Abstract
Theory suggests that sympatric speciation is possible; however, its prevalence in nature remains unknown. Because Neodiprion sawflies are host specialists and mate on their hosts, sympatric speciation via host shifts may be common in this genus. Here, we test this hypothesis using near-complete taxonomic sampling of a species group, comprehensive geographical and ecological data, and multiple comparative methods. Host-use data suggest that host shifts contributed to the evolution of reproductive isolation in Neodiprion and previous work has shown that gene flow accompanied divergence. However, geographical data provide surprisingly little support for the hypothesis that host shifts occurred in sympatry. While these data do not rule out sympatric host race formation in Neodiprion, they suggest that this speciation mode is uncommon in the genus and possibly in nature.
Keywords: Sympatric speciation, phytophagous insect, host shift, host race, age-range correlation test, Neodiprion
1. Introduction
Sympatric speciation has long been—and continues to be—a controversial topic in evolutionary biology. Initially, much of this controversy centred on whether speciation was possible without geographical barriers to gene flow. During a time when most evolutionary biologists believed sympatric speciation was exceedingly unlikely (Mayr 1963, 1970; Futuyma & Mayer 1980; Felsenstein 1981), for example, Guy Bush maintained that phytophagous insects could speciate in situ via host shifts (Bush 1969, 1975a,b). Evidence in favour of the latter view has accumulated over the last 25 years, and a large body of theoretical work now supports the plausibility of sympatric speciation (reviewed in Turelli et al. 2001; Gavrilets 2003; Coyne & Orr 2004; Bolnick & Fitzpatrick 2007). In addition, empirical studies in several now-well-known systems have confirmed that the essential features of Bush's sympatric host race formation (hereafter, SHRF) model are present in nature (e.g. Rhagoletis, Enchenopa and Eurosta; reviewed in Via 2001; Berlocher & Feder 2002; Dres & Mallet 2002; Coyne & Orr 2004; Bolnick & Fitzpatrick 2007; see also Sorenson et al. 2003).
Although most evolutionary biologists now agree that sympatric speciation is theoretically possible, debates regarding its prevalence in nature rage on. While some have argued that ‘a few promising cases…do not add up to strong support for the idea that this process is common’ (Coyne & Orr 2004, p. 178), others contend that ‘sympatric divergence may, in fact, be responsible for much of the incredible biodiversity in arthropods and other groups, such as nematodes’ (Bush 1998, p. 436). Current empirical estimates of the frequency of sympatric speciation suggest that it is rare (Lynch 1989; Barraclough & Vogler 2000; Coyne & Price 2000; Fitzpatrick & Turelli 2006; Bolnick & Fitzpatrick 2007; Phillimore et al. 2008; see also Kisel & Barraclough 2010); however, phytophagous insects, which comprise over one-quarter of all living species (Strong et al. 1984), are almost entirely absent from these studies (Bolnick & Fitzpatrick 2007; but see Berlocher 1998). In this study, we therefore evaluate the evidence for both sympatric speciation and its purported ecological mechanism—host shifts—in the pine-feeding sawfly genus Neodiprion.
Like many phytophagous insects, Neodiprion are intimately associated with their host plants throughout their life cycle: eggs are embedded within the host plant tissue, larvae often spend their entire feeding period on their natal host, cocoons are spun on or beneath the host, and mating occurs on the host plant (reviewed in Coppel & Benjamin 1965; Knerer & Atwood 1973). This tight association between Neodiprion and their hosts is thought to be responsible for the restricted host range of individual species (Knerer & Atwood 1973; Bjorkman & Larsson 1991; McCullough & Wagner 1993). Specifically, all Neodiprion species feed on host plants in the family Pinaceae, most species are found only on hosts in the genus Pinus, and many species will feed only on a single Pinus species. Even the more polyphagous Neodiprion species (i.e. those that feed on multiple Pinus species) tend to have preferences for particular pine species or habitat types (Coppel & Benjamin 1965; Smith 1979; Wilson et al. 1992; McMillin & Wagner 1993).
Because they are strict host specialists and mate on their hosts, many authors have hypothesized that Neodiprion sawflies undergo SHRF (Ghent & Wallace 1958; Alexander & Bigelow 1960; Knerer & Atwood 1972, 1973; Bush 1975a,b; Tauber & Tauber 1981; Strong et al. 1984). If speciation via SHRF has indeed been common in Neodiprion, we predict that: (i) speciation occurred in the absence of geographical barriers; (ii) speciation events were accompanied by changes in host use; and (iii) changes in host use occurred in sympatry. Here, we test these predictions using near-complete taxonomic sampling of a species group, comprehensive geographical and ecological data, and multiple comparative methods.
2. Material and methods
(a). Specimens and DNA sequence data
North American Neodiprion specimens were collected, reared and preserved, and sequence data were obtained for a large mitochondrial region (COI/COII, 1752 bp) and three nuclear genes (EF1α, 1094 bp; CAD, 916 bp; ANL43, 776 bp), as described by Linnen & Farrell (2007; GenBank accession numbers EF361837 to EF362376). Because mitochondrial genes introgress readily in this genus (Linnen & Farrell 2007), nuclear genes were used to estimate phylogenetic relationships. Also, because the ecology, geography and taxonomy of sertifer group species are poorly known, all analyses described here focus on the monophyletic lecontei group (Linnen & Farrell 2007, 2008b). In total, sequence data for 19 lecontei group species (1–14 populations per species) were used in this study. The only lecontei group species that were not sampled were two Cuban species: N. cubensis (Hochmut) and N. insularis (Cresson). The impact of missing species on inferences regarding the geography of speciation is considered in §3.
(b). Phylogenetic and dating analyses
For comparative tests of speciation mode, we used the ‘best estimate’ of the Neodiprion (lecontei group) species tree described in Linnen & Farrell (2008a; figure 1). Many nodes in this tree had high (more than 95%) posterior probabilities, were robust to taxonomic sampling and species-tree method, and were supported by independent morphological evidence (Linnen & Farrell 2008a). Nonetheless, we considered the impact of phylogenetic uncertainty on all comparative tests of speciation mode. Branch lengths for all trees were estimated in MrBayes v. 3.1 (Ronquist & Huelsenbeck 2003) and scaled to time using r8s v. 1.71 (Sanderson 2003), as described in the electronic supplementary material.
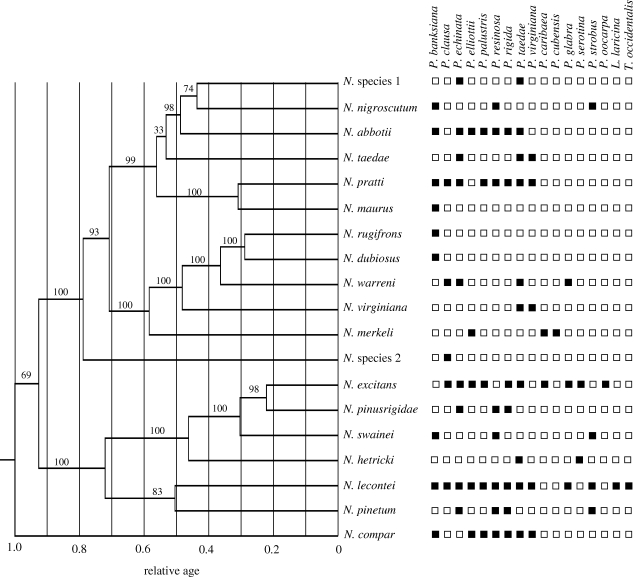
Figure 1.
Chronogram and host-use data for Neodiprion (lecontei group). Values above nodes are Bayesian posterior probabilities. Black squares indicate that a host plant (top) is used by a particular Neodiprion species; white squares indicate that a host is not used. Host plant genera are abbreviated as follows: P. = Pinus; L. = Larix; T. = Thuja. While Neodiprion are generally restricted to hosts in the genus Pinus, N. lecontei has been recorded on non-Pinus hosts during population outbreaks.
(c). Biogeography of speciation
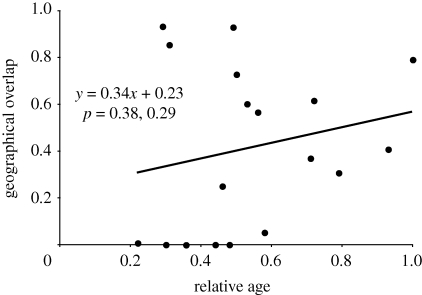
If SHRF has been prevalent in Neodiprion, sister species are expected to have overlapping ranges. To account for post-speciation range changes, we applied the age-range correlation (ARC) test to phylogenetic and geographic data from the genus. This test plots proportional geographic overlap among sister clades as a function of time since divergence and makes the following predictions: overlap between sister species (or clades) that originated in sympatry will tend to decrease over time (negative slope; y-intercept > 0.5), while sister species (or clades) that originated in allopatry will become more sympatric as time since divergence increases (positive slope; y-intercept < 0.5) (Lynch 1989; Barraclough et al. 1998; Berlocher 1998; Barraclough & Vogler 2000). The ARC test assumes that (i) there has been a single predominant speciation mode, and (ii) range changes have not eroded the relationship between geographical overlap and time (Barraclough & Vogler 2000; Losos & Glor 2003; Fitzpatrick & Turelli 2006). In §3, we consider the impact of violating these assumptions.
Geographic ranges for ARC tests were estimated from locality data, and total range areas for each species (19 total) and overlap areas for all species pairs (171 total) were estimated using ArcGIS v. 9. Next, following Fitzpatrick & Turelli (2006), we used PhyloCorrect (a Mathematica (Wolfram Research, Champaign, IL, USA) program provided by M. Turelli and B. Fitzpatrick) to calculate nested overlap averages for each node in the Neodiprion species tree (figure 1), to regress these values onto node age and to assess the significance of this linear regression. Additional details on range estimation and overlap analyses are available in the electronic supplementary material. To explore the impact of phylogenetic uncertainty, we repeated ARC analyses for species trees with alternative resolutions for nodes that were sensitive to taxonomic sampling/species-tree method and/or had posterior probabilities below 95 per cent (figure 1; Linnen & Farrell 2008a).
(d). Ecology of speciation
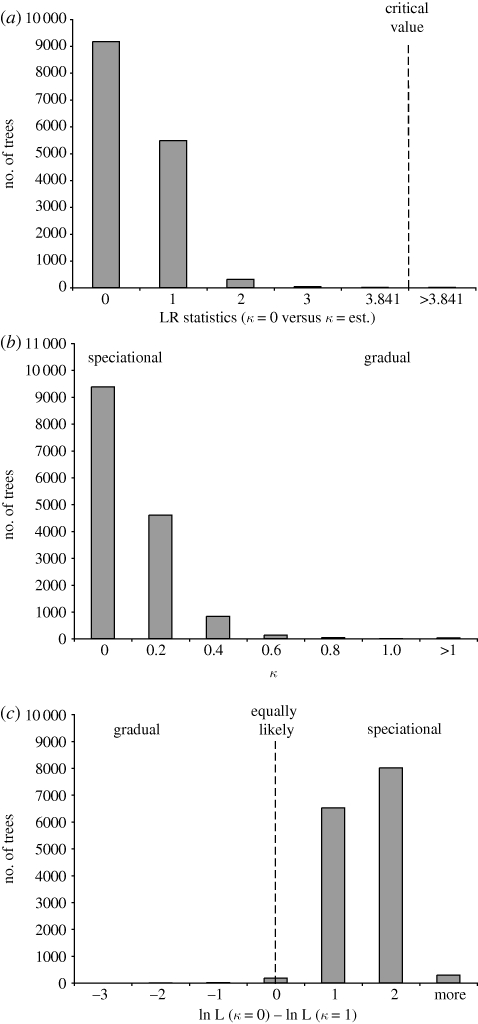
If speciation via SHRF has been prevalent in Neodiprion, host use is predicted to fit a speciational (punctuated) model of evolution, in which changes in host use occur during speciation events. To test this prediction, we used maximum likelihood and BayesMultiState to compare alternative models of host-use evolution (Pagel et al. 2004; implemented in BayesTraits v. 1.0, available at www.evolution.rdg.ac.uk). BayesMultiState assumes a continuous-time Markov model of evolution for discrete character data, and the tempo of trait evolution is characterized by the scaling parameter, κ, which defines the relationship between branch length and the probability of character change. A speciational model of evolution corresponds to κ = 0 (probability of change is unrelated to branch length), whereas a gradual model is represented by κ = 1 (change is directly proportional to branch length). We used BayesTraits to calculate the likelihood of the host-use data under each of these models and under a model in which κ was estimated from the data. We then used likelihood-ratio tests (LRT; LR = −2Δln L; LR is χ2-distributed with 1 d.f.) to compare the two simpler models to the model in which κ was estimated (Pagel 1994, 1997, 1999). To account for phylogenetic uncertainty, we calculated likelihoods for the three alternative models (κ = 0, κ = 1, κ = estimated) for all post-burn-in trees obtained in Bayesian Markov chain Monte Carlo analyses of two exemplar datasets (i.e. a single individual was sampled per species) as described in the electronic supplementary material.
For tests of host-use evolution, we compiled a list of hosts for each species and calculated pairwise host-use overlap as described in the electronic supplementary material. We then created a matrix with one representative for each Neodiprion species, and treated each recorded host as a separate character (17 total) with two character states (0 = host is used by species; 1 = host is not used by species). In BayesTraits, this translated into a Markov model in which hosts were gained and lost from a limited host pool over time. Separate rates were estimated for gains and losses, but these rates were set to be equal across hosts to reduce the number of parameters in the model (i.e. two transition rates instead of 34). While obviously simplified, this model accords with current hypotheses regarding host-use evolution in phytophagous insects (Janz & Nylin 1998; Thompson 1998; Janz et al. 2001; Janz 2002). This model also seems reasonable given Neodiprion biology: lecontei group species are restricted to Pinus hosts, but different Neodiprion species use different combinations of Pinus species.
3. Results and discussion
(a). Geography of speciation
The chronogram for our ‘best’ species tree is given in figure 1. The positive relationship between geographic overlap and time suggests that allopatric speciation may predominate in Neodiprion; however, this relationship was not significant (figure 2). These results were unaffected by alternative phylogenetic resolutions (electronic supplementary material, table S1). One potential source of bias in these plots is that taxonomic sampling was geographically biased—specifically, the Caribbean, Central America and eastern Mexico were not sampled. However, the number of missing species is small (two) and our non-significant ARC result is unlikely to have been rendered significant by their inclusion; nevertheless, these conclusions should be revisited as additional taxa are sampled.
Figure 2.
Geographical overlap plotted as a function of relative age (ARC plot) for Neodiprion. Each point represents a node in the species tree in figure 1. Regression line and two-tailed p-values (slope, intercept) were calculated using PhyloCorrect as described in the text.
Lack of a significant ARC trend in the lecontei group is probably explained by a combination of two biological processes: post-speciation range changes and mixed speciation modes (Barraclough & Vogler 2000; Losos & Glor 2003; Fitzpatrick & Turelli 2006; Phillimore et al. 2008). First, dramatic climatic oscillations over the past two million years are known to have affected Pinus distributions (MacDonald et al. 1998; Schmidtling 2003); thus, post-speciation range changes have clearly occurred in Neodiprion. The potential for range change over short periods of time in Neodiprion is further illustrated by the rapid expansion of N. sertifer following its introduction into North America in 1925 (Schaffner 1939; Lyons 1963). Second, the observation that there are an approximately equal number of sister-species pairs—which have had less time for extensive range changes—with sympatric and allopatric ranges (sympatric: N. rugifrons/N. dubiosus = 94% overlap; N. pratti/N. maurus = 85% overlap; N. lecontei/N. pinetum = 72% overlap; allopatric: N. nigroscutum/N. species 1 = 0% overlap; N. excitans/N. pinusrigidae = 1% overlap; figures 1 and 2) suggests that a mixture of speciation modes may also contribute to a lack of a significant ARC trend in Neodiprion.
In this study, we applied the ARC method to a group of organisms with features conducive to sympatric speciation; like most ARC studies to date (e.g. Fitzpatrick & Turelli 2006; Jiggins et al. 2006), our results were largely inconclusive. Thus, regardless of taxon and divergence mode, it appears that ARC assumptions (i.e. limited post-speciation range change and a single speciation mode) are rarely met. In fact, because there is no reason to expect that a sympatrically speciating lineage would forgo allopatric speciation, speciation modes are more likely to be mixed—and ARC results more likely to be inconclusive—in sympatrically speciating taxa than in taxa that speciate only in allopatry. To address this issue, Berlocher & Feder (2002) recommended partitioning taxa a priori into ‘host-shifters’ and ‘non-shifters’, and performing ARC tests separately for each group (see also Berlocher 1998). This could be done in two ways. First, one could partition sister-species pairs within a lineage into shifters (species are on different hosts) and non-shifters (species are on the same host)—the SHRF hypothesis predicts negative ARC slopes in the shifters, but not the non-shifters. Unfortunately, this test cannot be performed in Neodiprion, as there are only five sister-species pairs and only one of these is unambiguously a host-shifter pair (N. species 1 and N. nigroscutum; figure 1). It is worth noting, however, that this single host-shifter pair has 0 per cent geographic overlap—this pattern is not expected under the SHRF model.
Second, one could categorize entire lineages as ‘host-shifters’ (have features conducive to SHRF) or ‘non-shifters’ (lack features conducive to SHRF; Berlocher 1998; Berlocher & Feder 2002). In this context, ARC plots could be compared across taxa (even if inconclusive within taxa). Specifically, the SHRF hypothesis predicts that ARC y-intercepts—the average geographic overlap at the time of speciation—will be higher, on average, for host-shifter clades than for non-shifter clades. ARC slopes are probably not comparable across diverse taxa because these will be heavily influenced by competitive interactions between species and by differing propensities for range change. Two additional metrics for comparing shifters and non-shifters are the proportion of sister-species pairs with zero geographical overlap and the proportion with complete overlap. While these metrics do not make full use of the data implicit in phylogenies, Phillimore et al. (2008) demonstrated via simulation that they are robust indicators of the geography of speciation, whereas ARC correlation coefficients are not (ARC intercepts and slopes were not tested in their study). Thus, while the host-shifter/non-shifter test cannot be carried out within the lecontei group, the results presented here provide a rare host-shifter data point (ARC intercept = 0.23, proportion of allopatric sister species = 0.2, proportion of sympatric species = 0) for comparison with non-shifter taxa.
(b). Ecology of speciation
Table S2 in the electronic supplementary material lists inferred host ranges for each species. Figure 1 maps these data onto the Neodiprion phylogeny and figure 3 summarizes analyses of host-use evolution. Results were very similar for both sets of exemplars, so only one set is shown (ExemA). Consistent with the SHRF hypothesis, almost no trees (i.e. < 0.05%) were able to reject the speciational (κ = 0) model of host-use evolution (figure 3a). In contrast, nearly 50 times as many trees were able to reject the gradual (κ = 1) model of evolution using LRTs. While this is still a small number of trees (2.5%), two additional lines of evidence suggest that the host-use data is best explained by the speciational model: (i) maximum-likelihood estimates of κ were consistently low (e.g. 96% of all estimates were below 0.25; figure 3b); and (ii) likelihood scores for κ = 0 were consistently higher than for κ = 1 (figure 3c).
Figure 3.
Comparison of models of host-use evolution. Each graph is a frequency distribution of values estimated from 15 000 trees (post-burn-in trees obtained in a Bayesian analysis see text for details). (a) LRT statistics (LR = −2Δln L) for comparisons between models in which κ was estimated from the data (κ = est.) and κ was set to 0 (κ = 0; this corresponds to a speciational model of evolution). Nearly all comparisons failed to reject the simpler model (κ = 0). (b) Estimates of the parameter κ, which describes the relationship between branch length and the probability of character change. Values near 1 correspond to a gradual model; but most values clustered around 0, which corresponds to a speciational model. (c) Comparison of the likelihood of the data under a speciational model (κ = 0) to the likelihood of the data under a gradual model (κ = 1). Host-use data were almost always more probable under a speciational model of evolution.
Because characters that change predominantly at speciation events need not necessarily play a causal role in speciation (Barraclough & Nee 2001), additional evidence is needed to link host shifts to the evolution of reproductive isolation. Under the SHRF model, gene flow between species pairs that share hosts is predicted to be higher, on average, than between those that do not share hosts. Estimates of mitochondrial introgression from a previous study (Linnen & Farrell 2007) allowed us to test this prediction. As predicted, species that shared at least one host had significantly higher mitochondrial gene flow rates (2Nm) than pairs that shared no hosts (p = 0.00015; one-tailed Mann–Whitney U-test). One explanation for this result is that species that do not share hosts also have less geographical overlap (p < 1 × 10−6; one-tailed Mann–Whitney U-test), and therefore less opportunity for gene flow, than species that share hosts. However, when we examined only non-allopatric species pairs (i.e. greater than 0% geographic overlap), we found that mitochondrial gene flow rates still differed significantly between pairs with low host-use overlap (less than or equal to 20%) and those with high host-use overlap (greater than 20%; p = 0.0032; one-tailed Mann–Whitney U-test). In contrast, these groups did not differ significantly in geographical overlap (p = 0.12; one-tailed Mann–Whitney U-test), suggesting that divergent host use is a barrier to reproduction between species with overlapping geographic ranges. Taken together, our results suggest that host shifts have contributed to Neodiprion diversification and add to a rapidly growing body of literature that suggests that ecology in general (e.g. Schluter 2001; McKinnon et al. 2004; Funk et al. 2006; Langerhans et al. 2007) and host use in particular (e.g. Via 1999; Funk et al. 2002; Linn et al. 2004; Stireman et al. 2005; Nosil 2007) are important engines of diversity.
In addition to predicting that host-use evolution will fit a speciational model and that gene flow will be higher between species that share hosts, the SHRF hypothesis also predicts that sister species will use different hosts in sympatry. While our data support the first two predictions, they do not support the third. Specifically, host-use overlap and geographic overlap between sister clades are positively correlated (Kendall's τ = 0.52; p = 0.0025), four out of five sister-species pairs share at least one host, and the only sister-species pair with 0 per cent host-use overlap is completely allopatric (figure 1; electronic supplementary material, table S2). These observations suggest that, while host shifts sometimes play a role in Neodiprion speciation, they may occur primarily during periods of allopatry. Because ranges can change following speciation, however, additional data (e.g. Wang & Hey 2010) are needed to verify that contemporary overlap between each sister-species pair accurately reflects the geography of speciation.
An important caveat for the host-use data presented here is that we have not accounted for preferences for particular host species, host attributes or habitat differences, all of which have been reported for Neodiprion species (e.g. Coppel & Benjamin 1965; Smith 1979; Wilson et al. 1992; McMillin & Wagner 1993; electronic supplementary material, table S2). For example, N. pinetum is rarely found on any host other than Pinus strobus (Baker 1972; Smith 1979), while its sympatric sister species N. lecontei avoids this host except during outbreaks, when other hosts are unavailable (Atwood 1961; Wilson et al. 1992). Therefore, SHRF is still a plausible mechanism for this sister-species pair (as is allopatric speciation via a host shift followed by secondary contact). In contrast, N. maurus, N. rugifrons and N. dubiosus are all monophagous on jack pine, and N. pratti prefers this host in northern parts of its range (electronic supplementary material, table S2), suggesting that mechanisms other than host shifts are responsible for these two speciation events (figure 1). For example, N. maurus has evolved a novel overwintering mode that temporally isolates it from N. pratti (Knerer 1991). At present, there are no obvious barriers to reproduction between N. rugifrons and N. dubiosus, as these species overlap temporally (Becker et al. 1966), hybridize readily in the laboratory (Kraemer & Coppel 1983), do not have divergent preferences for host or habitat characteristics (C. Linnen 2002, 2004, personal observation) and have experienced substantial gene flow in nature (Linnen & Farrell 2007). One possibility is that these two species are in the process of merging after a period of divergence (without a host shift) in allopatry, but additional data are needed to test this hypothesis. In short, while detailed ecological studies are needed to better quantify host-use overlap and to assess its role in speciation, existing data imply that SHRF can be ruled out for all but one sister-species pair in Neodiprion (we note, however, that SHRF may still be responsible for older bifurcations in the Neodiprion tree).
(c). SHRF in Neodiprion and other insects
Host specialization and a tendency to mate on the host plant make Neodiprion sawflies prime candidates for testing the SHRF hypothesis. However, in spite of Neodiprion's long history in the SHRF literature, we found surprisingly little evidence that this speciation mode is common in the genus. Instead, one possible interpretation of our geographical, ecological and gene-flow data is that host shifts occur primarily in allopatry, and adaptation to novel hosts contributes to pre- and/or post-mating barriers to reproduction upon secondary contact (or, in some cases, prevents secondary contact altogether). Testing this hypothesis will require (i) determining the contribution of divergent host use to reproductive barriers between species (e.g. Nosil 2007); and (ii) documenting patterns of divergence and gene flow across selected (host use) and neutral loci (e.g. Beaumont 2005; Won & Hey 2005; Hey 2010). Nonetheless, existing data imply that simple labels such as ‘sympatric’ and ‘allopatric’ do not adequately describe the complex mix of geographical isolation, contact and gene flow apparent in Neodiprion and in other taxa (e.g. Feder et al. 2003; Coyne & Orr 2004; Mallet 2005; Xie et al. 2007).
While our data suggest that speciation via SHRF is rare in Neodiprion, and possibly in nature, it remains to be seen whether sympatric speciation, even if rare, is more prevalent in insects that specialize and mate on their hosts than in other taxa. Testing this hypothesis will require characterizing patterns of geographic overlap (e.g. ARC intercepts and the proportion of allopatric and sympatric sister-species pairs; see above) in additional host-shifter and non-shifter clades. Moreover, estimates of the amount and timing of gene flow accompanying divergence (e.g. Won & Hey 2005; Niemiller et al. 2008; Hey 2010) could also be used to determine whether speciation modes differ consistently between host-shifter and non-shifter taxa. If host-shifters speciate sympatrically more often and/or can tolerate more gene flow during divergence than non-shifters, this could explain, in part, why more than 25 per cent of all species are plant-feeding insects (Strong et al. 1984) and why independent origins of phytophagy are consistently associated with increased diversification rates (Mitter et al. 1988).
Acknowledgements
We thank the following individuals and institutions for access to museum specimens and collection data: S. Barudzija (Natural Resources Canada, Canadian Forest Service), D. Smith (National Museum of Natural History), C. Favret (Illinois Natural History Survey), L. Stange (Florida State Collection of Arthropods), H. Goulet (Agriculture Canada) and S. Krauth (University of Wisconsin). We are grateful to B. Fitzpatrick and M. Turelli for sharing their Mathematica code for ARC tests; and to B. Bossert, D. Haig, E. Kay, N. Pierce and J. Wakeley for discussions and/or comments on this manuscript. We also thank M. Noor, J. Feder and two anonymous reviewers for suggestions that greatly improved the manuscript. Funding for this research was provided by the National Science Foundation (Graduate Research Fellowship and Dissertation Improvement Grant DEB-0308815), the Environmental Protection Agency (Science to Achieve Results Graduate Fellowship), the American Museum of Natural History (Theodore Roosevelt Memorial Fund), the Museum of Comparative Zoology (Putnam Expeditionary Fund) and the Department of Organismic and Evolutionary Biology at Harvard University.
References
- Alexander R. D., Bigelow R. S.1960Allochronic speciation in field crickets, and a new species Acheta veletis. Evolution 14, 334–346 [Google Scholar]
- Atwood C. E.1961Present status of the sawfly family Diprionidae (Hymenoptera) in Ontario. Proc. Entomol. Soc. Ont. 91, 205–215 [Google Scholar]
- Baker W. L.1972Eastern forest insects. Miscellaneous publication number 1175. Washington, DC: US Department of Agriculture Forest Service [Google Scholar]
- Barraclough T. G., Nee S.2001Phylogenetics and speciation. Trends Ecol. Evol. 16, 391–399 [DOI] [PubMed] [Google Scholar]
- Barraclough T. G., Vogler A. P.2000Detecting the geographical pattern of speciation from species-level phylogenies. Am. Nat. 155, 419–434 (doi:10.1086/303332) [DOI] [PubMed] [Google Scholar]
- Barraclough T. G., Vogler A. P., Harvey P. H.1998Revealing the factors that promote speciation. Phil. Trans. R. Soc. Lond. B 353, 241–249 (doi:10.1098/rstb.1998.0206) [Google Scholar]
- Beaumont M. A.2005Adaptation and speciation: what can Fst tell us? Trends Ecol. Evol. 20, 435–440 [DOI] [PubMed] [Google Scholar]
- Becker G. C., Wilkinson R. C., Benjamin D. M.1966Taxonomy of Neodiprion rugifrons and N. dubiosus (Hymenoptera: Tenthredinoidea: Diprionidae). Ann. Entomol. Soc. Am. 59, 173–178 [Google Scholar]
- Berlocher S. H.1998Can sympatric speciation be proven from phylogenetic and biogeographic evidence? In Endless forms: species and speciation (eds Howard D. J., Berlocher S. H.), pp. 99–113 New York, NY: Oxford University Press [Google Scholar]
- Berlocher S. H., Feder J. L.2002Sympatric speciation in phytophagous insects: moving beyond controversy? Annu. Rev. Entomol. 47, 773–815 (doi:10.1146/annurev.ento.47.091201.145312) [DOI] [PubMed] [Google Scholar]
- Bjorkman C., Larsson S.1991Pine sawfly defense and variation in host plant resin acids: a trade-off with growth. Ecol. Entomol. 16, 283–289 (doi:10.1111/j.1365-2311.1991.tb00219.x) [Google Scholar]
- Bolnick D., Fitzpatrick B.2007Sympatric speciation: models and empirical evidence. Ann. Rev. Ecol. Evol. Syst. 38, 459–487 (doi:10.1146/annurev.ecolsys.38.091206.095804) [Google Scholar]
- Bush G. L.1969Sympatric host race formation and speciation in frugivorous flies of genus Rhagoletis (Diptera, Tephritidae). Evolution 23, 237–251 [DOI] [PubMed] [Google Scholar]
- Bush G. L.1975aModes of animal speciation. Annu. Rev. Ecol. Syst. 6, 339–364 (doi:10.1146/annurev.es.06.110175.002011) [Google Scholar]
- Bush G. L.1975bSympatric speciation in phytophagous parasitic insects. In Evolutionary strategies of parasitic insects and mites (ed. Price P. W.), pp. 187–207 New York, NY: Plenum [Google Scholar]
- Bush G. L.1998The conceptual radicalization of an evolutionary biologist. In Endless forms: species and speciation (eds Howard D. J., Berlocher S. H.), pp. 425–438 New York, NY: Oxford University Press [Google Scholar]
- Coppel H. C., Benjamin D. M.1965Bionomics of Nearctic pine-feeding diprionids. Annu. Rev. Entomol. 10, 69–96 (doi:10.1146/annurev.en.10.010165.000441) [Google Scholar]
- Coyne J. A., Orr H. A.2004Speciation. Sunderland, MA: Sinauer Associates, Inc [Google Scholar]
- Coyne J. A., Price T. D.2000Little evidence for sympatric speciation in island birds. Evolution 54, 2166–2171 (doi:10.1554/0014-3820(2000)054[2166:LEFSSI]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Dres M., Mallet J.2002Host races in plant-feeding insects and their importance in sympatric speciation. Phil. Trans. R. Soc. Lond. B 357, 471–492 (doi:10.1098/rstb.2002.1059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder J. L., et al. 2003Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl Acad. Sci. USA 100, 10 314–10 319 (doi:10.1073/pnas.1730757100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.1981Skepticism towards Santa Rosalia, or why are there so few kinds of animals. Evolution 35, 124–138 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick B. M., Turelli M.2006The geography of mammalian speciation: Mixed signals from phylogenies and range maps. Evolution 60, 601–615 (doi:10.1554/05-453.1) [PubMed] [Google Scholar]
- Funk D. J., Filchak K. E., Feder J. L.2002Herbivorous insects: model systems for the comparative study of speciation ecology. Genetica 116, 251–267 (doi:10.1023/A:1021236510453) [PubMed] [Google Scholar]
- Funk D. J., Nosil P., Etges W. J.2006Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc. Natl Acad. Sci. USA 103, 3209–3213 (doi:10.1073/pnas.0508653103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma D. J., Mayer G. C.1980Non-allopatric speciation in animals. Syst. Zool. 29, 254–271 [Google Scholar]
- Gavrilets S.2003Models of speciation: what have we learned in 40 years? Evolution 57, 2197–2215 (doi:10.1554/02-727) [DOI] [PubMed] [Google Scholar]
- Ghent A. W., Wallace D. R.1958Oviposition behavior of the Swaine jack-pine sawfly. For. Sci. 4, 264–272 [Google Scholar]
- Hey J.2010Isolation with migration models for more than two populations. Mol. Biol. Evol. 27, 905–920 (doi:10.1093/molbev/msp296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz N.2002Evolutionary ecology of oviposition strategies. In Chemoecology of insect eggs and egg deposition (eds Hilker M., Meiners T.), pp. 349–376 Berlin, Germany: Blackwell [Google Scholar]
- Janz N., Nylin S.1998Butterflies and plants: a phylogenetic study. Evolution 52, 486–502 [DOI] [PubMed] [Google Scholar]
- Janz N., Nyblom K., Nylin S.2001Evolutionary dynamics of host-plant specialization: a case study of the tribe Nymphalini. Evolution 55, 783–796 (doi:10.1554/0014-3820(2001)055[0783:EDOHPS]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Jiggins C. D., Mallarino R., Willmott K. R., Bermingham E.2006The phylogenetic pattern of speciation and wing pattern change in neotropical Ithomia butterflies (Lepidoptera: Nymphalidae). Evolution 60, 1454–1466 (doi:10.1554/05-483.1) [DOI] [PubMed] [Google Scholar]
- Kisel Y., Barraclough T. G.2010Speciation has a spatial scale that depends on levels of gene flow. Am. Nat. 175, 316–334 (doi:10.1086/650369) [DOI] [PubMed] [Google Scholar]
- Knerer G.1991Spontaneous speciation through an overwintering change in a sawfly. Naturwissenschaften 78, 328–330 [DOI] [PubMed] [Google Scholar]
- Knerer G., Atwood C. E.1972Evolutionary trends in subsocial sawflies belonging to Neodiprion abietis complex (Hymenoptera: Tenthredinoidea). Am. Zool. 12, 407–418 [Google Scholar]
- Knerer G., Atwood C. E.1973Diprionid sawflies: polymorphism and speciation. Science 179, 1090–1099 (doi:10.1126/science.179.4078.1090) [DOI] [PubMed] [Google Scholar]
- Kraemer M. E., Coppel H. C.1983Hybridization of jack pine feeding sawflies (Diproinidae: Neodiprion). Forestry Research Notes Madison, WI: University of Wisconsin [Google Scholar]
- Langerhans R. B., Gifford M. E., Joseph E. O.2007Ecological speciation in Gambusia fishes. Evolution 61, 2056–2074 (doi:10.1111/j.1558-5646.2007.00171.x) [DOI] [PubMed] [Google Scholar]
- Linn C. E., Darnbroski H. R., Feder J. L., Berlocher S. H., Nojima S., Roelofs W. L.2004Postzygotic isolating factor in sympatric speciation in Rhagoletis flies: reduced response of hybrids to parental host-fruit odors. Proc. Natl Acad. Sci. USA 101, 17 753–17 758 (doi:10.1073/pnas.0408255101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen C. R., Farrell B. D.2007Mitonuclear discordance is caused by rampant mitochondrial introgression in Neodiprion (Hymenoptera: Diprionidae) sawflies. Evolution 61, 1417–1438 (doi:10.1111/j.1558-5646.2007.00114.x) [DOI] [PubMed] [Google Scholar]
- Linnen C. R., Farrell B. D.2008aComparison of methods for species-tree inference in the sawfly genus Neodiprion (Hymenoptera: Diprionidae). Syst. Biol. 57, 876–890 (doi:10.1080/10635150802580949) [DOI] [PubMed] [Google Scholar]
- Linnen C. R., Farrell B. D.2008bPhylogenetic analysis of nuclear and mitochondrial genes reveals evolutionary relationships and mitochondrial introgression in the sertifer species group of the genus Neodiprion (Hymenoptera: Diprionidae). Mol. Phylogenet. Evol. 48, 240–257 (doi:10.1016/j.ympev.2008.03.021) [DOI] [PubMed] [Google Scholar]
- Losos J. B., Glor R. E.2003Phylogenetic comparative methods and the geography of speciation. Trends Ecol. Evol. 18, 220–227 (doi:10.1016/S0169-5347(03)00037-5) [Google Scholar]
- Lynch J. D.1989The gauge of speciation: on the frequencies of mode of speciation. In Speciation and its consequences (eds Otte D., Endler J. A.), pp. 527–553 Sunderland, MA: Sinauer [Google Scholar]
- Lyons L. A.1963The European Pine Sawfly, Neodiprion sertifer (Geoff.) (Hymenoptera: Diprionidae): a review with emphasis on studies in Ontario. Proc. Entomol. Soc. Ont. 94, 5–37 [Google Scholar]
- MacDonald G. M., Cwynar L. C., Whitlock C.1998The late Quaternary dynamics of pines in northern North America. In Ecology and biogeography of Pinus (ed. Richardson D. M.), pp. 122–136 Cambridge, UK: Cambridge University Press [Google Scholar]
- Mallet J.2005Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 (doi:10.1016/j.tree.2005.02.010) [DOI] [PubMed] [Google Scholar]
- Mayr E.1963Animal species and evolution. Cambridge, UK: Belknap [Google Scholar]
- Mayr E.1970Populations, species, and evolution: an abridgement of animal species and evolution. Cambridge, UK: Belknap [Google Scholar]
- McCullough D. G., Wagner M. R.1993Defusing host defenses: ovipositional adaptations of sawflies to pine resins. In Sawfly life history adaptations to woody plants (eds Wagner M., Raffa K. F.), pp. 157–172 San Diego, CA: Academic Press, Inc [Google Scholar]
- McKinnon J. S., Mori S., Blackman B. K., David L., Kingsley D. M., Jamieson L., Chou J., Schluter D.2004Evidence for ecology's role in speciation. Nature 429, 294–298 (doi:10.1038/nature02556) [DOI] [PubMed] [Google Scholar]
- McMillin J. D., Wagner M. R.1993Influence of stand characteristics and site quality on sawfly population dynamics. In Sawfly life history adaptations to woody plants (eds Wagner M. R., Raffa K. F.), pp. 333–361 San Diego, CA: Academic Press, Inc [Google Scholar]
- Mitter C., Farrell B., Wiegmann B.1988The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification. Am. Nat. 132, 107–128 [Google Scholar]
- Niemiller M. L., Fitzpatrick B. M., Miller B. T.2008Recent divergence with gene flow in Tennessee cave salamanders (Plethodontidae: Gyrinophilus) inferred from gene genealogies. Mol. Ecol. 17, 2258–2275 (doi:10.1111/j.1365-294X.2008.03750.x) [DOI] [PubMed] [Google Scholar]
- Nosil P.2007Divergent host plant adaptation and reproductive isolation between ecotypes of Timema cristinae walking sticks. Am. Nat. 169, 151–162 (doi:10.1086/510634) [DOI] [PubMed] [Google Scholar]
- Pagel M.1994Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. B 255, 37–45 (doi:10.1098/rspb.1994.0006) [Google Scholar]
- Pagel M.1997Inferring evolutionary processes from phylogenies. Zool. Scr. 26, 331–348 (doi:10.1111/j.1463-6409.1997.tb00423.x) [Google Scholar]
- Pagel M.1999Inferring the historical patterns of biological evolution. Nature 401, 877–884 (doi:10.1038/44766) [DOI] [PubMed] [Google Scholar]
- Pagel M., Meade A., Barker D.2004Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673–684 (doi:10.1080/10635150490522232) [DOI] [PubMed] [Google Scholar]
- Phillimore A., Orme C., Thomas G., Blackburn T., Bennett P., Gaston K., Owens I.2008Sympatric speciation in birds is rare: insights from range data and simulations. Am. Nat. 171, 646–657 (doi:10.1086/587074) [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P.2003MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- Sanderson M. J.2003r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19, 301–302 (doi:10.1093/bioinformatics/19.2.301) [DOI] [PubMed] [Google Scholar]
- Schaffner J. V., Jr1939Neodiprion sertifer (Geoff.), a pine sawfly accidentally introduced into New Jersey from Europe. J. Econ. Entomol. 32, 887–888 [Google Scholar]
- Schluter D.2001Ecology and the origin of species. Trends Ecol. Evol. 16, 372–380 (doi:10.1016/S0169-5347(01)02198-X) [DOI] [PubMed] [Google Scholar]
- Schmidtling R. C.2003The southern pines during the Pleistocene. Acta Hort. 615, 203–209 [Google Scholar]
- Smith D. R.1979Symphyta. In Catalog of Hymenoptera in America north of Mexico, vol. 1 (ed. Krombein K. V., et al.), pp. 3–137 Washington, DC: Smithsonian Institution Press [Google Scholar]
- Sorenson M. D., Sefc K. M., Payne R. B.2003Speciation by host switch in brood parasitic indigobirds. Nature 424, 928–931 (doi:10.1038/nature01863) [DOI] [PubMed] [Google Scholar]
- Stireman J. O., Nason J. D., Heard S. B.2005Host-associated genetic differentiation in phytophagous insects: general phenomenon or isolated exceptions? Evidence from a goldenrod-insect community. Evolution 59, 2573–2587 (doi:10.1554/05-222.1) [PubMed] [Google Scholar]
- Strong D. R., Lawton J. H., Southwood R.1984Insects on plants: community patterns and mechanisms. Cambridge, MA: Harvard University Press [Google Scholar]
- Tauber C. A., Tauber M. J.1981Insect seasonal cycles: genetics and evolution. Annu. Rev. Ecol. Syst. 12, 281–308 (doi:10.1146/annurev.es.12.110181.001433) [Google Scholar]
- Thompson J. N.1998The evolution of diet breadth: monophagy and polyphagy in swallowtail butterflies. J. Evol. Biol. 11, 563–578 (doi:10.1046/j.1420-9101.1998.11050563.x) [Google Scholar]
- Turelli M., Barton N. H., Coyne J. A.2001Theory and speciation. Trends Ecol. Evol. 16, 330–343 (doi:10.1016/S0169-5347(01)02177-2) [DOI] [PubMed] [Google Scholar]
- Via S.1999Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution 53, 1446–1457 [DOI] [PubMed] [Google Scholar]
- Via S.2001Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 16, 381–390 (doi:10.1016/S0169-5347(01)02188-7) [DOI] [PubMed] [Google Scholar]
- Wang Y., Hey J.2010Estimating divergence parameters with small samples from a large number of loci. Genetics 184, 363–379 (doi:10.1534/genetics.109.110528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L. F., Wilkinson R. C., Averill R. C.1992Redheaded pine sawfly: its ecology and management. Washington, DC: USDA [Google Scholar]
- Won Y. J., Hey J.2005Divergence population genetics of chimpanzees. Mol. Biol. Evol. 22, 297–307 (doi:10.1093/molbev/msi017) [DOI] [PubMed] [Google Scholar]
- Xie X., Rull J., Michel A. P., Velez S., Forbes A. A., Lobo N. F., Aluja M., Feder J. L.2007Hawthorn-infesting populations of Rhagoletis pomonella in Mexico and speciation mode plurality. Evolution 61, 1091–1105 (doi:10.1111/j.1558-5646.2007.00091.x) [DOI] [PubMed] [Google Scholar]