs from which we report natural rates of non-development and embryo mortality, and another 857 developing embryos that were culled after 4 days of artificial incubation for DNA analysis of whole-embryo tissue. Parents as well as offspring were subject to marker genotyping.
(b). Microsatellite genotyping
DNA was prepared from embryos and adults, and used for microsatellite genotyping with 10 species-specific hypervariable markers (Forstmeier et al. 2007a). Two markers (Tgu10, Tgu12) are located on the zebra finch chromosome 1, one (Tgu5) on chromosome 1A, two (Tgu8, Tgu9) on chromosome 2, three (Tgu1, Tgu6, and Tgu7) on chromosome 5, one (Tgu4) on chromosome 6 and one (Tgu3) on chromosome 9. Details on the alleles found in our population are presented in electronic supplementary material, table S1. All allele calls were verified by checking for Mendelian inheritance, and genotyping errors were corrected accordingly. Out of 2594 genotyped individuals, we initially found 19 that showed three alleles at one (n = 10), two (n = 6), three (n = 1) or five (n = 2) loci. Each of these cases was reanalysed twice starting from tissue or blood samples. One case (an adult bird with one seemingly three-allelic locus) proved to be a genotyping error, while the other 18 cases were confirmed.
(c). Detection probabilities
To quantify the power of detecting three alleles at a marker locus, we randomly drew 10 000 times three alleles from the population's pool of alleles at every marker locus and checked for non-identity of all the three alleles. For chromosomes with several markers, we assumed no linkage disequilibrium between those markers. Hence, the probability of non-detection of trisomy for a chromosome was estimated as the product of the non-detection probabilities of all marker loci on that chromosome.
(d). Interpretation of SNP genotype data
SNP markers were identified from transcriptome sequencing using Roche 454 Life Sciences massive parallel sequencing technology of pools of unrelated zebra finches (Warren et al. 2010), as described in Backström et al. (2010). They were mapped by BLAST analysis onto the zebra finch genome (Warren et al. 2010) and markers were selected to cover all assembled chromosomes with an approximately even density. SNP genotyping was performed using the Illumina GoldenGate assay on an Illumina BeadStation 500GX, at the Uppsala University SNP Technology Platform (http://www.medsci.uu.se/molmed/snpgenotyping/index.htm). See Backström et al. (2010) for a description of SNP validation and a quality criteria applied. Triploid or trisomic birds were identified from markers showing normalized fluorescence signal ‘ratios’ of the two alleles at approximately 0.33 and 0.67; normal heterozygous diploids show ratios at approximately 0.50 (electronic supplementary material, figure S1). As there is some overlap in the tails of the ratio distributions for AA homozygotes versus birds with the allelic combination AAB (and for BB homozygotes versus birds with ABB), and for AAB versus ABB birds (electronic supplementary material, figure S1), we inferred individuals to have two copies of the A allele and one copy of the B allele when the ratio was 0.25–0.45, and two copies of the B allele and one copy of the A allele when the ratio was 0.65–0.85. Given the overlap in allelic signal ratio distributions between diploid heterozygotes and individuals with two copies of one allele and one of the other, we cannot firmly assign trisomy when the number of non-homozygous markers on a chromosome is limited. The number of markers decreased with chromosome size and 10 of the smallest chromosomes had 10 or fewer markers. In addition, the assembled genome sequence does not include several microchromosomes and hence no markers are available from these.
We inferred the parental origin of extra chromosomes by focusing on markers where parents were homozygous for different alleles (i.e. AA and BB, respectively). For example, if a father was scored as CC, a mother as TT and an offspring as CCT, the extra chromosome was taken to have a paternal origin. Having in this way established which parent contributed an extra chromosome, we sought to determine whether this was in the form of that parent contributing two copies of the same chromosome or one copy each of its two different chromosomes.
3. Results and discussion
Single-locus microsatellites are expected to show a maximum of two alleles, however, we found several birds where one or more markers displayed three length variants. These cases were highly non-randomly distributed among the different categories of birds: none were found among 1210 adults, while 12 (3.6%) were found among 331 embryos that had died during development (Fisher's exact test, p < 0.0001). This over-representation of birds with three alleles among dead embryos clearly suggests that such deviant genotypes are lethal. To provide an independent assessment of the incidence of deviant genotypes, we analysed 857 embryos culled after 4 days of artificial incubation. We found 5 (0.6%) birds showing one or more loci with three alleles, which is significantly more frequent than in adult birds (Fisher's exact test, p = 0.012). Out of 214 chicks that died between hatching and fledging, one individual (0.5%) showed three alleles (individual 390_06 in electronic supplementary material, table S2).
Microsatellite loci with three alleles are suggestive of chromosomal abnormality, although in theory this could potentially also arise from methodological artefacts or, for example, gene conversion or locus-specific duplication. In all of the 18 cases (electronic supplementary material, table S1), the additional alleles could be traced back to at least two generations up the breeding pedigree, rendering most methodological artefacts unlikely.
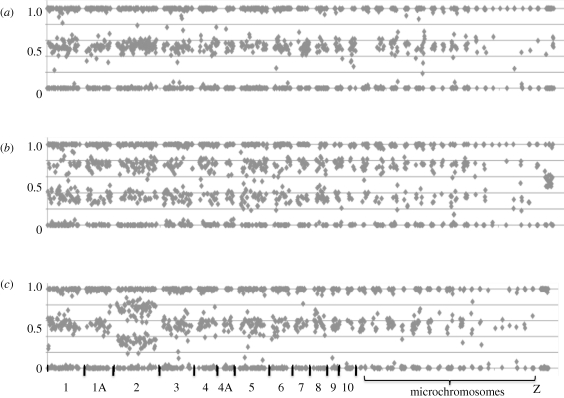
To reveal the genetic basis for these anomalies, we subjected the 18 birds showing deviant microsatellite genotypes to genotyping with a panel of 1920 di-allelic SNPs more or less evenly distributed across the zebra finch genome. The results from this effort are illustrative, as exemplified in figure 1. Normal individuals (more than 1000 individuals) show allele calls with either approximately equal signals from the two alleles (heterozygotes) or essentially only a signal from one of the two alleles (the two different homozygotes) (figure 1a). By contrast, birds which initially showed microsatellite loci with three alleles have, in addition to homozygous signals, numerous SNP allele calls across whole chromosomes where the ratio of the two alleles' signal intensities is approximately one-third or two-thirds, confirming that three copies of chromosomes are present. Eight birds show this pattern for a single or a limited number of chromosomes, with the remaining chromosomes showing the normally expected pattern of either equal signals from the two alleles or just a signal from one allele, which we interpret to represent cases of trisomies (figure 1c). Another eight birds show the pattern for all autosomes and we consider these to be triploid birds (figure 1b). For one bird we found scattered markers on different chromosomes with a pattern consistent with three chromosomal copies, however, as there also were heterozygous markers corresponding to normal diploidy within the same chromosomes, this may potentially represent a case of trisomy mosaicism (Tucker et al. 2007); we do not consider this individual further. For an additional bird we did not have sufficient amount of DNA for SNP genotyping.
Figure 1.
Normalized (from 0 to 1) allele call signal ratios of SNP markers across the zebra finch genome. Markers are sorted by chromosome and position within each chromosome, starting with chromosome 1 to the left and then in descending order according to chromosome number. Individual chromosomes are separated by a gap without marker data. (a) Normal adult individual; (b) triploid embryo; (c) trisomic embryo (three copies of chromosome 2).
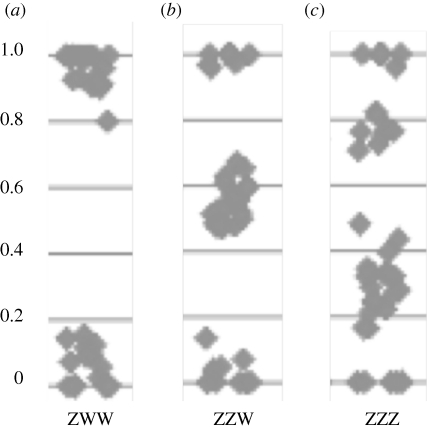
Our SNP array does not include markers on the female-specific W-chromosome as the incidence of polymorphism on the avian W chromosome is extremely low (Berlin & Ellegren 2004). However, among the eight apparently triploid birds, three showed a pattern consistent with having three copies also of the Z-chromosome (figure 2), suggesting an aberrant karyotype for sex chromosomes as well. The remaining triploids had either two or one copy of the Z-chromosome, presumably representing ZZW and ZWW birds, respectively.
Figure 2.
Allele call signal ratios from Z-linked markers on inferred triploid zebra finch embryos. (a) Shows a bird that is homozygous for all Z-linked loci; (b) shows a bird that is homozygous for some and heterozygous for other Z-linked markers; and (c) shows a bird with three copies of the Z-chromosome (ZZZ). Assuming that all birds carry three sex chromosomes, the inferred karyotypes of panels (a,c) birds are Z(WW) and ZZ(W), respectively (only indirect evidence for the presence of W chromosomes).
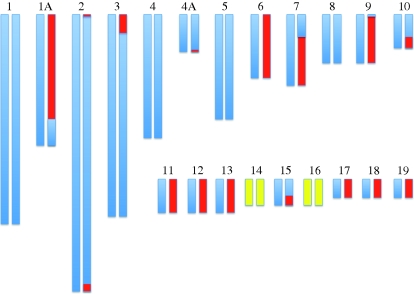
To address the mechanistic basis for the presence of three chromosome copies, we sought to identify the parental and allelic origin of extra chromosomes. The former was straightforwardly assessed by considering all markers that had the parental genotypes AA (one parent) and BB (the other parent); triploid offspring from these parents consistently either showed AAB or ABB genotypes, revealing the origin of the extra chromosome. Among the eight triploid offspring, there were four cases of a paternal origin and four cases of a maternal origin of extra alleles. For trisomies, the corresponding numbers were two and six, showing that both forms of aberration arise in spermatogenesis (or by polyspermy, see below) as well as in oogenesis. By inferring the allelic contribution from the parent transmitting extra chromosomes, we then asked if the two chromosomes contributed by this parent were two copies of the same chromosome or one copy each of the two different chromosomes. For each triploid individual some chromosomes were represented by two identical copies, some by two different copies and some constituted a combination of identical and different copies (figure 3). The latter could arise from recombination, and the pattern of genetic exchange between parental chromosomes being strongly biased towards chromosome ends is entirely consistent with the highly elevated rate of recombination towards telomeres in the zebra finch genome (Stapley et al. 2008; Backström et al. 2010). Crossing-over events take place in prophase I of meiosis, so this would suggest that extra chromosomes are the result of subsequent non-disjunction in either the first or the second meiotic cell division, resulting in diploid gametes (electronic supplementary material, figure S1). For the four cases of maternally inherited triploidy, we can differentiate between non-disjunction in the first versus second meiotic division based on sex chromosome patterns (electronic supplementary material, table S1). While the two cases of ZZ(W) point to the first meiotic division (since both the maternal Z and the (only indirectly inferred) maternal W are represented), the Z(WW) (118_05) and ZZZ (550_06) cases must have originated from the second meiotic division. We finally note that since the number of cases for which we could infer the mechanism of chromosomal aberration was low, it is difficult to draw firm conclusions on the generality of these observations.
Figure 3.
Example of the inferred allelic origin of a diploid gamete (individual 118_05 in electronic supplementary material, table S2), resulting in a triploid zebra finch offspring. Blue and red indicate the two different parental chromosomal copies, respectively, where combinations of the two colours suggest recombination events. Chromosomes without markers are in yellow. Note that recombination events in the ends of chromosomes, where the majority of recombination occurs in the zebra finch may have remained undetected owing to a lack of terminal markers.
While it is difficult to see an alternative and plausible explanation for triploids with extra chromosomes of maternal origin, paternally originating triploidy could potentially arise as a consequence of a single egg being fertilized by two sperm. Polyspermy, in which more than one sperm enters the ova, occurs frequently in birds (Okzmura & Nishiyama 1978) and could thus potentially result in dispermic fertilization. We cannot exclude that some offspring with extra chromosomes of paternal origin have arisen from dispermy rather than from fertilization by diploid sperm. Like among triploids, trisomic offspring showed either two copies of the same chromosome, one copy each of the two different chromosomes, or what looks like a product of recombination.
Five out of the seven clearly trisomic birds showed three copies of chromosome 2 (one of these birds also had an extra copy of chromosome 3). Clearly, the detection of trisomies is defined by the genomic distribution and level of allelic polymorphism of the set of 10 microsatellites used in the initial screening of embryos (located on six chromosomes, namely 1, 1A, 2, 5, 6 and 9). However, observed counts of the six different types of trisomies (1, 0, 5, 0, 0, 1) deviated significantly from expected counts (see §2c), reflecting the marker-based power to detect trisomies (1.36, 1.01, 1.40, 1.59, 0.82, 0.82; goodness-of-fit test, exact p = 0.022). One possible explanation for this observation is that to the extent trisomies for other chromosomes do arise, their lethal effect on developing embryos manifests at very early stages of zygote development, i.e. prior to our sampling. This would bear some resemblance to the situation in humans, where trisomy-21 dominates heavily among subjects born with an extra autosomal copy, clearly because most other trisomies are lethal (rare cases of trisomy-13 and trisomy-18 are seen among live-born). Yet, with only 8 per cent of the incubated eggs in our population not developing, there is limited room for such early deaths. It could thus be speculated that the incidence of trisomy is higher for chromosome 2 than for other zebra finch chromosomes. In humans, such heterogeneity in the incidence of different types of trisomies exists, with trisomy-16 being the most common form (usually lethal) (Hassold & Jacobs 1984).
While seven different fathers were involved in the seven cases of paternally originating triploidy/trisomy, there were three mothers producing two maternally triploid/trisomic offspring each, and five mothers producing one each. To test how often such repeated incidences would be expected by chance, we randomly picked 18 parents (7 fathers and 11 mothers) from the 1965 embryos or offspring that we had genotyped. We found that in only 8.9 per cent out of 10 000 simulations, there were fewer than 16 different parents involved. This suggests that there is some individual repeatability for producing triploid/trisomic offspring. Moreover, the 15 involved parents originated from only 11 different families (two pairs of sisters, a brother–sister pair and a father–son pair). Again, random sampling showed that in only 7.4 per cent out of 10 000 simulations, less than 12 different families are expected to be involved. This suggests some hereditary component to producing triploid/trisomic offspring, as has been found repeatedly (Bloom 1972; Fechheimer 1981; Zwick et al. 1999).
The background to producing triploid or trisomic offspring might be: (i) environmental; (ii) non-additive genetic; (iii) additive genetic; or (iv) age-related effects. Despite extensive research effort on humans, no environmental factors for producing trisomic offspring have been identified (McFadden & Friedman 1997; Hassold & Hunt 2001). While inbreeding is known to reduce hatching success (Briskie & Mackintosh 2004; Spottiswoode & Møller 2004) probably owing to the expression of recessive deleterious mutations (non-additive effect), inbreeding does not seem to increase the risk of producing trisomic offspring in humans (MacCluer 1980; Zlotogora 1997; Sayee & Thomas 1998). In our zebra finch population, inbreeding is relatively modest (Forstmeier et al. 2004, 2007b), and the 15 individuals that produced triploid or trisomic offspring were only non-significantly less heterozygous at microsatellite (two-samples t-test for equal variances, t197 = 1.75, p = 0.082) or SNP markers (t197 = 0.46, p = 0.65) than the 184 simultaneously breeding parents that apparently produced only diploid offspring. Essentially, all parents were of the same age. The reasons for the overall high rate of embryo mortality in our population (27%) remain uncertain. With an estimated genetic load of only 0.4 lethal mutations per haploid genome in this population (Bolund et al. 2010), inbreeding depression owing to recessive lethal alleles seems to be only a minor cause of embryo mortality. As we show, chromosomal anomalities contribute to the high rate of hatching failure, but do not fully explain it. Like for any captive population, suboptimal environmental conditions during breeding may be another factor to bear in mind.
Our results show that chromosomal abnormalities resulting from non-disjunction or failure of meiosis are a source of embryo mortality in the zebra finch, at least in captivity. The frequency of detected triploidy or trisomy among dead embryos was 3.6 per cent but the actual proportion of abnormalities may be much higher, in particular with regard to trisomy. First, power calculations indicate that, on average, the microsatellites used in this study would only detect 66 per cent of cases of three different alleles, a consequence of the finite number of allelic states of the markers. Second, our SNP analysis showed that trisomic chromosomes include regions with two copies of the same parental allele. Such cases would remain undetected in qualitative (rather than quantitative) microsatellite analysis. Third, and most importantly, our microsatellite markers covered only 6 out of the 40 chromosomes of the zebra finch. Hence, if the remaining 34 pairs of chromosomes showed similarly high rates of trisomy as the six chromosomes we covered, it may well be that trisomy is responsible for about a fourth of the embryo deaths, which would probably be the highest rate known among non-human species. However, such extrapolation may be invalid given the heterogeneity in rates of trisomy seen among the six chromosomes covered. Moreover, it could potentially be that trisomy for any of the small microchromosomes will have less severe consequences. Hence, large-scale screens of dead embryos with the SNP array, or with other markers, would be required for a more precise estimate of the overall rate of trisomy.
The relatively frequent occurrence of trisomy is puzzling for evolutionary biologists, since any additive genetic variance in propensity to produce such offspring should be constantly eroded by natural selection. It was therefore suggested (Axelrod & Hamilton 1981; Day & Taylor 1998) that trisomy might represent the unfortunate outcome of the conflict between the maternal and paternal chromosomes during oogenesis over migration to the oocyte versus the polar body (chromosome drive theory). As this conflict is absent in spermatogenesis, our finding of paternally inherited trisomy would in this case have to be explained as a non-adaptive by-product of a strong chromosome drive evolved within females. Also, it may very well be that there are multiple non-mutually excluding explanations for trisomy.
Acknowledgements
The study was approved by the animal care and ethics representative of the Max Planck Institute for Ornithology.
We thank Melanie Schneider for producing the microsatellite data, Harriet Mellenius for processing the SNP data, Niclas Backström and Holger Schielzeth for help with analyses, Elisabeth Bolund, Holger Schielzeth and Katrin Martin for the collection of samples, Edward Morrow for manuscript comments and Bart Kempenaers for support, discussion and manuscript comments. The SNP Technology Platform at Uppsala University is gratefully acknowledged. Financial support was obtained from the Swedish Research Council and the Max Planck Society.
References
- Anderson D. J.1990Evolution of obligate siblicide in boobies. 1. A test of the insurance-egg hypothesis. Am. Nat. 135, 334–350 (doi:10.1086/285049) [Google Scholar]
- Axelrod R., Hamilton W. D.1981The evolution of cooperation. Science 211, 1390–1396 (doi:10.1126/science.7466396) [DOI] [PubMed] [Google Scholar]
- Backström N., et al. 2010The recombinational landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 20, 485–495 (doi:10.1101/gr.101410.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S., Ellegren H.2004Chicken W: a genetically uniform chromosome in a highly variable genome. Proc. Natl Acad. Sci. USA 101, 15 967–15 969 (doi:10.1073/pnas.0405126101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkhead T. R., Hall J., Schut E., Hemmings N.2008Unhatched eggs: methods for discriminating between infertility and early embryo mortality. Ibis 150, 508–517 (doi:10.1111/j.1474-919X.2008.00813.x) [Google Scholar]
- Bloom S. E.1972Chromosome abnormalities in chicken (Gallus domesticus) embryos: types, frequencies and phenotypic effects. Chromosoma 37, 309 (doi:10.1007/BF00319873) [DOI] [PubMed] [Google Scholar]
- Bolund E., Martin K., Kempenaers B., Forstmeier W.2010Inbreeding depression of sexually selected traits and attractiveness in the zebra finch. Anim. Behav. 79, 947–955 (doi:10.1016/j.anbehav.2010.01.014) [Google Scholar]
- Briskie J. V., Mackintosh M.2004Hatching failure increases with severity of population bottlenecks in birds. Proc. Natl Acad. Sci. USA 101, 558–561 (doi:10.1073/pnas.0305103101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T., Taylor P. D.1998Chromosomal drive and the evolution of meiotic nondisjunction and trisomy in humans. Proc. Natl Acad. Sci. USA 95, 2361–2365 (doi:10.1073/pnas.95.5.2361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard W. G.1996Female control: sexual selection by cryptic female choice. Princeton, NJ: Princeton University Press [Google Scholar]
- Fechheimer N. S.1981Origins of heteroploidy in chicken embryos. Poultry Sci. 60, 1365–1371 [DOI] [PubMed] [Google Scholar]
- Forstmeier W., Coltman D. W., Birkhead T. R.2004Maternal effects influence the sexual behavior of sons and daughters in the zebra finch. Evolution 58, 2574–2583 (doi:10.1554/04-325) [DOI] [PubMed] [Google Scholar]
- Forstmeier W., Schielzeth H., Schneider M., Kempenaers B.2007aDevelopment of polymorphic microsatellite markers for the zebra finch (Taeniopygia guttata). Mol. Ecol. Notes 7, 1026–1028 (doi:10.1111/j.1471-8286.2007.01762.x) [Google Scholar]
- Forstmeier W., Segelbacher G., Mueller J. C., Kempenaers B.2007bGenetic variation and differentiation in captive and wild zebra finches (Taeniopygia guttata). Mol. Ecol. 16, 4039–4050 (doi:10.1111/j.1365-294X.2007.03444.x) [DOI] [PubMed] [Google Scholar]
- Hailu C., Wagner K. U., Saar W., Pingel H.1995Frequency of chromosome aberrations in association with embryonic mortality of hybrid ducks. In Proc. 10th Eur. Symp. on Waterfowl, 26–31 March 1995, pp. 304–308 Halle (Saale), Germany [Google Scholar]
- Hassold T., Hunt P.2001To ERR (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (doi:10.1038/35066065) [DOI] [PubMed] [Google Scholar]
- Hassold T. J., Jacobs P. A.1984Trisomy in man. Ann. Rev. Genet. 18, 69–97 (doi:10.1146/annurev.ge.18.120184.000441) [DOI] [PubMed] [Google Scholar]
- Koenig W. D.1982Ecological and social-factors affecting hatchability of eggs. Auk 99, 526–536 [Google Scholar]
- MacCluer J.1980Inbreeding and human fetal death. In Human embryonic and fetal death (eds Porter I. H., Hook E. B.), pp. 241–259 New York, NY: Academic Press [Google Scholar]
- McFadden D., Friedman J. M.1997Chromosome abnormalities in human beings. Mut. Res.-Fundam. Mol. Mech. Mutagen. 396, 129–140 (doi:10.1016/S0027-5107(97)00179-6) [DOI] [PubMed] [Google Scholar]
- Morrow E. H., Arnqvist G., Pitcher T. E.2002The evolution of infertility: does hatching rate in birds coevolve with female polyandry? J. Evol. Biol. 15, 702–709 (doi:10.1046/j.1420-9101.2002.00445.x) [Google Scholar]
- Okzmura F., Nishiyama H.1978The passage of spermatozoa through the vitelline membrane in the domestic fowl, Gallus gallus. Cell Tissue Res. 188, 497–508 (doi:10.1007/BF00219787) [DOI] [PubMed] [Google Scholar]
- Sayee R., Thomas I. M.1998Consanguinity, non-disjunction, parental age and Down's syndrome. J. Ind. Med. Assoc. 96, 335–337 [PubMed] [Google Scholar]
- Spottiswoode C., Møller A. P.2004Genetic similarity and hatching success in birds. Proc. R. Soc. Lond. B 271, 267–272 (doi:10.1098/rspb.2003.2605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J., Birkhead T. R., Burke T., Slate J.2008A linkage map of the zebra finch Taeniopygia guttata provides new insights into avian genome evolution. Genetics 179, 651–667 (doi:10.1534/genetics.107.086264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne M. H., Collins R. K., Sheldon B. L.1990Triploidy and other chromosomal abnormalities in a selected line of chickens. In 9th European Colloquium on Cytogenetics of Domestic Animals, pp. S212–S216 Toulouse, France: Editions Scientifiques Elsevier [Google Scholar]
- Tucker M. E., Garringer H. J., Weaver D. D.2007Phenotypic spectrum of mosaic trisomy 18: two new patients, a literature review, and counseling issues. Am. J. Med. Genet. Part A 143A, 505–517 (doi:10.1002/ajmg.a.31535) [DOI] [PubMed] [Google Scholar]
- Warren W. C., et al. 2010The genome of a songbird. Nature 464, 757–762 (doi:10.1038/nature08819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zann R.1996The zebra finch. New York, NY: Oxford University Press [Google Scholar]
- Zlotogora J.1997Genetic disorders among Palestinian Arabs. 1. Effects of consanguinity. Am. J. Med. Genet. 68, 472–475 (doi:10.1002/(SICI)1096-8628(19970211)68:4<472::AID-AJMG20>3.0.CO;2-O) [DOI] [PubMed] [Google Scholar]
- Zwick M. E., Salstrom J. L., Langley C. H.1999Genetic variation in rates of nondisjunction: association of two naturally occurring polymorphisms in the chromokinesin nod with increased rates of nondisjunction in Drosophila melanogaster. Genetics 152, 1605–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]