Calponin 3 (CNN3), an actin binding molecule, negatively regulates trophoblast differentiation and fusion. CNN3 phosphorylation modulates the actin-binding capacity of CNN3 and most probably regulates cytoskeleton remodeling that renders cells capable of undergoing fusion.
Abstract
Cell–cell fusion is an intriguing differentiation process, essential for placental development and maturation. A proteomic approach identified a cytoplasmic protein, calponin 3 (CNN3), related to the fusion of BeWo choriocarcinoma cells. CNN3 was expressed in cytotrophoblasts in human placenta. CNN3 gene knockdown promoted actin cytoskeletal rearrangement and syncytium formation in BeWo cells, suggesting CNN3 to be a negative regulator of trophoblast fusion. Indeed, CNN3 depletion promoted BeWo cell fusion. CNN3 at the cytoplasmic face of cytoskeleton was dislocated from F-actin with forskolin treatment and diffused into the cytoplasm in a phosphorylation-dependent manner. Phosphorylation sites were located at Ser293/296 in the C-terminal region, and deletion of this region or site-specific disruption of Ser293/296 suppressed syncytium formation. These CNN3 mutants were colocalized with F-actin and remained there after forskolin treatment, suggesting that dissociation of CNN3 from F-actin is modulated by the phosphorylation status of the C-terminal region unique to CNN3 in the CNN family proteins. The mutant missing these phosphorylation sites displayed a dominant negative effect on cell fusion, while replacement of Ser293/296 with aspartic acid enhanced syncytium formation. These results indicated that CNN3 regulates actin cytoskeleton rearrangement which is required for the plasma membranes of trophoblasts to become fusion competent.
INTRODUCTION
Cellular fusion is a dramatic biological event observed in a wide variety of organisms. The fusion process has been studied independently in different species and cells: yeast, C. elegans epidermal cells, myoblasts, macrophages, and trophoblasts, as well as during both physiological and pathological events such as fertilization, tumorigenesis, and tissue regeneration (Chen and Olson, 2005). Furthermore, virus- or chemical-induced cell–cell fusion is currently an indispensable tool for studying gene expression, chromosomal mapping, antibody production, and cancer immunotherapy. Although the mechanisms underlying cellular fusion are not fully understood, some fusogens and transcription factors participating in cell type–specific processes have been identified; e.g., a fusogenic membrane protein called syncytin and transcription factor GCMa (glial cell missing) are known to be required for placental development (Mi et al., 2000; Schreiber et al., 2000; Yu et al., 2002). In addition to Eff-1 and FUS-1 in C. elegans epithelial cell fusion, Duf, Rst, and other immunoglobulin (Ig) domain-containing transmembrane proteins are essential for muscle cell fusion and development (Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001; Mohler et al., 2002; Kontani et al., 2005; Sohn et al., 2009). While most of these molecules are specific to certain systems, a few such as the guanine-nucleotide exchange factors (e.g., Dock180 and Brag2) are involved in myoblast as well as macrophage fusion (Pajcini et al., 2008). However, the dynamics of cell–cell adhesion, alignment, and membrane mixing are apparently quite similar among the fusion processes of these cell types, and it is therefore conceivable that cytoskeletal rearrangement is finely regulated during the course of cell fusion.
Calponin was originally identified as a molecule binding to F-actin, calmodulin, and tropomyosin and to be involved in regulating the contraction/relaxation cycle in smooth muscle cells (Takahashi et al., 1988; Takahashi and Nadal-Ginard, 1991). Three CNN family proteins are characterized by the N-terminal calponin homology domain (CHD) and a middle region containing actin binding site 1 (ABS1) and ABS2, and are distinguished from each other by their unique C-terminal tails (Takahashi and Nadal-Ginard, 1991; Strasser et al., 1993; Applegate et al., 1994; Morgan and Gangopadhyay, 2001; Rozenblum and Gimona, 2007): basic, neutral, and acidic CNNs, or CNN1, CNN2, and CNN3, respectively. In this study we carried out a proteomic search for fusion-related molecules in BeWo choriocarcinoma cells and found calponin3 (CNN3) to be required for cellular fusion. This novel function of CNN3 was associated with actin cytoskeletal rearrangement regulated by phosphorylation of the CNN3-specific C-terminal region.
MATERIALS AND METHODS
The following reagents and antibodies were used: mouse monoclonal anti-E-cadherin IgG from BD Bioscience (Franklin Lakes, NJ); mouse monoclonal anti-desmoplakin 1/2 IgG from Progen (Heidelberg, Germany); mouse polyclonal anti-human CNN3 IgG from Abnova (Taipei, Taiwan), mouse monoclonal anti-FLAG (M2) IgG, and mouse monoclonal anti-actin IgG from Sigma (St. Louis, MO); rabbit polyclonal anti-CNN3 IgG and anti–syncytin-1 from Santa Cruz (Santa Cruz, CA); rabbit polyclonal anti-FLAG IgG from Cell Signaling (Danvers, MA); ProQ-Diamond Phosphoprotein Gel Stain and SYPRO Ruby from Molecular Probes (Eugene, OR); lysylendopeptidase (Achromobacter lyticus protease I) from Wako (Osaka, Japan); trypsin (Sequence Grade Modified Trypsin, from porcine pancreas) from Promega (Madison, WI).
Phospho-Specific CNN3 Antibodies
Anti-CNN3 pS293 and pS296 rabbit antibodies were raised against phosphorylated peptides: N′-CQGTGTNG(phos)SEI; and N′-EISD(phos)SDYQAEC (MBL, Nagoya, Japan). Antibodies were affinity-purified from serum by using the corresponding phosphorylated peptide-coupled agarose beads. The phospho-specific antibodies were then affinity-purified by immunoadsorption with nonphosphorylated peptides. The specificities of the resulting antibodies were verified by ELISA.
Cloning and Site-Directed Mutagenesis of Human CNN3
Human CNN3 cDNA was amplified from the random-primed in-house cDNA library of BeWo cells (American Type Culture Collection, Manassas, VA) and inserted into a XhoI/EcoRI site of pENTR/flag to generate N-terminal Flag-tagged CNN3, or a XhoI/BamHI site of EYFP-C1 (Clontech, Mountain View, CA) to generate EYFP-CNN3. C-terminal deletion (ΔC) or site-directed mutagenesis was performed using a KOD-Plus Mutagenesis kit (TOYOBO, Osaka, Japan) according to the manufacturer's protocol. For the ΔC mutant, a stop codon followed by an EcoRI site was introduced by PCR.
Cell Culture, Treatment, Transfection, and Transduction of Lentivirus Vectors
BeWo cells constitutively expressing fluorescent protein (CFP-Nuc or DsRed) were maintained in an undifferentiated state in F12 Ham medium (Wako) supplemented with 10% fetal bovine serum (FBS). Differentiation was induced by treatment with 50 μM forskolin (Wako), for up to 96 h (Wice et al., 1990; Lyden et al., 1993; Keryer et al., 1998) with daily exchange of the forskolin-containing medium. Recombinant lentiviruses kindly provided by Dr. Hiroyuki Miyoshi (RIKEN BRC, Ibaraki, Japan) were produced by transient transfection of HEK293T cells according to reported protocols (Zufferey et al., 1997; Miyoshi, 2004). Briefly, subconfluent HEK293T cells were cotransfected with 20 μg of a self-inactivating (SIN) vector, 10 μg of a pCAG-HIVgp and 10 μg of a pCMV-VSV-G-RSV-Rev by calcium phosphate precipitation. The medium was changed 18h later, and the recombinant lentivirus vectors were harvested after an additional 48h incubation.
Detection of CNN3 in Placenta
Human placentas at 21 and 37 wk gestation were obtained, under informed consent, after elective abortion and Caesarian section, respectively, and transferred to our laboratory after anonymization according to the Ethical Guidelines for Clinical Research in Osaka Medical Center and Research Institute for Maternal and Child Health. The placentas were cut into small pieces, which were then fixed with formaldehyde and paraffin-embedded. Mouse placentas were isolated at 8.5–12.5 dpc and subjected to the same fixation and embedding procedures as the human placentas. For immunohisotochemistry, rabbit polyclonal IgG against human CNN3 was used for these human and mouse tissues, and peroxidase-based color development was performed using ABC and AEC (3-amino-9-ethylcarbazole) kits (Vector, Burlingame, CA).
Detection of Cell Fusion in BeWo Cells
After fixation with 4% paraformaldehyde, cells were immunostained for E-cadherin and desmoplakin with monoclonal antibodies, and cell nuclei were counterstained with DAPI. Photographs were taken with an Olympus microscope IX51 (Tokyo, Japan) equipped with a cooled charge-coupled device camera, and the images were prepared for presentation using Q-Capture Pro. For BeWo or BeWo/HEK293 cell fusion, the cells constitutively expressing DsRed and CFP-Nuc were harvested by trypsinization, mixed at a ratio of 1:1 for BeWo or 2:1 for BeWo/HEK293 and then seeded in microplate wells (22 mm diameter) at a density of 2 × 104 cells/ml. After cell adhesion, the cells were fed daily with forskolin-containing media. The fusion frequency was counted as fusion induction, represented by the number of nuclei in DsRed and CFP-Nuc double positive syncytia against the total number of nuclei in the DsRed-positive cells, as determined by microscopic observation (×100 magnification). The degree of maturation was calculated as the fusion index, which was represented by the average number of nuclei per DsRed and CFP-Nuc double positive cells, as determined by microscopic observation (×100 magnification).
shRNA-Induced Degradation of CNN3
The shRNA-encoding lentiviral vectors were created by inserting the annealed complementary oligonucleotides into the BamHI and HindIII site located at the 3′ end of a human U6 promoter-containing the pENTER4 vector. The shRNAs in pENTR4 were then subcloned into a CSII-RfA-CMV-GFP or a CSII-RfA-CMV-CFP-Nuc vector by an L-R clonase reaction (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Purified lentiviruses allow simultaneous expression of both shRNA and marker genes. The sequences for the coding strand of shRNA were (5′–3′) GATCCGTCGACTGTGGATTGGCATCTGTGAAGCCACAGATGGGATGCCAATCCACAGTCGACTTTTTTA and (5′–3′) AGCTTAAAAAAGTCGACTGTGGATTGGCATCCCATCTGTGGCTTCACAGATGCCAATCCACAGTCGACG for controls, (5′–3′) GATCCGCACATGCTCGGAAGGAAACTGTGAAGCCACAGATGGGTTTCCTTCCGAGCATGTGCTTTTTTA and (5′–3′) AGCTTAAAAAAGCACATGCTCGGAAGGAAACCCATCTGTGGCTTCACAGTTTCCTTCCGAGCATGTGCG for hCNN3 shRNA1, (5′–3′) GATCCGGCTCAACACCTTGCTCATCTGTGAAGCCACAGATGGGATGAGCAAGGTGTTGAGCCTTTTTTA and (5′–3′) AGCTTAAAAAAGGCTCAACACCTTGCTCATCCCATCTGTGGCTTCACAGATGAGCAAGGTGTTGAGCCG for hCNN3 shRNA2.
Immunoblot Analysis
Cells were washed twice with ice-cold PBS, scraped, and lysed at 4°C in a buffer containing 20 mM Tris-HCl pH 7.2, 150 mM NaCl, 0.1% NP-40, 0.5% Triton-X 100, 5 mM EDTA, 1 mM Na3VO4, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml aprotinin, and 1 mM dithiothreitol (DTT) and then centrifuged at 15,000 × g for 15 min. The supernatants were collected and the protein concentrations were determined by the Bradford method (Bio-Rad, Hercules, CA). Equal amounts of proteins were loaded on a 10% SDS-PAGE gel, and then transferred to PVDF membranes (Schleicher & Schuell, Dassel, Germany). The membrane was incubated with primary and secondary antibodies for 1h each and detection was performed using an ECL kit (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions.
Purification of CAPMPs from the Apical-PM Protein Fraction
PMs from BeWo cells were isolated using a cationic colloidal silica method (Chaney and Jacobson, 1983; Ghitescu et al., 1997; Stolz et al., 1999; Ghitescu et al., 2001; Rahbar and Fenselau, 2004). The cells were washed twice with ice-cold PBS (−), then with PM coating (PMC) buffer (0.5 mM CaCl2, 1 mM MgCl2, 20 mM MES, 135 mM NaCl, pH 5.3). The cells were next incubated in a 5% suspension of cationic colloidal silica (Sigma-Aldrich, St. Louis, MO) in PMC buffer on ice for 1 min. The solution phase was removed, and the cells were washed with PMC buffer to remove excess silica. The cells were then incubated in a 10 mg/ml polyacrylic acid (Sigma) solution with PMC buffer at pH 6.1 on ice for 1 min. The polyacrylic acid solution was removed, and the cells were washed with PMC buffer. The cells were washed briefly in lysis buffer (2.5 mM imidazole, pH 7.0) and then incubated with lysis buffer containing a protease inhibitor (1 mM PMSF and 50 μg/ml aprotinin) on ice for 30 min. The cells were scraped and the cell suspension was recovered and homogenized by 20 strokes with a Dounce homogenizer. The lysate was mixed with an equal volume of 100% Nycodenz and then placed over a 1.0 ml layer of 70% Nycodenz. The tube was topped off with lysis buffer and then centrifuged in a swing bucket at 20,000 × g for 30 min. After removal of the layer containing nuclei, the pellet containing silica-coated PMs was washed three times with lysis buffer. CAPMPs were extracted from the silica-coated PMs by incubation in 100 mM Na2CO3, at pH 11.4 on ice for 30 min followed by centrifugation at 12,000 × g for 10 min (Hubbard and Ma, 1983; Ghitescu et al., 1997). Back extraction was performed once with the same procedure except for omission of the 10 min centrifugation.
Two-Dimensional Electrophoresis (2-DE)
The CAPMPs were precipitated with trichloroacetic acid (TCA) and then washed with ethanol. The pellet was solubilized in 2-DE buffer (7M urea, 2M thiourea and 4% CHAPS), and a 40 μg sample was subjected to 2-DE using an IPG strip (13 cm, pH4-7L) (GE Healthcare) for the first dimension separation. Fluorescent detection was carried out for phosphoproteins using ProQ-Diamond Phosphoprotein Gel Stain according to the manufacturer's protocol, and the images were acquired with a FluoroPhoreStar 3000 (Anatech, Tokyo, Japan). Subsequently, the gels were washed and stained with SYPRO Ruby to visualize total proteins.
RESULTS
Identification of Calponin 3 as a Fusion-Related Protein
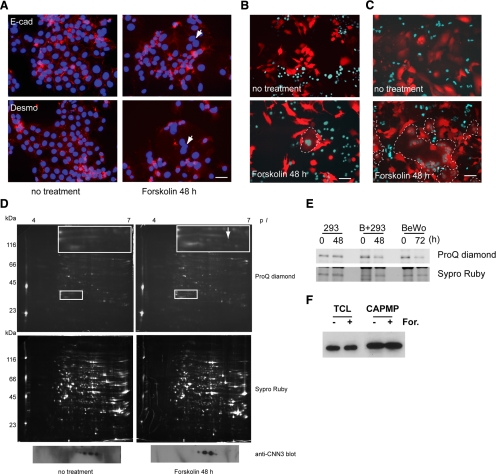
BeWo cells differentiate to form syncytiotrophoblasts via a cyclic AMP-dependent pathway after forskolin treatment (Wice et al., 1990; Keryer et al., 1998). Syncytium formation is usually observed by immunostaining for E-cadherin or desmoplakin localized at intercellular boundaries (Keryer et al., 1998), but discriminating between multi- and mononuclear cells is often quite difficult (Figure 1A). To overcome the problem, we developed a differential coloring system by using coculture [e.g., red fluorescent protein in the cytoplasm (DsRed cells) and enhanced-cyan fluorescent protein with a nuclear localization signal (ECFP-Nuc cells)] to visualize fusion by detecting DsRed/ECFP-Nuc-double positive cells (Figure 1B). DsRed/ECFP-Nuc-double positive syncytia were detected after 2-d forskolin treatment, but fusion efficiency was <5% for homotypic BeWo/BeWo cell fusion (Figure 1B). In contrast, heterotypic fusion of BeWo with HEK293 cells, which formed a huge hybrid syncytium, was found to be more efficient (Figure 1C), allowing robust proteomic analysis.
Figure 1.
Identification of CNN3 as a fusion-related protein. (A) BeWo cell fusion visualized by the conventional staining with anti–E-cadherin or anti-desmoplakin in combination with nuclear staining with DAPI. Cells were treated with or without 50 μM forskolin for 48 h. Arrows indicate a syncytium in which these marker proteins were translocated and diffused into the cytoplasm. Scale bar: 50 μm. (B) Detection of forskolin-induced syncytium between DsRed- and CFP-Nuc-expressing BeWo cells. A syncytium is easily recognized by the cyan-colored multiple nuclei within the homogeneously red-colored cytoplasm, whereas unfused cells have either red cytoplasm or cyan nuclei. Scale bar: 100 μm. (C) Heterologous fusion of CFP-Nuc–expressing HEK293 cells with BeWo cells expressing DsRed. The incidence of BeWo/HEK293 fusion was higher than that of homologous BeWo cell fusion, and the hybrid syncytium was much larger than that of BeWo-derived homologous syncytium. Scale bar: 250 μm. The broken lines in B and C show the periphery of the syncytium. (D) Cytoplasmically attached peripheral membrane proteins (CAPMPs) isolated from BeWo/HEK293 coculture were separated by 2-DE. The gels were stained with ProQ Diamond phospho-specific staining (top panels) or SYPRO Ruby protein staining (middle panels). A spot (arrow) among a series of protein spots in inset was subjected to in-gel digestion followed by peptide mass fingerprinting, and CNN3 was identified. All these spots in line were confirmed to be CNN3 by Western blotting (bottom panels). (E) Endogenous CNN3 was immunopurified using anti-CNN3 antibody from total cell lysate (TCL) of the cells incubated with 50 μM forskolin for the indicated times. The phosphorylation status of CNN3 was analyzed by ProQ Diamond phospho-specific staining, and the same gel was stained by Sypro Ruby to verify each sample equal in the amount of CNN3. “B + 293” indicates a BeWo/HEK293 mixed culture. (F) Western blot of CNN3 in the TCL or the CAPMP fraction from BeWo cells in the presence or absence of 50 μM forskolin for 72 h.
To identify fusion-related molecules in the plasma membrane (PM), “cytoplasmically attached peripheral membrane proteins” (CAPMPs) were isolated from the PM fraction using cationic colloidal silica (Chaney and Jacobson, 1983; Ghitescu et al., 1997). Two-dimensional electrophoresis (2-DE) followed by ProQ Diamond staining and peptide mass fingerprinting identified several cytoskeletal, transmembrane protein-associated molecules showing altered expressions, isoelectric points, and/or phosphorylation levels in response to forskolin treatment (Supplemental Table 1). Among them, we focused on CNN3, or acidic CNN (Supplemental Table 2), because this molecule was believed to be involved in cytoskeletal rearrangement and thereby to be related to membrane mixing and fusion. CNN3 was identified in at least five spots by 2-DE followed by Western blotting (Figure 1D, bottom panels). After forskolin treatment, the intensity of acidic spots was decreased, and ProQ Diamond staining suggested this change to be due to dephosphorylation (Figure 1D). To confirm the change in CNN3 phosphorylation levels with fusion induction, endogenous CNN3 molecules were immunoprecipitated and phospho-CNN3 was detected by ProQ-Diamond staining. The overall phosphorylation level of CNN3 was decreased in both homotypic BeWo/BeWo and heterotypic BeWo/HEK293 fusion, while CNN3 protein levels were unchanged (Figure 1, E and F). In the absence of BeWo cells, HEK293 cells did not fuse with each other (data not shown) and there were no other associated changes in CNN3 phosphorylation even with forskolin treatment (Figure 1E). These results suggest CNN3 to have multiple phosphorylation sites and that CNN3 undergoes dephosphorylation during BeWo cell fusion. RT-PCR analysis showed no expression of other members of the CNN family (i.e., CNN1 and CNN2, in BeWo or HEK293 cells) (Supplemental Fig. S1A).
In Vivo CNN3 Expression in the Placental Tissues of Humans and Mice
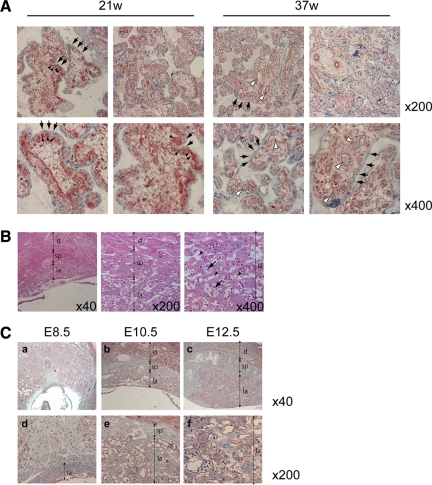
BeWo is a human choriocarcinoma cell line. In the human placenta, cytotrophoblasts fuse together to form syncytiotrophoblasts or fuse with syncytiotrophoblasts, replenishing the syncytium that covers the surfaces of chorionic villi. To investigate CNN3 expression in vivo, chorionic villi in human placenta were analyzed by immunohistchemistry. As shown in Figure 2A, CNN3 was expressed in cytotrophoblasts beneath the syncytiotrophoblast layer but not in this layer itself. In term placenta, the population of cytotrophoblasts is quite small, and CNN3 expression was observed only in endothelial cells in chorionic villi. This histological distribution indicated that CNN3 expression disappears during the course of tropoblastic cell differentiation.
Figure 2.
CNN3 expression in human and mouse placental tissues. (A) Immunohistochemistry of chorionic villi of human placentas at 21 and 37 wk of gestation. CNN3 was detected in the cytotrophoblasts (black arrowheads) and fetal endothelial cells (white arrowheads) but not in the syncytiotrophoblast layer (arrows). (B) Histology of mouse placenta. Transverse sections (4 μm) were stained with hematoxylin/eosin. A few maternal blood sinuses (arrowheads) and fetal blood vessels (arrows) are found within the labyrinth region. Localizations of the labyrinth layer (la), spongiotrophoblast layer (sp), and maternal decidua (d) are indicated. (C) Immunohistochemistry of mouse placental tissues at 8.5 dpc (a, d), 10.5 dpc (b, e), and 12.5 dpc (c, f). CNN3 was detected in the labyrinth layer and maternal deciduas.
In mice, the labyrinth is a tissue in which trophoblasts differentiate and fuse to form a syncytiotrophoblast layer (for review see Rossant and Cross, 2001). As shown in Figure 2, B and C, CNN3 was detected in both labyrinth and maternal deciduas at 10.5 dpc but not in the spongiotrophoblast layer at any time during placental development. CNN3 was expressed by fetal endothelial cells as well (Figure 2C, panel f). These observations suggested CNN3 to play a role in placental development in both human and mouse.
CNN3 Depletion by RNAi Enhances BeWo Cell Fusion
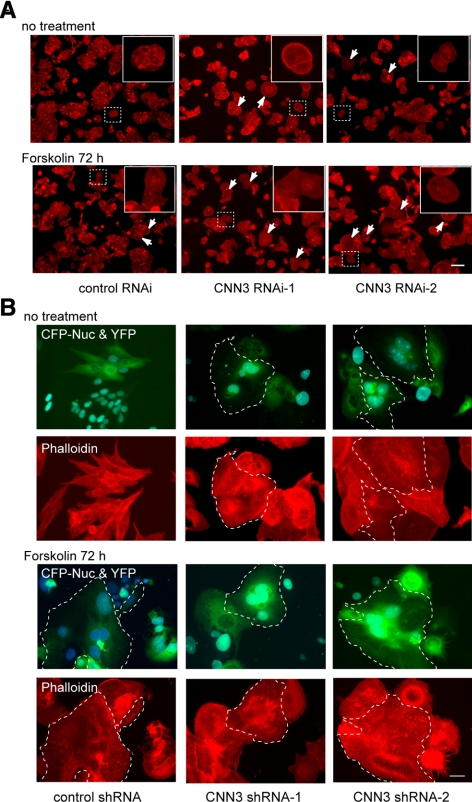
To study the role of CNN3 in trophoblastic cell fusion, lentivirus vector-based RNA-interference (RNAi) constructs, which allowed simultaneous expressions of a short hairpin RNA (shRNA) and a marker protein, either enhanced yellow-green fluorescent protein (EYFP) or ECFP-Nuc, were introduced into BeWo cells. The resulting depletion with either CNN3 RNAi sequence was at least 90%, as demonstrated by Western blotting (Supplemental Fig. S1B). Phalloidin staining revealed that the CNN3 gene knockdown induced morphological change in BeWo cells. In contrast to the cubic and island-like morphology of control cells, the CNN3-depleted cells had a broad, flat cytoplasm (Figure 3A). This observation of morphological change was supported by Phalloidin staining for F-actin. As shown in Figure 3A, actin bundles disappeared from CNN3-depleted cells even in the absence of forskolin. Furthermore, CNN3 depletion promoted syncytium formation even in the absence of forskolin and was further augmented by forskolin treatment (Figure 3, A and B), as were the morphological features described above. These results indicated that CNN3 prevents BeWo cells from undergoing cell–cell fusion.
Figure 3.
CNN3 gene knockdown. (A) Control or CNN3-specific shRNA was introduced into BeWo cells expressing EYFP or CFP-Nuc. After washing, different marker expressing cells were trypsinized and mixed in the culture in the absence or presence of 50 μM forskolin for 72 h. After fixation, cells were stained with Alexa Fluor 568-conjugated phalloidin (red). Scale bar: 40 μm. Arrows indicate the multinucleated cells identified by an EYFP/CFP-Nuc double-color system. (B) Higher magnification images of A. The cell morphology was visualized by Alexa Fluor 568-conjugated phalloidin staining. Broken lines show the periphery of the EYFP/CFP-Nuc double positive syncytium. Note that CNN3-depletion induced syncytium formation without forskolin treatment and increased the number of multinucleated cells after forskolin treatment. Scale bar: 100 μm.
Syncytin is a fusogenic protein playing a central role in trophoblast fusion in both humans and mice, and the expression is regulated by GCMa, a cAMP-responsible transcription factor (Mi et al., 2000; Frendo et al., 2003; Dupressoir et al., 2005; Dupressoir et al., 2009). In this study, syncytin-1 expression levels were decreased by forskolin treatment for BeWo cell fusion as reported by Vargas et al. (Vargas et al., 2009), and this syncytin-1 down-regulation was suppressed by CNN3 knockdown (Supplemental Fig. S1C). These findings suggested that CNN3 might regulate syncytin-1 expression or affect stabilization of this membrane protein. Only trophoblastic cell lines are able to induce GCMa and form syncytium in response to cAMP signaling (Yu et al., 2002), and thus neither morphological change nor syncytium formation was induced by CNN3 depletion in HEK293 cells (data not shown).
CNN3 Phosphorylation Sites Involved in Cell Fusion
The actin-binding property of CNN1 is regulated by kinase- and phosphatase-dependent phosphorylation/dephosphorylation of serine, threonine, or tyrosine residues. Two phosphorylation sites at Ser175 and Thr184 of CNN1 are conserved in this family (Figure 4A), and phosphorylation of these residues dissociates CNN1 from F-actin in vitro (Nakamura et al., 1993; Tang et al., 1996; Kaneko et al., 2000; Abouzaglou et al., 2004).
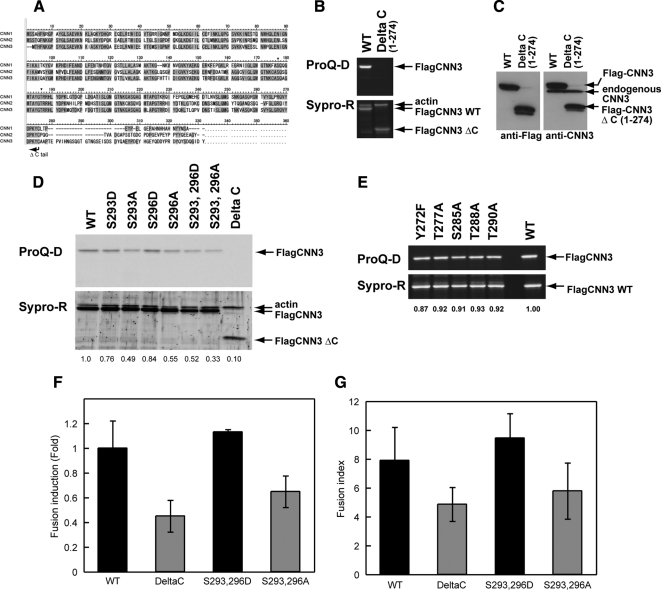
Figure 4.
CNN3 phosphorylation in BeWo cell fusion. (A) Comparison of the amino acid sequences of human CNN family proteins. CNN1 phosphorylation sites are indicated by asterisks. The sequence of ΔC mutant is truncated after Cys274 (arrow). (B) HEK293 cells transiently expressing WT or ΔC CNN3. Flag-CNN3 with a wild or Δ C sequence was recovered by immunoprecipitation from cell lysate using anti-Flag-agarose beads. The eluate was subjected to SDS-PAGE, and the phosphorylated CNN3s were visualized by ProQ Diamond phospho-specific staining (upper panel) followed by Sypro Ruby protein staining (lower panel). Note that the ΔC mutant was not phosphorylated but retained the actin-binding property. (C) Flag-tagged WT or ΔC CNN3 identified by Western blotting using anti-FLAG (left panel) or anti-CNN3 (right panel) antibodies. Each 20-μg sample of HEK293 cell lysate was loaded. (D and E) Phosphorylation levels of WT or mutant Flag-CNN3 transiently expressed in HEK293 cells. Flag-CNN3 was immunopurified using anti–Flag-agarose beads and was subjected to SDS-PAGE followed by ProQ Diamond phospho-specific staining. The ratio of phosphorylated and total CNN3 was calculated and indicated below each lane, showing decreased phosphorylation of S293A and S296A CNN3s. (F and G) BeWo cell fusion at 96 h after forskolin treatment in various mutants. The fusion frequency was counted as “fusion induction” (F), and the degree of maturation was calculated as “fusion index” (G). Data from three independent experiments were averaged, and standard deviations are indicated by error bars.
To determine CNN3 phosphorylation sites, Flag-CNN3 was recovered from HEK293 cells and subjected to tryptic digestion. Mass spectrometry (MS) of the phosphopeptides, enriched by TiO2 (Sugiyama et al., 2007), identified at least three, two major and one minor, phosphorylation sites in the peptide corresponding to residues 272–318 (Supplemental Fig. S2, A–E) and two phosphorylation sites in a smaller peptide corresponding to residues 272–298 (Supplemental Fig. S2, F and G). These peptides are located in the acidic tail which is unique to CNN3 in the CNN family, suggesting this specific region to be involved in the regulation of fusion. Indeed, the mutant (ΔC) lacking the C-terminal sequence from residue 274 was negative for ProQ Diamond phospho-specific staining (Figure 4, B and D), indicating the phosphorylation sites to be located exclusively within the C-terminal region. Sypro Ruby staining revealed similar amounts of Flag-tagged WT or ΔC CNN3 to be recovered by immunoprecipitation using anti-Flag-agarose beads. In Figure 4B, a 40-kDa protein coprecipitated with Flag-tagged WT or ΔC CNN3 was identified as actin by peptide mass fingerprinting (PMF) analysis. The ΔC mutant retained its actin-binding property, because the ABS1 and ABS2 remained intact (Danninger and Gimona, 2000). Indeed, actin amounts in the immunoprecipitates were similar in cells transfected with wild-type or ΔC CNN3 before forskolin treatment.
Subsequently, to identify the phosphorylation sites of CNN3 (Supplemental Fig. S2, G and H for sequence), various Flag-CNN3 mutants were transiently expressed in HEK293 cells, and Flag-CNN3 was recovered by immunoprecipitation using anti-Flag agarose beads. The phosphorylation levels of each CNN3 mutant were evaluated by ProQ Diamond staining. Among the putative phosphorylation sites within the 272–298 region, the substitution of Ser293 or Ser296 with Ala decreased phosphorylation levels of CNN3 and these mutants also retained actin binding properties (Figures 4D and 4E). S293/296A double mutation drastically reduced CNN3 phosphorylation (Figure 4D and Supplemental Fig. S2J). S293/296 phosphorylation was also identified in BeWo cells (data not shown). None of the point mutations at other putative S/T/Y phosphorylation sites altered phosphorylation status (Figure 4E), indicating Ser293 and Ser296 to be the major phosphorylation sites in the C-terminal region of CNN3.
To address whether CNN3 phosphorylation is involved in cell fusion and syncytium formation, ΔC, S293/296A, or S293/296D was introduced into BeWo cells expressing DsRed or CFP-Nuc, and the ratio of the numbers of DsRed/CFP-Nuc double positive cells to total DsRed positive cells was calculated. As shown in Figure 4F, ΔC or S293/296A mutation significantly suppressed forskolin-induced cell fusion via a dominant negative effect, while the cells expressing S293/296D showed retention of fusion efficiency. The number of nuclei in the syncytium, or fusion index, decreased by 30–40% in cells expressing ΔC or S293/296A and increased by 20% in those expressing S293/296D (Figure 4G). The effects of S293A and S296A mutations were additive (data not shown), and ΔC exerted the most prominent suppression on cell fusion.
Several lines of evidence indicate that CNN1 is phosphorylated by multiple kinases including Rho-associated kinase (ROCK), Erk, CaMK, and PKC (Winder and Walsh, 1990; Kaneko et al., 2000). To identify CNN3-specific kinases, we performed a search using NetPosK (http://www.cbs.dtu.dk/services/) and found casein kinase (CK) 1/2 to be a candidate kinase for Ser296 phosphorylation. We then focused on ROCK as well as CK and carried out an in vitro phosphorylation assay using a rCNN3 substrate. ProQ Diamond phospho-specific staining revealed that the active forms of both ROCK2 and CK1 were capable of phosphorylating CNN3. Ser293 was preferentially phosphorylated by ROCK rather than CK1, while the overall phosphorylation activity of CK1 was higher than that of ROCK2 (Supplemental Fig. S1G). However, pretreatment with specific inhibitors against these kinases did not change the constitutive CNN3 phosphorylation in unstimulated BeWo cells (data not shown).
Phosphorylation Status and Subcellular Distribution of Endogenous CNN3 in Cell Fusion
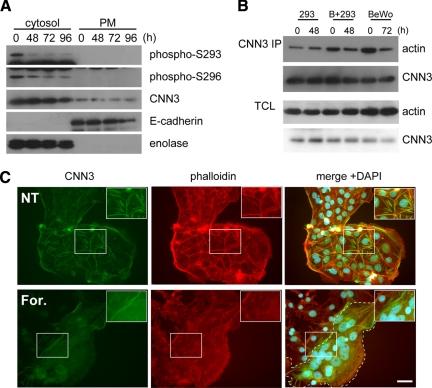
We prepared antibodies separately recognizing phosphoserine at either Ser293 or Ser296 (See Supplemental Fig. S3 for properties of these antibodies). The anti-pSer293 antibody was much less reactive to Ser296A mutant than Ser296D mutant or wild-type CNN3 (Supplemental Figure 3E), suggesting that Ser296 phosphorylation preceded Ser293 phosphorylation. Next, we analyzed site-specific phosphorylation of endogenous CNN3 from different subcellular fractions. With forskolin treatment, cytosolic CNN3 was dephosphorylated at these residues (Figure 5A), and phospho-S293 spots recovered from CAPMP fraction were sifted to the basic side (Supplemental Figure 1H). These results were consistent with the reduction of total CNN3 phosphorylation levels shown in Figure 1E.
Figure 5.
Phosphorylation status and subcellular distribution of endogenous CNN3. (A) Site-specific phosphorylation of CNN3. Each 20-μg sample of the cytosolic or plasma membrane (PM) fraction from BeWo cells was subjected to SDS-PAGE followed by Western blotting using antibodies against specific phosphorylation site at S293 or S296 (see Supplemental Fig. S3 for their specificity). Enolase (cytosol) and E-cadherin (PM) were used as the control for cell fractionation. (B) Coimmunoprecipitation of actin with CNN3. Endogenous CNN3 was immunopurified from BeWo and/or HEK293 cells after forskolin treatment. Immuno-purified CNN3 or total cell lysates (TCL) were analyzed by Western blotting for actin or CNN3. (C) Dissociation of endogenous CNN3 from actin cytoskeleton in BeWo cell fusion. CNN3 was immunostained using rabbit anti-CNN3 IgG, and the cells were visualized by Alexa Fluor 488-conjugated secondary antibody (green). Subsequently, Alexa Fluor 568-conjugated phalloidin (red) and DAPI staining was performed (blue). Endogenous CNN3 in multinucleated BeWo cells was dissociated from actin cytoskeleton after forskolin treatment. Broken line shows the periphery of syncytium. Scale bar: 50 μm.
In Figure 5B, the amount of actin bound to endogenous CNN3 was markedly decreased in both homotypic (BeWo/BeWo) and heterotypic (BeWo/HEK293) cell fusion. This result was consistent with that of Flag-tagged CNN3 shown in Figure 4. Indeed, endogenous CNN3 was well colocalized with F-actin before forskolin treatment and diffused into the cytoplasm in the syncytium after stimulation, as demonstrated by microscopic observations (Figure 5C).
Binding of CNN3 to Actin Cytoskeleton Is Regulated by CNN3 C-Terminal Region
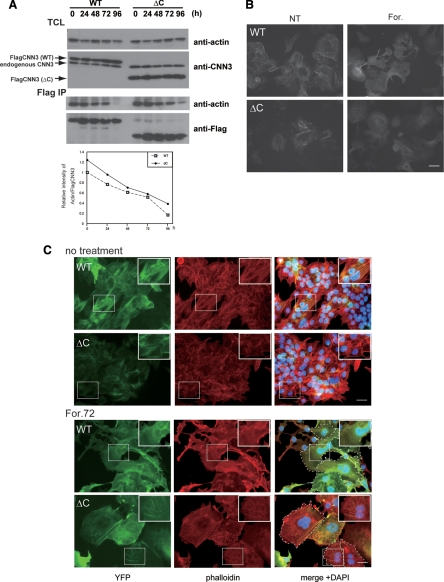
Implications of S293/296 phosphorylation in cytoskeletal rearrangement and cell fusion were then investigated using BeWo cells expressing ΔC mutant. Compared with the wild-type CNN3 transfectant, a significant amount of actin remained bound to ΔC CNN3 at 96 h after forskolin treatment, though dissociated from actin in a time-dependent manner (Figure 6A). The levels of actin and ΔC CNN3 in the total cell lysate were unchanged (Figure 6A, upper panels). This result was consistent with the previous reports showing the tail region of CNNs to negatively regulate actin binding capacity (Danninger and Gimona, 2000; Burgstaller et al., 2002). Microscopic observations of living cells revealed the YFP-tagged ΔC mutant as well as the wild-type CNN3 to be localized at stress fiber-like bundles and the cytoplasmic face of the PM before forskolin treatment (Figure 6B left panels). After stimulation, wild-type CNN3 diffused into the cytoplasm, but the ΔC mutant remained at the cytoskeletal bundles (Figure 6B right panels). Phalloidin staining demonstrated colocalization of wild or ΔC CNN3 with F-actin before forskolin treatment (Figure 6C). The wild-type CNN3 was not colocalized with F-actin in multinucleated cells after stimulation, indicating dissociation from the actin cytoskeleton. The S293/296D CNN3 underwent a similar dislocation (Supplemental Fig. S4). In contrast, ΔC CNN3 remained at the actin cytoskeleton even in the multinucleated cells, which were found only a few among the ΔC CNN3-expressing cells and retained F-actin bundles (Figure 6C). The Ser293/296A CNN3 behaved in a similar manner to the ΔC mutant, and the F-actin cytoskeleton remained after forskolin treatment (Supplemental Fig. S4). These results indicated that the phosphorylated Ser293/296 are required for dissociation of CNN3 from actin cytoskeleton and thus for efficient trophoblastic cell fusion.
Figure 6.
Phosphorylation-dependent association of CNN3 with actin cytoskeleton. (A) Association of ΔC CNN3 with actin. Wild-type (WT) or ΔC Flag-CNN3 expressing BeWo cells were treated with forskolin for the indicated times, and association of CNN3 and actin was analyzed by co-IP assay using anti-Flag agarose beads. Note that a significant amount of ΔC CNN3 remained in association with actin at 96h after forskolin treatment, compared with WT CNN3. (B) Subcellular localization of EYFP-CNN3. WT or ΔC EYFP-CNN3 was introduced into BeWo cells, and the localization of EYFP-CNN3 was visualized in the presence or absence of forskolin in living cells. (C) Colocalization of CNN3 and F-actin. BeWo cells expressing EYFP-CNN3 were cultivated in the presence (lower panels) or absence (upper panels) of 50 μM forskolin for 72 h. After fixation, the cells were stained by Alexa fluor 568-conjugated phalloidin (red) and DAPI (blue). Note that WT EYFP-CNN3 expressing syncytium did not colocalize with F-actin after forskolin treatment, but ΔC EYFP-CNN3 still overlapped with the F-actin even in multinucleated cells. The dotted lines show the periphery of the syncytium. Scale bar: 50 μm.
DISCUSSION
Three CNN family proteins have an N-terminal CHD followed by a region containing actin binding sites, and the C-terminal region specifies each isoform. CNN1 is involved in regulating the contraction/relaxation cycle in smooth muscle cells. Binding of CNN1 to the actin cytoskeleton inhibits actomyosin ATPase activity (el-Mezgueldi et al., 1996), and is regulated by phosphorylation at the conserved residues in the ABS region by protein kinase C or Ca2+/calmodulin dependent protein kinase II (Winder and Walsh, 1990; Kaneko et al., 2000). Gene targeting of CNN1 results in ectopic bone formation, suggesting a negative role of CNN1 in osteogenesis (Yoshikawa et al., 1998). CNN2 is involved in macrophage differentiation, and other CNN isoforms do not compensate for loss of this CNN2 function (Huang et al., 2008). These reports indicate the CNN family to be involved in differentiation as well as contractile mechanics and cellular motility. In the present study, CNN3 was shown to participate in the cytoskeletal reorganization necessary for trophoblastic fusion, playing a role similar to that of CNN2 in cytoskeletal rearrangement and migration of endothelial cells during vascular development (Fukui et al., 1997; Tang et al., 2006). Depletion of CNN3 by RNAi enhanced cell fusion of BeWo cells, suggesting that CNN3 impedes the cytoskeletal rearrangement necessary for cell fusion by binding F-actin.
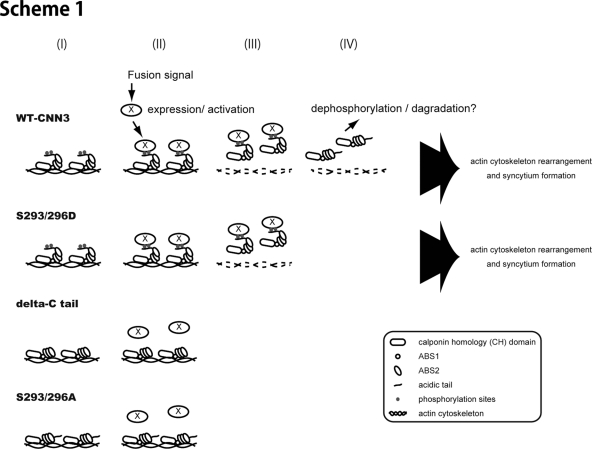
CNN binding to the actin cytoskeleton is conferred by ABS1 and ABS2 and the C-terminal region negatively regulates this basal property of CNN as demonstrated for CNN1 and CNN2 (Danninger and Gimona, 2000; Burgstaller et al., 2002). We examined the C-terminal region of CNN3 and found the phosphorylation status at S293/296 to regulate CNN3 binding to the actin cytoskeleton and thereby cytoskeletal remodeling, which renders the PM flexible and cells competent to fuse. In fact, cells expressing ΔC and S293/296A mutants which lack this regulatory system displayed decreased fusion efficiency. The phenotype of cells expressing S293/296A CNN3 was mild compared with those expressing ΔC CNN3, probably due to minor phosphorylation sites remaining in the C-terminal region. On the other hand, cells expressing the S293/296D mutant displayed a fusion efficiency similar to that of those expressing wild-type CNN3. The S293/296D CNN3 was localized at F-actin bundle and actin cytoskeleton. After forskolin treatment, the cells expressing S293/296D CNN3 underwent normal processes including F-actin bundle disappearance, and the actin cytoskeleton rearrangement. In unstimulated BeWo cells, the role of Ser293/296 phosphorylation would be to keep actin cytoskeleton flexible for de novo actin polymerization and/or treadmilling. Phosphorylated CNN3 is colocalized with F-actin and undergoes release, recycling, and rebinding, while lack of C-terminal phosphorylation sites, or the regulatory tail, enhances and stabilizes the CNN3 binding to actin and reduces actin dynamics and membrane flexibility. On stimulation by forskolin, factor (or signal) “X” recognizes or binds to the phosphorylated C-terminal region, which regulates the affinity of the actin binding region to actin, and thereby CNN3 is released from F-actin. The overall process involving S293/296 phosphorylation is illustrated in Scheme 1; (i) In unstimulated cells, CNN3 is constitutively phosphorylated at Ser293 and Ser296 and is associated with F-actin cytoskeleton via actin-binding sites. (ii, iii) After fusion induction, factor (or signal) “X” is induced/activated and binds to phosphorylated CNN3, which is then released from actin cytoskeleton. (iv) Dephosphorylation of S293/296 or degradation of CNN3 occurs after dissociation from actin. Up-regulation of phosphatases might be involved in the last phase. The dephosphorylated CNN3 is not recycled, because F-actin bundle, or the binding target, rapidly disappears in the forskolin-treated, fused, or unfused, BeWo cells. In summary, CNN3 phosphorylation probably plays distinct roles of regulating actin binding in a steady-state condition and of receiving factor “X” after fusion induction.
Scheme 1.
Model of CNN3-dependent trophoblast fusion (I) In unstimulated cells, CNN3 is constitutively phosphorylated at Ser293 and Ser296 in the regulatory tail and is associated with F-actin cytoskeleton via actin-binding sites. (II) When cells receive a fusion signal, factor (or signal) “X” is induced/activated and binds to (or recognizes) the phosphorylated regulatory tail and triggered CNN3 dissociation from actin. The phosphomimetic S293/296D mutant responds to the signal, while ΔC and S293/296A are unresponsive and remain attached to F-actin. (III) After CNN3 release, actin bundles are rearranged and then disappeared. (IV) Released CNN3 is dephosphorylated or degradaded in cytoplasm.
In human placenta, CNN3 expression was markedly decreased in the course of trophoblastic cell differentiation (Figure 2A), indicating CNN3 undergoes down-regulation by decreased gene expression or protein degradation after cell fusion in vivo. It has been reported that CNN2 synthesis and degradation were regulated by mechanical tension (Hossain et al., 2006) and that CNNs 1 and 3 were cleaved by μ-calpain (Tsunekawa et al., 1989; Yoshimoto et al., 2000). In BeWo cells, CNN3 might be cleaved and down-regulated in mature syncytium, although we did not study the fate of CNN3 after cell fusion in vitro.
In the present study, forskolin promoted dissociation of CNN3 from actin in BeWo cells, but had the opposite effect in HEK293 cells (Figure 4A). This discrepancy is probably due to a difference in motility between these cells. The cellular motility and membrane ruffling of HEK293 cells in culture are inherently much faster than those in BeWo cells, and forskolin accelerates the movement of HEK293 cells while decelerating that of BeWo cells (our unpublished observations with time-lapse imaging).
ROCK, protein kinase C, CaM kinase, and Src family kinases have been reported to phosphorylate Ser, Thr, or Tyr residues of CNN1 (Nakamura et al., 1993; Tang et al., 1996; Kaneko et al., 2000; Abouzaglou et al., 2004). In addition, a line of studies demonstrated the involvement of MAP kinase family or ROCK in labyrinth formation (for review see Rossant and Cross, 2001; Hatano et al., 2003; Thumkeo et al., 2003). In our preliminary study, ROCK1/2 and ERK1/2 were capable of phosphorylating S293 of CNN3 in vitro, while casein kinase phosphorylated other residues (Supplemental Fig. S1H). On the other hand, it is likely that specific phosphatases are also involved in this mechanism, as protein phosphatases 1 and 2B are responsible for CNN1 dephosphorylation (Ichikawa et al., 1993; Fraser and Walsh, 1995). Indeed, Vargas et al. reported acceleration of trophoblast fusion with inhibition of tyrosine phosphatase (Vargas et al., 2008), and we identified altered expressions of protein phosphatase(s) alpha/beta according to BeWo cell fusion (Supplemental Table 1).
Membrane flexibility is necessary for membrane fusion, though it is not a sufficient condition. In the present study, a novel function of CNN3 in trophoblastic cell fusion was involved in cytoskeletal dynamics, which impacts on plasma membrane flexibility and mixing, consequently on cell fusion. This novel function of CNN3 was identified in myoblasts as well as trophoblasts and works in myotube formation (our unpublished observations), suggesting that regulation of actin cytoskeletal rearrangement by CNN3 is required for these cells to become fusion competent. This regulatory mechanism probably underlies a wide range of cellular events and manipulations including virus-cell fusion, hybridoma production, and stem cell–mediated tissue regeneration.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Katsuhito Takahashi for helpful insights on properties of the CNN family and Dr. Masahiro Nakayama for helpful comments on the histology of the human placenta. This work was supported in part by Grants-in-Aid for Young Scientists (B) (17790273) from Japan Society for the Promotion of Science and from Takeda Science Foundation.
Abbreviations used:
- ABS
Actin binding site
- CAPMP
cytoplasmically attached peripheral membrane protein
- CHD
calponin homology domain
- CK
casein kinase
- CNN3
calponin 3
- DMEM
Dulbecco's modified Eagle's medium
- DsRed
red fluorescent protein
- DTT
dithiothreitol
- ECFP
enhanced cyan fluorescent protein
- EYFP
enhanced yellow fluorescent protein
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HS
horse serum
- PBS
phosphate-buffered saline
- PM
plasma membrane
- PMSF
phenylmethylsulfonyl fluoride
- shRNA
short hairpin RNA.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0261) on September 22, 2010.
REFERENCES
- Abouzaglou J., Benistant C., Gimona M., Roustan C., Kassab R., Fattoum A. Tyrosine phosphorylation of calponins: inhibition of the interaction with F-actin. Eur. J. Biochem. 2004;271:2615–2623. doi: 10.1111/j.1432-1033.2004.04190.x. [DOI] [PubMed] [Google Scholar]
- Applegate D., Feng W., Green R. S., Taubman M. B. Cloning and expression of a novel acidic calponin isoform from rat aortic vascular smooth muscle. J. Biol. Chem. 1994;269:10683–10690. [PubMed] [Google Scholar]
- Burgstaller G., Kranewitter W. J., Gimona M. The molecular basis for the autoregulation of calponin by isoform-specific C-terminal tail sequences. J. Cell Sci. 2002;115:2021–2029. doi: 10.1242/jcs.115.10.2021. [DOI] [PubMed] [Google Scholar]
- Chaney L. K., Jacobson B. S. Coating cells with colloidal silica for high yield isolation of plasma membrane sheets and identification of transmembrane proteins. J. Biol. Chem. 1983;258:10062–10072. [PubMed] [Google Scholar]
- Chen E. H., Olson E. N. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Danninger C., Gimona M. Live dynamics of GFP-calponin: isoform-specific modulation of the actin cytoskeleton and autoregulation by C-terminal sequences. J. Cell Sci. 2000;113:3725–3736. doi: 10.1242/jcs.113.21.3725. [DOI] [PubMed] [Google Scholar]
- Dupressoir A., Marceau G., Vernochet C., Benit L., Kanellopoulos C., Sapin V., Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A., Vernochet C., Bawa O., Harper F., Pierron G., Opolon P., Heidmann T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. USA. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Mezgueldi M., Strasser P., Fattoum A., Gimona M. Expressing functional domains of mouse calponin: involvement of the region around alanine 145 in the actomyosin ATPase inhibitory activity of calponin. Biochemistry. 1996;35:3654–3661. doi: 10.1021/bi952027e. [DOI] [PubMed] [Google Scholar]
- Fraser E. D., Walsh M. P. Dephosphorylation of calponin by type 2B protein phosphatase. Biochemistry. 1995;34:9151–9158. doi: 10.1021/bi00028a026. [DOI] [PubMed] [Google Scholar]
- Frendo J. L., Olivier D., Cheynet V., Blond J. L., Bouton O., Vidaud M., Rabreau M., Evain-Brion D., Mallet F. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Masuda H., Takagi M., Takahashi K., Kiyokane K. The presence of h2-calponin in human keratinocyte. J. Dermatol. Sci. 1997;14:29–36. doi: 10.1016/s0923-1811(96)00545-2. [DOI] [PubMed] [Google Scholar]
- Ghitescu L. D., Crine P., Jacobson B. S. Antibodies specific to the plasma membrane of rat lung microvascular endothelium. Exp. Cell. Res. 1997;232:47–55. doi: 10.1006/excr.1997.3490. [DOI] [PubMed] [Google Scholar]
- Ghitescu L. D., Gugliucci A., Dumas F. Actin and annexins I and II are among the main endothelial plasmalemma-associated proteins forming early glucose adducts in experimental diabetes. Diabetes. 2001;50:1666–1674. doi: 10.2337/diabetes.50.7.1666. [DOI] [PubMed] [Google Scholar]
- Hatano N., Mori Y., Oh-hora M., Kosugi A., Fujikawa T., Nakai N., Niwa H., Miyazaki J., Hamaoka T., Ogata M. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells. 2003;8:847–856. doi: 10.1046/j.1365-2443.2003.00680.x. [DOI] [PubMed] [Google Scholar]
- Hossain M. M., Smith P. G., Wu K., Jin J. P. Cytoskeletal tension regulates both expression and degradation of h2-calponin in lung alveolar cells. Biochemistry. 2006;45:15670–15683. doi: 10.1021/bi061718f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q. Q., Hossain M. M., Wu K., Parai K., Pope R. M., Jin J. P. Role of H2-calponin in regulating macrophage motility and phagocytosis. J. Biol. Chem. 2008;283:25887–25899. doi: 10.1074/jbc.M801163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard A. L., Ma A. Isolation of rat hepatocyte plasma membranes. II. Identification of membrane-associated cytoskeletal proteins. J. Cell. Biol. 1983;96:230–239. doi: 10.1083/jcb.96.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa K., Ito M., Okubo S., Konishi T., Nakano T., Mino T., Nakamura F., Naka M., Tanaka T. Calponin phosphatase from smooth muscle: a possible role of type 1 protein phosphatase in smooth muscle relaxation. Biochem. Biophys. Res. Commun. 1993;193:827–833. doi: 10.1006/bbrc.1993.1700. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Amano M., Maeda A., Goto H., Takahashi K., Ito M., Kaibuchi K. Identification of calponin as a novel substrate of Rho-kinase. Biochem. Biophys. Res. Commun. 2000;273:110–116. doi: 10.1006/bbrc.2000.2901. [DOI] [PubMed] [Google Scholar]
- Keryer G., Alsat E., Tasken K., Evain-Brion D. Cyclic AMP-dependent protein kinases and human trophoblast cell differentiation in vitro. J. Cell. Sci. 1998;111:995–1004. doi: 10.1242/jcs.111.7.995. [DOI] [PubMed] [Google Scholar]
- Kontani K., Moskowitz I. P., Rothman J. H. Repression of cell-cell fusion by components of the C. elegans vacuolar ATPase complex. Dev. Cell. 2005;8:787–794. doi: 10.1016/j.devcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Lyden T. W., Ng A. K., Rote N. S. Modulation of phosphatidylserine epitope expression by BeWo cells during forskolin treatment. Placenta. 1993;14:177–186. doi: 10.1016/s0143-4004(05)80259-0. [DOI] [PubMed] [Google Scholar]
- Mi S., Lee X., Li X., Veldman G. M., Finnerty H., Racie L., LaVallie E., Tang X. Y., Edouard P., Howes S., Keith J. C., Jr, McCoy J. M. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Miyoshi H. Gene delivery to hematopoietic stem cells using lentiviral vectors. Methods Mol Biol. 2004;246:429–438. doi: 10.1385/1-59259-650-9:429. [DOI] [PubMed] [Google Scholar]
- Mohler W. A., Shemer G., del Campo J. J., Valansi C., Opoku-Serebuoh E., Scranton V., Assaf N., White J. G., Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Morgan K. G., Gangopadhyay S. S. Invited review: cross-bridge regulation by thin filament-associated proteins. J. Appl. Physiol. 2001;91:953–962. doi: 10.1152/jappl.2001.91.2.953. [DOI] [PubMed] [Google Scholar]
- Nakamura F., Mino T., Yamamoto J., Naka M., Tanaka T. Identification of the regulatory site in smooth muscle calponin that is phosphorylated by protein kinase C. J. Biol. Chem. 1993;268:6194–6201. [PubMed] [Google Scholar]
- Pajcini K. V., Pomerantz J. H., Alkan O., Doyonnas R., Blau H. M. Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. J. Cell. Biol. 2008;180:1005–1019. doi: 10.1083/jcb.200707191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbar A. M., Fenselau C. Integration of Jacobson's pellicle method into proteomic strategies for plasma membrane proteins. J. Proteome Res. 2004;3:1267–1277. doi: 10.1021/pr040004t. [DOI] [PubMed] [Google Scholar]
- Rossant J., Cross J. C. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Rozenblum G. T., Gimona M. Calponins: adaptable modular regulators of the actin cytoskeleton. Int. J. Biochem. Cell. Biol. 2007;31:31. doi: 10.1016/j.biocel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M., Coutts N., Price A., Taylor M. V., Bate M. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 2000;102:189–198. doi: 10.1016/s0092-8674(00)00024-6. [DOI] [PubMed] [Google Scholar]
- Schreiber J., Riethmacher-Sonnenberg E., Riethmacher D., Tuerk E. E., Enderich J., Bosl M. R., Wegner M. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol. Cell. Biol. 2000;20:2466–2474. doi: 10.1128/mcb.20.7.2466-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn R. L., Huang P., Kawahara G., Mitchell M., Guyon J., Kalluri R., Kunkel L. M., Gussoni E. A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc. Natl. Acad. Sci. USA. 2009;106:9274–9279. doi: 10.1073/pnas.0904398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz D. B., Ross M. A., Salem H. M., Mars W. M., Michalopoulos G. K., Enomoto K. Cationic colloidal silica membrane perturbation as a means of examining changes at the sinusoidal surface during liver regeneration. Am. J. Pathol. 1999;155:1487–1498. doi: 10.1016/S0002-9440(10)65464-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser P., Gimona M., Moessler H., Herzog M., Small J. V. Mammalian calponin. Identification and expression of genetic variants. FEBS Lett. 1993;330:13–18. doi: 10.1016/0014-5793(93)80909-e. [DOI] [PubMed] [Google Scholar]
- Strunkelnberg M., Bonengel B., Moda L. M., Hertenstein A., de Couet H. G., Ramos R. G., Fischbach K. F. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128:4229–4239. doi: 10.1242/dev.128.21.4229. [DOI] [PubMed] [Google Scholar]
- Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics. 2007;6:1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Hiwada K., Kokubu T. Vascular smooth muscle calponin: a novel troponin T-like protein. Hypertension. 1988;11:620–626. doi: 10.1161/01.hyp.11.6.620. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Nadal-Ginard B. Molecular cloning and sequence analysis of smooth muscle calponin. J. Biol. Chem. 1991;266:13284–13288. [PubMed] [Google Scholar]
- Tang D. C., Kang H. M., Jin J. P., Fraser E. D., Walsh M. P. Structure-function relations of smooth muscle calponin. The critical role of serine 175. J. Biol. Chem. 1996;271:8605–8611. doi: 10.1074/jbc.271.15.8605. [DOI] [PubMed] [Google Scholar]
- Tang J., Hu G., Hanai J., Yadlapalli G., Lin Y., Zhang B., Galloway J., Bahary N., Sinha S., Thisse B., Thisse C., Jin J. P., Zon L. I., Sukhatme V. P. A critical role for calponin 2 in vascular development. J. Biol. Chem. 2006;281:6664–6672. doi: 10.1074/jbc.M506991200. [DOI] [PubMed] [Google Scholar]
- Thumkeo D., Keel J., Ishizaki T., Hirose M., Nonomura K., Oshima H., Oshima M., Taketo M. M., Narumiya S. Targeted disruption of the mouse rho-associated kinase 2 gene results in intrauterine growth retardation and fetal death. Mol. Cell. Biol. 2003;23:5043–5055. doi: 10.1128/MCB.23.14.5043-5055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunekawa S., Takahashi K., Abe M., Hiwada K., Ozawa K., Murachi T. Calpain proteolysis of free and bound forms of calponin, a troponin T-like protein in smooth muscle. FEBS Lett. 1989;250:493–496. doi: 10.1016/0014-5793(89)80783-5. [DOI] [PubMed] [Google Scholar]
- Vargas A., Moreau J., Landry S., LeBellego F., Toufaily C., Rassart E., Lafond J., Barbeau B. Syncytin-2 plays an important role in the fusion of human trophoblast cells. J. Mol. Biol. 2009;392:301–318. doi: 10.1016/j.jmb.2009.07.025. [DOI] [PubMed] [Google Scholar]
- Vargas A., Moreau J., Le Bellego F., Lafond J., Barbeau B. Induction of trophoblast cell fusion by a protein tyrosine phosphatase inhibitor. Placenta. 2008;29:170–174. doi: 10.1016/j.placenta.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Wice B., Menton D., Geuze H., Schwartz A. L. Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp. Cell. Res. 1990;186:306–316. doi: 10.1016/0014-4827(90)90310-7. [DOI] [PubMed] [Google Scholar]
- Winder S. J., Walsh M. P. Smooth muscle calponin. Inhibition of actomyosin MgATPase and regulation by phosphorylation. J. Biol. Chem. 1990;265:10148–10155. [PubMed] [Google Scholar]
- Yoshikawa H., Taniguchi S. I., Yamamura H., Mori S., Sugimoto M., Miyado K., Nakamura K., Nakao K., Katsuki M., Shibata N., Takahashi K. Mice lacking smooth muscle calponin display increased bone formation that is associated with enhancement of bone morphogenetic protein responses. Genes Cells. 1998;3:685–695. doi: 10.1046/j.1365-2443.1998.00214.x. [DOI] [PubMed] [Google Scholar]
- Yoshimoto R., Hori M., Ozaki H., Karaki H. Proteolysis of acidic calponin by mu-calpain. J. Biochem. 2000;128:1045–1049. doi: 10.1093/oxfordjournals.jbchem.a022832. [DOI] [PubMed] [Google Scholar]
- Yu C., Shen K., Lin M., Chen P., Lin C., Chang G. D., Chen H. GCMa regulates the syncytin-mediated trophoblastic fusion. J. Biol. Chem. 2002;277:50062–50068. doi: 10.1074/jbc.M209316200. [DOI] [PubMed] [Google Scholar]
- Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.