Abstract
A pathogen can readily mutate to infect new host types, but this does not guarantee successful establishment in the new habitat. What factors, then, dictate emergence success? One possibility is that the pathogen population cannot sustain itself on the new host type (i.e. host is a sink), but migration from a source population allows adaptive sustainability and eventual emergence by delivering beneficial mutations sampled from the source's standing genetic variation. This idea is relevant regardless of whether the sink host is truly novel (host shift) or whether the sink is an existing or related, similar host population thriving under conditions unfavourable to pathogen persistence (range expansion). We predicted that sink adaptation should occur faster under range expansion than during a host shift owing to the effects of source genetic variation on pathogen adaptability in the sink. Under range expansion, source migration should benefit emergence in the sink because selection acting on source and sink populations is likely to be congruent. By contrast, during host shifts, source migration is likely to disrupt emergence in the sink owing to uncorrelated selection or performance tradeoffs across host types. We tested this hypothesis by evolving bacteriophage populations on novel host bacteria under sink conditions, while manipulating emergence via host shift versus range expansion. Controls examined sink adaptation when unevolved founding genotypes served as migrants. As predicted, adaptability was fastest under range expansion, and controls did not adapt. Large, similar and similarly timed increases in fitness were observed in the host-shift populations, despite declines in mean fitness of immigrants through time. These results suggest that source populations are the origin of mutations that drive adaptive emergence at the edge of a pathogen's ecological or geographical range.
Keywords: emergence, experimental evolution, migration, host shift, Pseudomonas, virus
1. Introduction
Experimental evolution studies with pathogens show that mutations conferring the ability to infect novel hosts arise frequently (Crill et al. 2000; Duffy et al. 2006, 2007; Ferris et al. 2007). However, despite some dramatic examples (Plowright et al. 2008), successful pathogen emergence in nature seems to occur rarely. One plausible hypothesis is most emerging pathogens are initially maladapted to novel hosts and fail to attain positive population growth through constraints either on within-host reproduction or on between-host transmission or both (Dennehy et al. 2006; Dennehy 2009). If sufficiently strong, this initial maladaptation lowers the likelihood that the pathogen will persist on the novel host long enough to improve its fitness through the incorporation of additional mutations (Kirkpatrick & Barton 1997; Ronce & Kirkpatrick 2001; Holt & Hochberg 2002; Holt et al. 2004). However, an emerging pathogen population in a novel host may be maintained by migration, setting up the familiar source–sink paradigm of ecology. That is, a new host environment where a pathogen cannot sustain itself by in situ infection but nonetheless persists because of spillover infection from its initial host (the ‘source’) constitutes a demographic ‘sink’ for the pathogen (Antia et al. 2003; Sokurenko et al. 2006; Chattopadhyay et al. 2007). In such sinks, persistence and prevalence necessarily reflect the rate and temporal pattern of recurrent migration from source populations (Holt et al. 2004). As a concrete example, it has been suggested that human influenza A viruses in temperate regions exist as sink populations, seeded by persistent immigration from a source population of viruses in the tropics (Rambaut et al. 2008; Russell et al. 2008).
There is a growing appreciation of the diverse, and at times contradictory, impacts that migration can have upon local adaptation (Lenormand 2002; Holt et al. 2005). Early models suggested that gene flow constrains local adaptation to sinks (Antonovics 1976; Kirkpatrick & Barton 1997; Lenormand 2002). A conspicuous example occurs when selection pressures diverge among pathogen populations. Here, alleles beneficial or neutral in sources may be deleterious in sinks. The movement into sinks of immigrant individuals bearing such alleles may impact resident fitness in at least two ways. First, migration increases population size, potentially increasing competition for resources and thus lowering the fitness of adapted alleles (Gomulkiewicz et al. 1999). Second, mating between migrants and residents in sexual species can result in outbreeding depression if migrants are maladapted to local conditions. One quantitative genetic model of range evolution attributes range limitation along geographical gradients to outbreeding depression in peripheral populations (Kirkpatrick & Barton 1997).
However, the converse is also true. Because alleles deleterious or neutral in sources may be beneficial in sinks, migration may at times facilitate adaptation in sink environments by providing a source for adaptive genetic variation. This possibility may be particularly crucial in the early stages of pathogen emergence when sink population sizes are very low, de novo mutations are thus rare and residents in any case, on average, are no better adapted than are migrants (so minimizing the impact of migrational load). Analytical studies (Gomulkiewicz et al. 1999) and individual-based simulations (Holt et al. 2003a) demonstrate that, in such circumstances, mutation in the sink population itself has relatively little effect on the rate of sink adaptation, making a strong case for the importance of migration from sources as a conduit of genetic variation in the emergence process (e.g. in harsh sinks, population size is very low, providing little opportunity for a local mutational origin of variation).
Results from recent experimental evolution studies suggest that the effect of migration on adaptive evolution depends on the nature of the selective landscape. For instance, adaptation was positively associated with migration when Pseudomonas aeruginosa populations were evolved under antibiotic selection, particularly in harsh sinks containing multiple antibiotics (Perron et al. 2007). Similarly, when vesicular stomatitis virus (VSV) migration occurred between flasks containing a constant host-cell type grown in tissue culture, adaptation was positively correlated with migration rate (Miralles et al. 1999). However, when VSV was permitted to evolve with migration on heterogeneous hosts (i.e. a mix of three different host-cell types), adaptation was negatively correlated with migration rate (Cuevas et al. 2003). We conjectured from these prior studies that migration may be beneficial for pathogen adaptation when sources and sinks constitute the same host type, and less beneficial when selection on sources and sinks is incongruent, in effect pulling populations in different directions across an adaptive landscape.
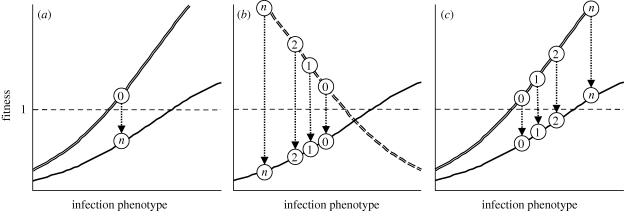
Theoretically, the role of migration as a conduit of genetic variation in pathogen emergence should apply, albeit in varying degree, to any selective landscape. To illustrate this point, we consider three distinct emergence scenarios that capture important generalities of source–sink evolution (illustrated in figure 1). First, in the absence of variation and evolution in the source population (figure 1a), the expected migrant genotype remains constant through time regardless of the selective landscape. Second, emergence may be onto a novel host type, constituting a host shift. We imagine that there is ongoing mutation and evolution in the source host, leading to improved adaptation there. We use the host-shift scenario to represent the general case in which the mean fitness of sink immigrants in the novel host is likely to decrease over time, owing either to evolution of genetic tradeoffs across hosts (as illustrated in figure 1b) or to mutation accumulation (Cooper & Lenski 2000). Third, an equally important scenario is emergence on a host type to which the virus is adapted, but under novel conditions, leading to range expansion. In range expansion, selection on host infection per se may actually be parallel in sources and sinks, even though other environmental factors depress sink population fitness. Of course, real-world host shifts and range expansions involve more complex selective landscapes; we use these terms only to indicate broad trends in mean immigrant fitness.
Figure 1.
Three emergence scenarios. Heavy curves represent simplified fitness landscapes in two environments. Circles represent phenotypic states at times 0, 1, 2,…, n. Evolution is indicated by shifts along the phenotype (x) axis. For the initial phenotype, one environment is a source (fitness ≥1, at or above the dashed horizontal line) and the other a sink. Arrows indicate migration from sources to sinks. (a) A static source population. Migration from the source (double curve) draws on a fixed pool of mutations and is unlikely to aid adaptation to the sink (single curve). (b) The host-shift scenario. The native host (dashed double curve) is a source for migrants to a novel host sink. Owing to disruptive selection, migrants are expected to be increasingly maladapted over time. (c) The range-expansion scenario. The same host type exists in both source and sink populations, but sink fitness is lower owing to environmental heterogeneity. In this case, migration delivers increasingly beneficial mutations to the sink.
The central purpose of this study is to test whether mutations that lead to adaptation in sink populations originate in situ or are delivered from larger source populations via migration. Migration should be most immediately facilitative in the range-expansion case, since mutations beneficial to the sink are also presumably being selected in the source. In the host-shift case, while selection in the native host environment is hypothesized to make migrants increasingly maladapted to a novel host type over time, repeated sampling of novel genetic variation in source populations may nonetheless eventually deliver beneficial mutations that promote successful emergence.
While theory regarding evolution of pathogens on new host populations and species (and pathogen emergence by extension) is elegant and well developed (e.g. Antia et al. 2003; André & Day 2005; Andre & Hochberg 2005; Yates et al. 2006), experimental tests still remain sparse. We tested the degree to which the emergence scenarios in figure 1 influence the process of emergence using experimental evolution of the dsRNA bacteriophage (phage) Φ6 and two different species of Pseudomonas host bacteria in laboratory microcosms. We used a host-range mutant of phage Φ6 to found replicate lineages that were experimentally evolved on the novel host under sink conditions. Sink populations periodically received a fixed number of migrants to maintain population size. Migrants were drawn from one of three sources: a frozen ancestral stock, a population evolving on the native host type (i.e. emergence in the sink represents a host shift) or a population evolving on the novel host type under permissive (source) conditions (i.e. emergence in the sink represents a range expansion on the same or similar hosts).
Our results indicated that the mean fitness of the immigrant pool on the novel host decreased over time in the host-shift treatment, and increased under the range-expansion regime. As expected, adaptation was greatest for the range-expansion scenario, in which the host type did not differ between source and sink populations. Importantly, adaptation did not occur when migrants were drawn from a frozen ancestral stock, indicating that genetic variation in the source was a dominant driver of sink pathogen emergence. In the host-shift treatment, sink populations adapted with a large and punctuated increase in fitness, consistent with the predictions of some analytical and numerical models of source–sink evolution under disruptive selection (Gomulkiewicz et al. 1999; Holt et al. 2003a). Overall, our results suggest that migration can be a key component of pathogen emergence, and that source genetic variation and the direction of selection in the source can strongly impact pathogen adaptability in sink environments.
2. Material and methods
(a). Overview
The experimental design tested the effects of source genetic variation on the adaptation of phage emerging in sink habitats. Sink habitats were established using published methods (Dennehy et al. 2006). If R0 denotes the expected number of offspring left behind by an individual phage, then if R0 < 1 the habitat is a sink. The quantity R0 combines the number of phage particles produced per infected cell, when that host cell has been infected by a single phage (so reflects within-host phage replication); the survival of those particles; the probability of infection of an uninfected host cell when it encounters a phage; and the number of host cells likely to be encountered. A novel host could be a sink because any of these quantities are low. We specifically used serial passages combined with dilution to create sinks; a high rate of dilution in effect is a high rate of mortality imposed on the phage. Briefly, replicate lineages derived from a phage Φ6 mutant were serially passaged on a strain of the novel host Pseudomonas pseudoalcaligenes on which virus populations were diluted 1 000 000-fold when passaged. Under these conditions, the virus population is unable to sustain itself on the novel host, and migration is required to prevent extinction (Dennehy et al. 2006). Treatments manipulated the genetic character of the source of migrants, contrasting three kinds of sources: (i) frozen stock identical to the founding ancestor, (ii) phage evolving on the native host species in persistent source populations, or (iii) phage evolving on the novel host species (again, maintained as persistent populations). We assayed fitness periodically over approximately 75 generations (15 days) to examine fitness trajectories in sources and sinks.
In each treatment, replicate sink populations received immigrants from a common source population. An alternative experimental design would have established replicate source populations, one for each sink, but this scenario was rejected because it would prohibit inferences regarding the source of beneficial mutation. By contrast, our design allowed strong inferences about the origin of genetic variation fuelling adaptation. Synchronous and similar changes in sink fitness within treatments would indicate an origin in the common pool of immigrants. To demonstrate this point, consider a mutant with a single representative in the inoculum of a source population plate. Owing to the strong spatial structure of viral populations grown on plates at low densities, a viable mutant present in the inoculum will only face selection after about five generations, when all virions in the population are removed from the plate and mixed in solution. At this point, a random sample is taken to inoculate the next plate in the serial passage experiment. This is also the point at which migration occurs. Our experimental design was such that the mutant, if nearly neutral, would be expected to contribute one representative in a sample of migrants to a sink population (approx. 106 progeny of the mutant on a plate with a sampling probability of approx. 10−6). The conditional Poisson probability of there being at least one such representative in each of three separate samples is (1 − e−1)3 ≈ 0.25. For comparison, consider a beneficial mutant arising in one of three independent replicate sink populations. The probability of the same mutation arising in the two other sinks on the same day would be of the order of the square of the per-nucleotide mutation rate, or approximately 10−12. While these arguments are crude, it remains far more likely that similar fitness trajectories among replicate sink populations indicate a single common source of beneficial mutations, than parallel but independent evolutionary trajectories.
Treatment (i) is set up to ensure that there is no novel genetic variation entering via the immigrant stream. The immigrants can potentially still have demographic consequences on evolution, for instance by competing with residents for host cells. By contrast, in treatments (ii) and (iii), genetic variation can arise via mutation in the source, and be introduced into the sink by immigration. However, in (iii), if there are tradeoffs in adaptation to the two hosts, the alleles that are positively selected in the source host might be expected to, on average, be disfavoured in the sink. As adaptation to the source host occurs, it should become increasingly unlikely that variants potentially adaptive in the sink will be retained there at high frequency, and so become part of an immigrant stream. This could describe the scenario common in nature, where pathogens ‘spill over’ between unrelated host species (Power & Mitchell 2004). By contrast, in our scenario (ii), there is a commonality in the selective regime, and adaptation in one host environment should generate variants that are also advantageous in the other host environment. In nature, this could describe host populations in a single species distributed across an environmental gradient, where some populations are demographic sources, and the other demographic sinks (e.g. near the edge of a species' geographical range). It could also conceivably pertain to ‘spillover’ between host species that are phylogenetically closely related, and so likely to represent comparable selective milieus.
(b). Study organisms
The dsRNA phage Φ6 (family Cystoviridae) is a virus that causes lytic (lethal) infection in certain plant-pathogenic Pseudomonas bacteria (Semancik et al. 1973; Vidaver et al. 1973; Mindich 2004). The typical laboratory host for phage Φ6 is Pseudomonas syringae pathovar phaseolicola (ATCC no. 21781; hereafter PP). Here, we used Φ6h (strain PT590), a previously described spontaneous mutant of wild-type Φ6 (Turner & Chao 1998). This virus differs from the wild-type by a non-synonymous point mutation (E8K) in the gene for attachment protein P3 on the M segment, which allows the virus to infect the novel host species used in our experiments: P. pseudoalcaligenes East River isolate A strain (from Leonard Mindich, Public Health Research Institute, Newark, NJ, USA; hereafter ERA). This mutation reduces virus fitness (growth relative to wild-type) on the native host by approximately 5 per cent (Turner & Chao 1998).
(c). Culture conditions
All phage and bacteria were reared, plated, incubated and diluted at 25°C in LC medium (10 g NaCl, 10 g Bacto tryptone and 5 g Bacto yeast extract per litre) at pH 7.5. Bacterial cultures were inoculated by placing a single colony in 10 ml LC medium in a sterile flask. Culture flasks were incubated with shaking (120 r.p.m.) at 25°C for 24 h, allowing bacteria to attain stationary-phase density (approx. 4 × 109 cells ml−1 for PP, and approx. 5 × 1010 cells ml−1 for ERA). All bacterial stocks were stored in a 4 : 6 glycerol/LC (v/v) solution at −80°C.
High-titre lysates of phage Φ6h were prepared by mixing phage drawn from frozen stock with 200 µl of stationary-phase PP in 3 ml top agar (0.7%), and pouring onto bottom agar (1.5%) in a sterile Petri dish. After 24 h, plaques formed in the top agar were harvested and resuspended in 4 ml of LC broth, followed by 10 min centrifugation at 3000 r.p.m. A bacteria-free lysate was obtained by filtering (0.22 µm, Durapore; Millipore, Bedford, MA, USA) the supernatant to remove bacteria. To determine population density, plaque-forming units per millilitre (pfu ml−1) were counted using dilution series on PP lawns. Lysates were stored at −20°C in a 1 : 1 glycerol/LC (v/v) solution.
(d). Source evolution
Phage Φ6h was used to found independent lineages on PP and on ERA; these two lineages were then experimentally evolved via serial passage for 15 days (approx. 75 generations; i.e. five generations per passage; Turner & Chao 1998). Each lineage was founded by approximately 104 particles of phage Φ6h mixed with approximately 109 PP cells or ERA cells in 3 ml top agar. Thus, the lineages were similar in terms of initial virus to cell ratio of 10−5. During 24 h incubation, the viruses formed plaques in the top agar as described above. The plaques were harvested to obtain a lysate that was then filtered and diluted 1 000 000-fold for the PP-evolved lineage and 1000-fold for the ERA-evolved lineage. To initiate the next passage, 100 µl from the respective dilution was transferred to fresh media. Dilution rates differed to allow populations to thrive as sources while avoiding overcrowding; per capita growth is higher on PP than on ERA (Dennehy et al. 2006). Source populations were large (greater than 1010 pfu ml−1), allowing the accrual of genetic variation through mutations. Lysates from each daily passage were stored in the freezer for later use.
(e). Sink evolution
Phage Φ6h was used to found three replicate lineages in each of three migration treatments as prescribed by the emergence scenarios in figure 1. As with source populations, each emerging population was initiated with approximately 104 pfu of phage Φ6h mixed with approximately 109 ERA cells in 3 ml top agar. After 24 h, the resulting plaques were harvested to obtain a lysate from each population, and the population was then diluted 1 000 000-fold and 100 µl of this dilution was used to initiate the next passage. The loss of individuals incurred by dilution exceeded gains from host infection for a ratio of population increase of approximately 10−0.5 (Dennehy et al. 2006), about two-thirds reduction per passage. Thus, these populations experienced sink conditions and were expected to rapidly go extinct in the absence of migration. A previously published study using these identical virus and bacteria strains propagated without immigration served as a basis for comparison to the current experiments, demonstrating the failure of in situ adaptation to rescue populations from extinction under these conditions (Dennehy et al. 2006).
At each passage, migrants from the source lineages were added to the corresponding sink lineage to rescue it from extinction. In the ancestral treatment, these migrants were sampled from the frozen stock of the founding ancestor: phage Φ6h. In contrast, host-shift and range-expansion lineages received migrants from source populations evolving on PP and ERA, respectively. For ease of experimentation, source populations were evolved before the sink evolution experiment; migrants were drawn from samples frozen at corresponding time points to simulate concurrent evolution of sources and sinks. To simulate migration, source migrants were removed from the freezer, titred and 104 phages were added to the treatment. While the exact ratio of viruses to cells at each passage cannot be calculated, it was initially of the order of 10−5. The plaques harvested at each passage were each assumed to have been initiated by a single virus. But as plaques grow, it is expected that the ratio of viruses to cells within a plaque increases greatly. Under these conditions, co-infection and reassortment of genetically distinct phage may occur, but these genetic exchange events should only involve mutants arising during plaque growth, since plaques are initially isogenic (but see §4).
(f). Paired-growth assays
Relative fitness was measured using paired-growth assays (Chao 1990) on the novel host, ERA. The common competitor in our relative fitness assays was strain PT88, a virus derived from phage Φ6h. The L segment of PT88 was modified by incorporating the alpha subunit of the Escherichia coli beta-galactosidase (β-gal) gene (Froissart et al. 2004). On LC indicator agar containing X-gal (0.4% w/v) and a lawn of P. phaseolicola bacteria harbouring the beta subunit of the β-gal gene (strain LM1034, kindly provided by L. Mindich), PT88 produces blue plaques whereas non-marked phages produce colourless plaques. To measure relative fitness, a test phage and PT88 were mixed at a 1 : 1 volumetric ratio, and then a dilution of this mixture containing approximately 200 viruses was plated on an ERA lawn. After 24 h incubation, approximately 200 plaques were harvested and filtered to obtain a cell-free lysate. The ratios of competing genotypes in the starting mixture, R0, and in the harvested lysate, R1, were obtained by plating on three indicator agar plates per sample, where the test phage and the marked competitor were distinguished by plaque colour. Thus, relative fitness was assayed on an ERA lawn, but the relative ratios were tracked using indicator agar. The number of plaques per plate was kept at approximately 200, to minimize plaque overlap and, hence, genetic exchange (segment reassortment) between plaques. We defined relative fitness, w, as (R1/R0) − w′, where w′ adjusts relative fitness to 1.0 at the beginning of the experiment.
(g). Absolute fitness assays
We defined absolute fitness, W, as the net rate of increase for a virus population under the sink conditions imposed in our study. Absolute fitness was measured by mixing an initial inoculum of approximately 104 particles, NI, of a test phage population with approximately 109 ERA cells in 3 ml top agar. After 24 h, the resulting plaques were harvested and titred using dilution series to estimate the final number of progeny viruses produced, NF. For analysis, the log10 ratio of progeny viruses to initial inoculum was calculated, and log10 absolute fitness was estimated as the net ratio of increase by subtracting the log10 dilution rate imposed during experimental evolution: log10W = log10 (NF/NI) − log10 (106).
(h). Analysis
Evolution of source population fitness on the novel host was measured by linear regression of relative fitness from the first to the 15th day of the experiment. Repeated-measures ANOVA of relative fitness in sink populations considered treatment × time and lineage-within-treatment × time interactions (in addition to main effects). The former interaction tested differences in the course of adaptation among treatments. The latter gave quantitative support to judgement by eye of the synchrony of adaptation within treatments and was followed by independent contrasts to infer the origin of beneficial mutations in each treatment. Results reported in this paper used unadjusted univariate tests, but all appropriate tests gave similar results. Final log10 W in sink populations was tested for difference from 0 using a t-test for each treatment. Emergence was equated with adaptation, the ability to persist locally without immigration, a condition obtained when log10W ≥ 0 (Holt 1985; Pulliam 1988; Gomulkiewicz et al. 1999).
3. Results
(a). Evolution in source populations
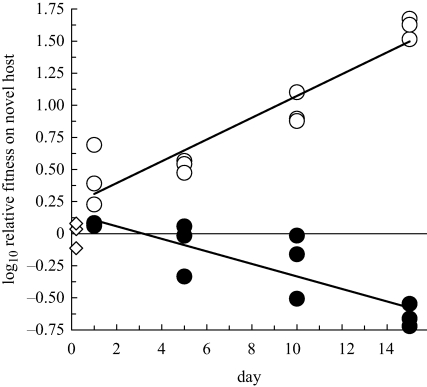
The two evolving source populations showed significant changes in log10 relative fitness on ERA, thus demonstrating changes through time in the average fitness of would-be migrants entering sinks (figure 2). Relative fitness was measured on ERA in paired-growth assays versus a common, unevolved competitor (§2). Performance (log10 relative fitness) of the ERA-evolved source lineage on ERA increased at an average rate of 0.085 (±0.01 s.e.) per day (r2 = 0.88, F1,10 = 73, p < 0.0001), corresponding to a doubling of relative fitness every 3.5 days. The log10 relative fitness of the PP-evolved lineage decreased by 0.049 (±0.01 s.e.) per day (r2 = 0.74, F1,10 = 28, p = 0.0003), corresponding to a halving of relative fitness every 6 days. This result is consistent with a tradeoff in performance between the two host types.
Figure 2.
Relative fitness trajectories of would-be migrants from source populations, assayed on the novel host. Two lineages founded by phage Φ6h were evolved for 15 days (approx. 75 generations) on either the native host (filled circles) or the novel host (open circles). Fitness relative to a marked genotype of the common ancestor was assayed at 0, 1, 5, 10 and 15 days of serial passage. All relative fitnesses were adjusted to set mean ancestral relative fitness to 1 (diamonds). Heavy lines are linear regressions through the data (not including the initial values).
(b). Emergence experiment
The changes in source population adaptation to the novel host allowed us to compare the process of emergence under the host-shift and range-expansion scenarios defined in figure 1b,c, in which migrant fitness decreases or increases over time, respectively. We allowed replicate virus lineages to evolve on ERA under sink conditions while being rescued by daily immigration of viruses drawn from the ERA-evolved source, PP-evolved source or a frozen stock of the founding ancestor phage Φ6h. We then measured the relative fitness on ERA. Data met the assumption of sphericity, and unadjusted univariate approximate F-tests were used to gauge the significance of responses. Similar results were obtained with other univariate and multivariate tests (data not shown).
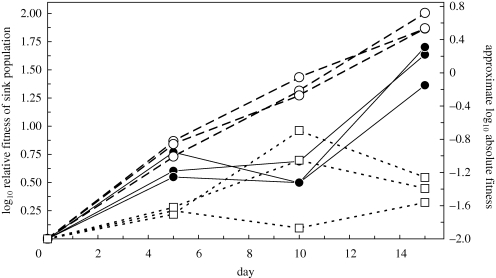
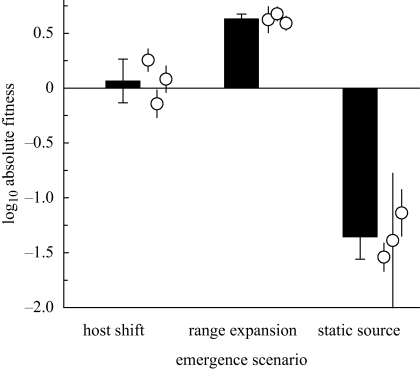
Relative fitness on ERA improved for lineages in all three emergence scenarios, but to different extents (time × treatment, F4,36 = 32.7, p < 0.0001). Not surprisingly, sink lineages receiving migrants from a source population experiencing parallel selection on ERA (i.e. range-expansion lineages) showed the greatest increases in sink fitness (figure 3) and had the highest mean fitness over the course of the experiment (treatment main effect, F2,18 = 75, p < 0.0001). In general, we noted that the fitness improvement of these sink lineages mirrored adaptation in the source population (cf. figures 2 and 3), although the increase was actually larger. Absolute fitness in the range-expansion treatment (0.63 ± 0.03 s.e.) was significantly greater than replacement at the end of the experiment (t2 = 25, p = 0.002; figure 4), indicating successful emergence.
Figure 3.
Relative fitness trajectories in emerging lineages. Populations growing on the novel host received a fixed supply of daily migrants from a source evolving on the native host (‘host shift’), a source evolving on the novel host (‘range expansion’) or from a frozen stock of the founding ancestor (‘static source’). Three replicate lineages were propagated independently in each treatment. Each point represents the mean of three estimates of fitness relative to a marked genotype of the common ancestor. The right axis indicates approximate absolute fitness obtained from orthogonal regression of the data in figure 4 on final relative fitness (absolute = 1.34 * relative − 1.97, r2 = 0.93, p < 0.0001). Filled circles, host shift; open circles, range expansions; open squares, static source.
Figure 4.
Final absolute fitness, measured as the net log10-transformed ratio of increase in sink populations in three emergence scenarios after 15 days. Bars show the grand mean (±1 s.d.) of three independent lineages per treatment. Open circles indicate the mean (±1 s.d.; n = 3) absolute fitness for each independent population.
Emergence failed in the controls where migrants were drawn from a frozen ancestral stock. Relative fitness did show an increasing trend over the course of the experiment, but adaptation was sporadic and some early fitness gains were later lost (figure 3). Absolute fitness after 15 days (−1.36 ± 0.12 s.e.) was significantly below replacement (t2 = −12, p = 0.007; figure 4), indicating that the novel host environment was still a sink.
Relative fitness in the host-shift treatment increased at nearly the same pace as the range-expansion treatment, although the tempo of adaptation was more punctuated (figure 3) and final relative fitness was lower. Absolute fitness in the host-shift treatment was not significantly different from replacement at the end of the experiment (0.065 ± 0.12 s.e.), with replicates straddling zero (t2 = 0.57, p = 0.63; figure 4). We cautiously take this result to indicate successful emergence according to our definition as absolute fitness permitting replacement, and hence population persistence in a given interaction. With observed variation among lineages in the host-shift treatment, the least maladapted treatment mean fitness we could detect with 80 per cent power was log10 W = 0.46. Of the three replicate lineages in the treatment, the most maladapted had log10 W = −0.14 (±0.07 s.e.); the most adapted had log10 W = 0.26 (±0.06 s.e.). Emergence in the host-shift treatment demonstrated that mutation in source populations provides a pool of variation for selection in the sink even when the dominant alleles have been selected in a direction that is on average disadvantageous in the sink environment. The contrast between scenarios (i) and (ii) shown in figure 3 demonstrates the importance of having some source of genetic variation.
(c). Origin of beneficial mutations
In the electronic supplementary material, we provide a quantitative argument suggesting that beneficial mutations that permit adaptation in the sink are likely to have arisen in the source and been sampled during immigration. Beyond the argument sketched there, it is worth noting that if sink adaptation is driven by mutations arising in the independent sinks, adaptive walks would take independent paths in each treatment. Inspection of figure 3, by contrast, shows a clear pattern of similarity in the timing and magnitude of changes in relative fitness within both of the treatments where lineages received migrants from an evolving source. This result indicates that the common source that provided immigrants to the sink populations within each of these treatments was the most likely origin of beneficial mutations that improved sink fitness. Only the frozen ancestor treatment (control) showed evidence for independent origin of beneficial mutations among replicate lineages, particularly as inferred by the data visible on day 10. Accordingly, a significant interaction of lineage-within-treatment × time (F12,36 = 2.89, p = 0.0068) was attributed to the frozen ancestor treatment by independent contrasts (F4,36 = 5.46, p = 0.0015). Lineages did not differ in mean fitness within treatments over the course of the experiment (lineage-within-treatment main effect, F6,18 = 1.40, p = 0.27).
4. Discussion
Infectious viruses have emerged to cause some of the most deadly pandemics in human history, and are regarded as major threats for future disease emergence. While ecological change is a likely driver of many emergence events (Plowright et al. 2008), evolution may also be responsible for some of the most dramatic host shifts and range expansions observed (Antia et al. 2003). A gap exists between our theoretical understanding of the factors contributing to the success of pathogen emergence and the experimental tests of that theory, and experimental evolution studies can reveal phenomena not yet addressed by theory. Using a mutant of the phage Φ6 able to infect two host species, we have examined the source–sink evolutionary dynamic that might describe emergence on novel hosts or in novel habitats where pathogens are initially maladapted.
One set of theory predicts that adaptation in emerging populations is fuelled by genetic variation transported from source populations via migration (Holt & Gomulkiewicz 1997; Gomulkiewicz et al. 1999). Other models predict that adaptation in novel environments is a largely local process constrained rather than aided by migration, when migration can impose a load via reproduction and recombination (Kirkpatrick & Barton 1997). Much of the theoretical and experimental focus in the debate between these predictions has been on the impact of migration itself (Miralles et al. 1999; Kawecki 2000; Cuevas et al. 2003; Holt et al. 2003b; Perron et al. 2007), and both positive and negative effects of immigration on local adaptation in a sink are a priori likely.
Our experiment has had a somewhat different focus, namely contrasting sources that differ in their own evolutionary trajectories, and then examining the consequences for sink evolution. We designed our experiment specifically to make strong inferences about the source of genetic variation in source–sink evolution. Each experimental group of three sink lineages shared a common source of immigrants. If adaptation resulted from in situ mutations in the sinks alone, then the three lineages would probably take independent paths on the adaptive landscape, represented by asynchronous and dissimilar increases in fitness over time.
To some extent, we did observe uncoordinated evolution in the control group receiving migrants from frozen ancestral stock (figure 3), fitting this conjecture of independent evolutionary trials. In contrast, lineages sharing evolving source populations showed synchronous patterns of adaptation, suggesting that most genetic variation arrived by migration. The use of a different, independent source population as the immigrant pool for each replicate sink population would have led to a different expectation about evolutionary synchrony. Our results therefore suggest that correlated evolution across multiple novel environments may arise because each population is coupled to a common source population, whose palette of genetic variation determines in large measure the range of variation accessible to evolution in each environment.
To test whether evolution in source populations is relevant to pathogen emergence, we measured sink adaptation using immigrants from different selective environments. Geographical range expansion on an existing host type might involve the same selective pressures everywhere along an environmental gradient, meaning that migrants from evolving source populations are likely to be pre-adapted to conditions at the range margin. Indeed, lineages in the range-expansion treatment showed the greatest improvement in relative fitness over the 15-day study (figure 3) and had the highest absolute fitness at its conclusion (figure 4).
A likely alternative emergence scenario involves incongruent selection pressures in source and sink environments as might occur during host shifts. Many ecological and evolutionary models assume tradeoffs in local adaptation of pathogens across hosts (Holt & Gomulkiewicz 1997; Kawecki & Ebert 2004). The source lineage that evolved on the native host suffered a fitness decline on the novel host species (figure 2). Despite this increasing maladaptation of immigrants over time, the sink lineages in the host-shift treatment nonetheless had adapted, or nearly so, after 15 days. The emergent population would be likely to persist and adapt further in the absence of continued migration. Our results show that coupling to a genetically shifting source population can fuel evolution and potential disease emergence even when migrants are drawn from a quite different selective environment. (The source origin of beneficial mutations in the host-shift treatment is supported by a mathematical model available as the electronic supplementary material.)
It is important to note the statistical limits on inference imposed by our experimental design. With only one source population per treatment, our results strictly provide conclusions only regarding the particular source populations we used. However, experimental evolution has often provided insights into general phenomena using replicates founded from common ancestors (e.g. the long-term E. coli evolution studies beginning with Lenski et al. 1991). Future extensions of our experiment, with replicate sets of common sources and independent sinks, will be needed to determine the frequency with which sink adaptation is probably driven by mutation in source populations, compared with in situ mutation. Our brief argument in §2 and our model in the electronic supplementary material suggest the primacy of source genetic variation in driving the evolution of pathogen emergence in our system.
It is intriguing that relative fitness increased so rapidly in the host-shift treatment and that the change was similarly timed among replicate lineages. Assuming a constant number of immigrants each census (as used in this experiment), sink populations reach and maintain a constant population size. Despite low fitness in the sink, the dominant migrant genotype has a population growth rate equilibrated precisely at replacement. Only a mutant that achieves a local population growth rate greater than replacement can invade. This ‘absolute fitness criterion’ (Holt & Gomulkiewicz 1997) suggests that large and rapid increases in fitness may be observed when selection differs strongly between source and sink environments, because if immigrants have low fitness in the sink, mutants of large effect may be required to surmount the demographic threshold of replacement. The order of magnitude increase in absolute fitness of the host-shift treatment between passages 10 and 15 represents a leap to fitness approximately 1 (as indicated by the approximate absolute fitness axis in figure 3) and is therefore roughly the smallest mutational effect expected to invade. As mentioned above, the synchrony of sink evolution points to the source as the origin of relevant genetic variation, but such synchrony might not always be expected even with a common pool of mutants shared among lineages. The standard model of source–sink evolution with disruptive selection predicts a punctuated pattern of adaptation, as observed, but this pattern arises owing to the rarity of beneficial mutations. If these beneficial mutants are indeed deleterious in the source population, the probability of sampling them repeatedly by migration decreases as selection progressively depletes the source pool of such mutants that, though disadvantageous there, are potentially beneficial in the sink environment.
We should note that the phage used in our study can in some circumstances experience a form of sexuality via the reshuffling of genotypes within host cells. Co-infection of the same bacterial cell by differing phage genotypes allows for reassortment: formation of hybrid progeny that inherit RNA segments from the different co-infecting parent viruses (Turner & Chao 1998; Turner et al. 1999). Reassortment similarly creates genetic variation impacting evolution of pathogens such as influenza A virus (Rambaut et al. 2008). Although our experiments generally took place under low-multiplicity conditions (i.e. ratios of viruses to cells much less than 1.0), reassortment in phage Φ6 can occur under relatively low multiplicities (Turner et al. 1999). Furthermore, even the rare occurrence of sex in biological populations can be profoundly important for adaptation if sex creates beneficial variants that are unlikely to be generated through mutation alone, particularly if gene action is non-additive. The determination of which process (in situ mutation, migrational input or recombinational shuffling) more often leads to adaptive evolutionary changes in sink populations of pathogens provides an intriguing avenue for future research in phage Φ6 and other virus systems featuring sexual exchange, and indicates a direction needed for future theoretical studies. It also places additional importance on the rate of source–sink migration, as the frequency of reassortment increases with population size in the sink, which in turn should be an increasing function of the number of migrants per generation.
Our experimental results support a conceptualization of pathogen range evolution, whether on a landscape defined by the parameters of host infection or in the sense of geographical range expansion. However the space of evolutionary possibilities is envisioned, our model provides a scenario wherein central persistent populations are able to dictate natural selection in peripheral sink populations, leading to a widespread distribution that coalesces back into a spatially circumscribed source (e.g. a scenario that may be important in flu virus evolution; Rambaut et al. 2008; Russell et al. 2008). Efforts towards vigilance against emergent pathogens should consider not only ecological changes that might lift impediments to geographical spread but also the processes generating and maintaining genetic variation in established populations. With increasing interest in pathogen emergence, the goals of basic and applied science have rarely been so congruent.
Acknowledgements
The authors thank Aashish Jethra and Yul Yang for assistance with experiments, Michael Barfield, Troy Day, Siobain Duffy, Dan Dykhuizen, Santiago Elena, Trevor Price and several anonymous reviewers for helpful comments and Leonard Mindich and Lin Chao for providing biological materials. This work was conducted in part with equipment in the Core Facility for Imaging, Cellular and Molecular Biology at Queens College. J.J.D. was supported by funding from the National Science Foundation (DBI-03-10205 and DEB-08-04039) and the Professional Staff Congress of the City University of New York. PET was supported by funding from the National Science Foundation (DEB-04-52163). N.A.F. was supported by National Institutes of Health funding to Dan Dykhuizen. R.D.H. was supported by the University of Florida Foundation and the National Science Foundation (DEB-0515565 and EF-0525751).
References
- André J. B., Day T.2005The effect of disease life history on the evolutionary emergence of novel pathogens. Proc. R. Soc. B 272, 1949–1956 (doi:10.1098/rspb.2005.3170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre J. B., Hochberg M. E.2005Virulence evolution in emerging infectious diseases. Evolution 59, 1406–1412 [PubMed] [Google Scholar]
- Antia R., Regoes R. R., Koella J. C., Bergstrom C. T.2003The role of evolution in the emergence of infectious diseases. Nature 426, 658–661 (doi:10.1038/nature02104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics J.1976Nature of limits to natural selection. Ann. Mo. Bot. Gard. 63, 224–247 (doi:10.2307/2395303) [Google Scholar]
- Chao L.1990Fitness of RNA virus decreased by Muller's Ratchet. Nature 348, 454–455 (doi:10.1038/348454a0) [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Feldgarden M., Weissman S. J., Dykhuizen D. E., van Belle G., Sokurenko E. V.2007Haplotype diversity in ‘source–sink’ dynamics of Escherichia coli urovirulence. J. Mol. Evol. 64, 204–214 (doi:10.1007/s00239-006-0063-5) [DOI] [PubMed] [Google Scholar]
- Cooper V., Lenski R.2000The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407, 736–739 (doi:10.1038/35037572) [DOI] [PubMed] [Google Scholar]
- Crill W. D., Wichman H. A., Bull J. J.2000Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154, 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas J. M., Moya A., Elena S. F.2003Evolution of RNA virus in spatially structured heterogeneous environments. J. Evol. Biol. 16, 456–466 (doi:10.1046/j.1420-9101.2003.00547.x) [DOI] [PubMed] [Google Scholar]
- Dennehy J. J.2009Bacteriophages as model organisms for virus emergence research. Trends Microbiol. 17, 450–457 (doi:10.1016/j.tim.2009.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy J. J., Friedenberg N. A., Holt R. D., Turner P. E.2006Viral ecology and the maintenance of novel host use. Am. Nat. 167, 429–439 [DOI] [PubMed] [Google Scholar]
- Duffy S., Turner P. E., Burch C. L.2006Pleiotropic costs of niche expansion in the RNA bacteriophage Φ6. Genetics 172, 751–757 (doi:10.1534/genetics.105.051136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Burch C. L., Turner P. E.2007Evolution of host specificity drives reproductive isolation among RNA viruses. Evolution 61, 2614–2622 (doi:10.1111/j.1558-5646.2007.00226.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris M. T., Joyce P., Burch C. L.2007High frequency of mutations that expand the host range of an RNA virus. Genetics 176, 1013–1022 (doi:10.1534/genetics.106.064634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froissart R., Wilke C. O., Montville R., Remold S. K., Chao L., Turner P. E.2004Co-infection weakens selection against epistatic mutations in RNA viruses. Genetics 168, 9–19 (doi:10.1534/genetics.104.030205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomulkiewicz R., Holt R. D., Barfield M.1999The effects of density dependence and immigration on local adaptation and niche evolution in a black-hole sink environment. Theor. Popul. Biol. 55, 283–296 (doi:10.1006/tpbi.1998.1405) [DOI] [PubMed] [Google Scholar]
- Holt R. D.1985Population dynamics in 2-patch environments—some anomalous consequences of an optimal habitat distribution. Theor. Popul. Biol. 28, 181–208 (doi:10.1016/0040-5809(85)90027-9) [Google Scholar]
- Holt R. D., Gomulkiewicz R.1997How does immigration influence local adaptation? A reexamination of a familiar paradigm. Am. Nat. 149, 563–572 [Google Scholar]
- Holt R. D., Hochberg M. E. (eds) 2002Virulence on the edge: a source–sink perspective. In Adaptive dynamics of infectious diseases: in pursuit of virulence management. (eds Dieckmann U., Metz J. A. J., Sabelis M. W., Sigmund K.), pp. 197–209 Cambridge, UK: Cambridge University Press [Google Scholar]
- Holt R. D., Dobson A. P., Begon M., Bowers R. G., Schauber E. M.2003aParasite establishment in host communities. Ecol. Lett. 6, 837–842 (doi:10.1046/j.1461-0248.2003.00501.x) [Google Scholar]
- Holt R. D., Gomulkiewicz R., Barfield M.2003bThe phenomology of niche evolution via quantitive traits in a ‘black-hole' sink. Proc. R. Soc. Lond. B 270, 215–224 (doi:10.1098/rspb.2002.2219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R. D., Barfield M., Gomulkiewicz R.2004Temporal variation can facilitate niche evolution in harsh sink environments. Am. Nat. 164, 187–200 [DOI] [PubMed] [Google Scholar]
- Holt R. D., Barfield M., Gomulkiewicz R.2005Theories of niche conservatism and evolution: could exotic species be potential tests? In Species invasions: insights into ecology, evolution, and biogeography (eds Sax J. S. D., Gaines S. D.), pp. 259–290 Sunderland, MA: Sinauer Associates [Google Scholar]
- Kawecki T. J.2000Adaptation to marginal habitats: contrasting influence of the dispersal rate on the fate of alleles with small and large effects. Proc. R. Soc. Lond. B 267, 1315–1320 (doi:10.1098/rspb.2000.1144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki T. J., Ebert D.2004Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241 (doi:10.1111/j.1461-0248.2004.00684.x) [Google Scholar]
- Kirkpatrick M., Barton N. H.1997Evolution of a species' range. Am. Nat. 150, 1–23 [DOI] [PubMed] [Google Scholar]
- Lenski R. E., Rose M. R., Simpson S. C., Tadler S. C.1991Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341 [Google Scholar]
- Lenormand T.2002Gene flow and the limits to natural selection. Trends Ecol. Evol. 17, 183–189 (doi:10.1016/S0169-5347(02)02497-7) [Google Scholar]
- Mindich L.2004Packaging, replication and recombination of the segmented genomes of bacteriophage Φ6 and its relatives. Virus Res. 101, 83–92 (doi:10.1016/j.virusres.2003.12.008) [DOI] [PubMed] [Google Scholar]
- Miralles R., Moya A., Elena S. F.1999Effect of population patchiness and migration rates on the adaptation and divergence of vesicular stomatitis virus quasispecies populations. J. Gen. Virol. 80, 2051–2059 [DOI] [PubMed] [Google Scholar]
- Perron G. G., Gonzalez A., Buckling A.2007Source–sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc. R. Soc. B 274, 2351–2356 (doi:10.1098/rspb.2007.0640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright R. K., Sokolow S. H., Gorman M. E., Daszak P., Foley J. E.2008Causal inference in disease ecology: investigating ecological drivers of disease emergence. Front. Ecol. Environ. 6, 420–429 (doi:10.1890/070086) [Google Scholar]
- Power A. G., Mitchell C. E.2004Pathogen spillover in disease epidemics. Am. Nat. 164, S79–S89 [DOI] [PubMed] [Google Scholar]
- Pulliam H. R.1988Sources, sinks and population regulation. Am. Nat. 132, 652–661 [Google Scholar]
- Rambaut A., Pybus O. G., Nelson M. I., Viboud C., Taubenberger J. K., Holmes E. C.2008The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 (doi:10.1038/nature06945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronce O., Kirkpatrick M.2001When sources become sinks: migrational meltdown in heterogeneous habitats. Evolution 55, 1520–1531 [DOI] [PubMed] [Google Scholar]
- Russell C. A., et al. 2008The global circulation of seasonal influenza A (H3N2) viruses. Science 320, 340–346 (doi:10.1126/science.1154137) [DOI] [PubMed] [Google Scholar]
- Semancik J. S., Vidaver A. K., Van Etten J. L.1973Characterization of a segmented double helical RNA from bacteriophage phi6. J. Mol. Biol. 78, 617 (doi:10.1016/0022-2836(73)90283-0) [DOI] [PubMed] [Google Scholar]
- Sokurenko E. V., Gomulkiewicz R., Dykhuizen D. E.2006Opinion—source–sink dynamics of virulence evolution. Nat. Rev. Microbiol. 4, 548–555 (doi:10.1038/nrmicro1446) [DOI] [PubMed] [Google Scholar]
- Turner P. E., Chao L.1998Sex and the evolution of intrahost competition in RNA virus phi 6. Genetics 150, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. E., Burch C. L., Hanley K. A., Chao L.1999Hybrid frequencies confirm limit to coinfection in the RNA bacteriophage phi 6. J. Virol. 73, 2420–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaver A. K., Koski R. K., Vanetten J. L.1973Bacteriophage Φ6—lipid containing virus of Pseudomonas phaseolicola. J. Virol. 11, 799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A., Antia R., Regoes R. R.2006How do pathogen evolution and host heterogeneity interact in disease emergence? Proc. R. Soc. B 273, 3075–3083 (doi:10.1098/rspb.2006.3681) [DOI] [PMC free article] [PubMed] [Google Scholar]