Ablation of nonmuscle myosin (NM) II-B and II-C in mice results in a defect in cardiac myocyte karyokinesis. More than 90% of the double knockout cardiac myocytes demonstrate defects in chromatid segregation and mitotic spindle formation. The requirement for NM II in karyokinesis is demonstrated in both the intact heart and in HL-1 atrial myocytes in culture.
Abstract
Ablation of nonmuscle myosin (NM) II-A or NM II-B results in mouse embryonic lethality. Here, we report the results of ablating NM II-C as well as NM II-C/II-B together in mice. NM II-C ablated mice survive to adulthood and show no obvious defects compared with wild-type littermates. However, ablation of NM II-C in mice expressing only 12% of wild-type amounts of NM II-B results in a marked increase in cardiac myocyte hypertrophy compared with the NM II-B hypomorphic mice alone. In addition, these hearts develop interstitial fibrosis associated with diffuse N-cadherin and β-catenin localization at the intercalated discs, where both NM II-B and II-C are normally concentrated. When both NM II-C and II-B are ablated the B−C−/B−C− cardiac myocytes show major defects in karyokinesis. More than 90% of B−C−/B−C− myocytes demonstrate defects in chromatid segregation and mitotic spindle formation accompanied by increased stability of microtubules and abnormal formation of multiple centrosomes. This requirement for NM II in karyokinesis is further demonstrated in the HL-1 cell line derived from mouse atrial myocytes, by using small interfering RNA knockdown of NM II or treatment with the myosin inhibitor blebbistatin. Our study shows that NM II is involved in regulating cardiac myocyte karyokinesis by affecting microtubule dynamics.
INTRODUCTION
Nonmuscle myosin (NM) IIs are ubiquitous proteins present in all eukaryotic cells that have been shown to play important roles in vertebrate development and in the normal functioning of the adult organism (Tullio et al., 1997; Conti et al., 2004; Donaudy et al., 2004; Pecci et al., 2008; Bhatt et al., 2009). These roles include, but are not limited to cell–cell adhesion, cell migration, and cytokinesis (Bresnick, 1999; Robinson and Spudich, 2004; Matsumura, 2005; Vicente-Manzanares et al., 2009). The myosin II molecule is composed of one pair of heavy chains (230 kDa) and two pairs of light chains (20 and 17 kDa). The nonmuscle myosin II heavy chains (NMHC IIs) are encoded by three genes located on three different chromosomes in humans and mice (Berg et al., 2001; Leal et al., 2003; Golomb et al., 2004). It is the heavy chain isoform that provides the protein name (NM II-A, II-B, and II-C) to the hexameric NM II molecule. Among the three NMHC II genes, NMHC II-B and II-C have been shown to undergo alternative splicing that results in the introduction of one or two additional exons into the mRNA region that encodes the globular head of the molecule (Itoh and Adelstein, 1995; Golomb et al., 2004; Jana et al., 2009). One alternative exon called B1 or C1 is spliced into the ATP-binding region and a second exon called B2 or C2 into the actin binding region. These additions affect the actin-activated MgATPase activity as well as the binding of actin to myosin (Golomb et al., 2004; Kim et al., 2005, 2008; Jana et al., 2009).
Our laboratory has been interested in the in vivo functions of the various NM II isoforms; ablation of two of the three isoforms, NM II-A and NM II-B, in mice showed markedly different phenotypes. Ablation of NM II-A resulted in lethality by embryonic day (E)6.5, with a defect in cell adhesion and visceral endoderm formation (Conti et al., 2004). Ablation of NM II-B resulted in survival to E14.5, but with marked abnormalities in the heart including a ventricular septal defect, abnormal positioning of the aorta, and abnormalities in cardiac myocyte cytokinesis, as well as brain abnormalities including hydrocephalus and the abnormal migration of certain groups of neurons (Tullio et al., 1997; Takeda et al., 2003; Ma et al., 2004).
NMHC II-C is the most recently identified member of the NMHC II family (Berg et al., 2001), and its function has not yet been fully investigated. Similarly to NMHC II-B, two alternative exons, C1 and C2, can be spliced in the mRNA, and this splicing increases the number of NM II-C isoforms to four (NM II-C0, NM II-C1, NM II-C2, and NM II-C1C2). Unlike the alternatively spliced NM II-B isoforms (NM II-B1 and NM II-B2), where expression is confined to the nervous system, NM II-C1 is expressed in many tissues, including the liver, kidney, lung, and brain (Golomb et al., 2004). Small interfering RNA (siRNA) knockdown of NM II-C in the A549 human lung tumor cell line led to the failure of the final abscission step of cytokinesis (Jana et al., 2006). Interestingly, this cell line only expressed the C1-inserted isoform of NM II-C. Replacement of NM II-C1 with the noninserted isoform did not completely rescue this phenotype. Of note is a report that knockdown NM II-C in the Neuro-2A cell line causes defects in neurite outgrowth (Wylie and Chantler, 2008).
To understand the function of NM II-C in vivo, we ablated NMHC II-C in mice, thereby ablating NM II-C (NMHC II-C along with its myosin light chains). Finding no obvious phenotype in NM II-C ablated mice, we crossed these mice with mice expressing relatively small amounts of NM II-B compared with wild-type mice, to generate NM II-B hypomorphic/II-C ablated mice. We also generated mice that were ablated for both NM II-B and II-C, which developed severe defects in cardiac myocyte karyokinesis.
MATERIALS AND METHODS
Generation of NMHC II-C–ablated Mice
A mouse (129/Sv) genomic clone containing the entire NMHC II-C gene (Myh14) locus was obtained from a RPCI-22 Mouse BAC Library (ResGen, Invitrogen, Carlsbad, CA). An ∼6.8-kb SalI-HindIII fragment including exons 3–5 and C1 and flanking introns was used to generate the targeting construct. A 0.3-kb fragment including the 157-base pair exon 3 was replaced by a neomycin resistance cassette (Neor) flanked by a pair of loxP sites. The Neor cassette includes a HindIII site denoted by HindIII* (Supplemental Figure S1A, b and c). A herpes simplex virus thymidine kinase expression cassette (TK) was inserted at the 3′ end of the construct for negative selection. The resulting targeting construct contains 2.2- and 4.3-kb regions homologous to the native gene at the 5′ and 3′ ends, respectively (Supplemental Figure S1Ab). The linearized plasmids were electroporated into CMT-1 embryonic stem (ES) cells (Specialty Media, Division of Cell and Molecular Technologies, Phillipsburg, NJ) and selected with Geneticin (0.35 mg/ml G418 Clontech, Mountain View, CA) and ganciclovir. Genomic DNA was isolated from drug-resistant ES cell clones, digested with HindIII, and analyzed by Southern blotting using the 5′ external probe indicated in Supplemental Figure S1Aa to identify targeted ES cell clones. The probe detects a 4.5-kb band for the targeted C− allele (Supplemental Figure S1Ac) and a 7.4-kb band for the wild-type allele (Supplemental Figure S1Aa). To generate chimeric mice, targeted ES cells were injected into blastocysts derived from C57BL/6 mice. Mice were maintained on a 129/Sv and C57BL/6 mixed background. Genotyping of progeny was determined by Southern blot or polymerase chain reaction (PCR) using genomic DNA isolated from mouse tails. All experiments were carried out according to the guidelines approved by National Heart, Lung, and Blood Institute Animal Care and Use Committee.
Tissue Preparation and Histological Analysis
Histological analysis of hearts was performed as described previously (Ma et al., 2009). In brief, hearts were fixed with 4% paraformaldehyde in phosphate-buffered saline, pH 7.4. They were then embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). To measure the cross-sectional area of myocytes, transverse sections were stained with Alexa 594 linked to wheat germ agglutinin (WGA) that binds to the cell membrane. Measurements (n > 100 cells from each animal) were obtained from confocal images using a Zeiss LMS 510 embedded measuring tool. The mean and SD of these measurements were calculated from 3 animals of each genotype.
Cell Culture and siRNA Knockdown
HL-1 cells were cultured in 0.02% gelatin/12.5 mg/ml fibronectin-coated culture dishes in Claycomb medium (JRH Biosciences) containing 10% fetal bovine serum with exchange of fresh media every three days (White et al., 2004). siRNA treatment was carried out as previously reported (Bao et al., 2005). To examine the mitotic spindles, cells were stained with β-tubulin to reveal the spindles and counterstained with 4,6-diamidino-2-phenylindole (DAPI) for chromosome alignment. Dividing cells containing other than one spindle (half or more than one) were counted as abnormal.
Immunoblot and Immunohistochemistry
Immunoblot (IB) analysis and immunofluorescence (IF) confocal microscopy were carried out as described previously (Ma et al., 2007). The following primary antibodies were used in this study: rabbit polyclonal antibodies NMHC II-A, II-B, and II-C (IF, 1:1000; Phillips et al., 1995; Golomb et al., 2004); connexin43 (IF, 1:100; Cell Signaling Technology, Danvers, MA); and desmin (IF, 1:200; Abcam, Cambridge, MA); and mouse monoclonal antibodies desmin (IF, 1:200; Dako North America, Carpinteria, CA), MF20 (IF, 1:30; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), E-cadherin (IF, 1:1000; BD Biosciences, San Jose, CA), N-cadherin (IF, 1:200; Zymed Laboratories, South San Francisco, CA), β-catenin (IF, 1:200; Zymed Laboratories), β-actin (IB, 1:5000; Sigma-Aldrich, St. Louis, MO), β-tubulin (IF, 1:1000; Sigma-Aldrich), acetylated α-tubulin (IF, 1:200; Sigma-Aldrich), γ-tubulin (IF, 1:1000; Sigma-Aldrich), and p21 (IB, 1:500; IF, 1:50; BD Biosciences). For confocal imaging, the fluorescence secondary antibodies used were Alexa 488 goat anti-rabbit immunoglobulin (Ig)G or Alexa 594 goat anti-mouse IgG (1:250; Invitrogen). The images were collected using an LSM510 META confocal microscope (Carl Zeiss). In all cases, when possible, comparison was made among littermates.
Quantification of NM II Isoform Expression
Quantification of relative amounts of NM II isoform was carried out as described previously (Babbin et al., 2009). The cell or tissue extract was separated by SDS-polyacrylamide gel electrophoresis. After the Coomassie Blue staining of the gel, bands near 230 kDa were excised, destained, reduced and alkylated, and digested with trypsin. Tryptic peptides were purified and concentrated on C18 resin Zip Tips (Millipore, Billerica, MA) and submitted to National Heart, Lung, and Blood Institute Proteomics Core Facility for analysis by liquid chromatography tandem mass spectroscopy. Peptide numbers for each of the NMHC II isoforms were counted, and the percentage contribution to total amount of NMHC II peptides was calculated.
Statistical Analysis
Data are expressed as the mean ± SD. Statistical significance was tested with a one-way analysis of variance followed by the Bonferroni test.
RESULTS
Generation of NM II-C–ablated Mice
NM II-C was ablated by replacing exon 3 of the NMHC II-C gene (Myh14) with a floxed Neor cassette by homologous recombination in ES cells (Supplemental Figure S1Ac). Deletion of exon 3 disrupts the normal reading frame of NMHC II-C mRNA and results in complete loss of NM II-C expression (Supplemental Figure S1Ba).
Nonmuscle Myosin II-C Is Dispensable for Embryonic Mouse Development
Homozygous mice ablated for NM II-C survive to adulthood and show no obvious difference compared with wild-type littermates. This may reflect the delayed expression of NM II-C during mouse embryonic development compared with NM II-A and II-B. In vitro studies using cell lines have proposed roles for NM II-C in cell division and neurite outgrowth (Jana et al., 2006; Wylie and Chantler, 2008). We examined the NM II-C ablated mice for these abnormalities but found no obvious defects in brain structure and no evidence for defective cell division in these mice, raising the possibility of compensatory NM II isoforms in vivo.
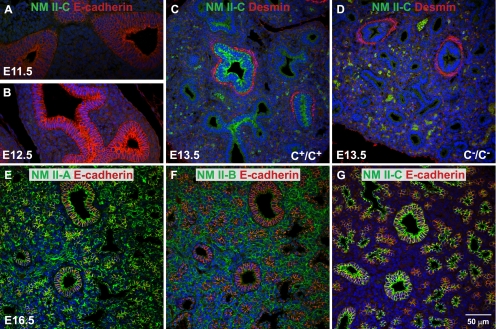
Previous analysis by immunoblot and immunofluorescence microscopy showed that NM II-C was expressed in various mouse tissues (Golomb et al., 2004). However, the relative abundance of the various NM II isoforms has only been reported to date for a few cell lines (Babbin et al., 2009; Smutny et al., 2010). To estimate the relative abundance of NM II-C in mouse tissues and cell lines, mass spectroscopy was performed to quantify the various peptides for each of the NM II isoforms. As shown in Table 1, except for the lung, where NM II-C accounts for 37 ± 4% of the total NM II, no measurable amounts of NM II-C are detected in the adult heart, brain, spinal cord, spleen, and kidney by using this technique, suggesting that relative to NM II-A and II-B, significantly less NM II-C is expressed. In contrast, as Table 1 shows, there are relatively large amounts of NM II-C in a number of cell lines. Distribution of the NM II isoforms was further analyzed by immunofluorescence confocal microscopy in the developing mouse lung. Figure 1, A–D, shows airway epithelial cells at E11.5–13.5 during lung development. Note the appearance of NM II-C starting at E13.5 (Figure 1C, green) and its absence in the II-C ablated mouse (Figure 1D). Figure 1, E–G, shows that in the E16.5 mouse lung, NM II-A (Figure 1E, green) is widely expressed and NM II-B (Figure 1F, green) is enriched in the parenchymal cells, whereas NM II-C (Figure 1G, green) is confined to airway epithelial cells. Because NM II-C is often coexpressed along with NM II-A and/or II-B and is usually expressed at a lower level compared with NM II-A or II-B, this raises the possibility that NM II-A or NM II-B could functionally replace NM II-C in NM II-C null mice. Importantly, the ablation of NM II-C does not interfere with lung development in a tissue that seems to contain relatively large amounts of the isoform (Table 1). Ablation of NM II-C in lungs and other tissues also does not increase or alter the pattern of NM II-A or II-B expression (Supplemental Figure S2).
Table 1.
Mass spectroscopy analysis of the relative abundance of nonmuscle myosin II heavy chain isoforms in mouse tissues, human platelets, and cell lines
| Tissues/cell lines | NMHC II-A (%) | NMHC II-B (%) | NMHC II-C (%) | n (of samples) |
|---|---|---|---|---|
| Mouse heart (P2) | 63 ± 3 | 37 ± 3 | ND | 2 |
| Mouse heart | 96 ± 4 | 5 ± 3 | ND | 2 |
| Mouse cerebral cortex | 29 ± 9 | 67 ± 13 | 4 ± 8 | 3 |
| Mouse cerebellum | 14 ± 16 | 81 ± 19 | 6 ± 5 | 4 |
| Mouse spinal cord | 29 ± 9 | 65 ± 5 | 6 ± 9 | 5 |
| Mouse kidney | 92 ± 4 | 7 ± 1 | 2 ± 2 | 2 |
| Mouse lung | 41 ± 2 | 22 ± 1 | 37 ± 4 | 2 |
| Mouse spleen | 100 | ND | ND | 2 |
| Mouse ES cells | 81 ± 7 | 19 ± 7 | ND | 4 |
| Human platelets | 100 | ND | ND | 3 |
| 3T3 cells (ms) | 83 ± 7 | 17 ± 7 | ND | 3 |
| Caco2-BBE cells (hu) | 89 ± 4 | 5 ± 2 | 6 ± 2 | 2 |
| COS-7 cells (mk) | ND | 86 ± 19 | 15 ± 19 | 2 |
| HeLa cells (hu) | 97 ± 3 | 3 ± 3 | ND | 4 |
| HFF cells (hu) | 95 ± 4 | 4 ± 4 | 1 ± 1 | 5 |
| HL-1 cells (ms) | 95 ± 3 | 5 ± 3 | ND | 3 |
| HT29 cells (hu) | 54 ± 3 | 1 ± 1 | 45 ± 3 | 4 |
| MDCK cells (dog) | 96 ± 3 | 1 ± 1 | 3 ± 2 | 2 |
| RBL cells (rat) | 99 ± 1 | 1 ± 1 | 1 ± 2 | 4 |
ND, not detected; P2, postnatal day 2; HFF, human foreskin fibroblasts; ms, mouse; hu, human; and mk, monkey.
Figure 1.
Expression of NM II-C in developing mouse lungs. (A–D) Immunofluorescence confocal images of E11.5–E13.5 mouse lungs stained for NMHC II-C (green). E-Cadherin is a marker for epithelial cells (A and B, red). Desmin is a marker for (smooth) muscle cells (C and D, red). No NM II-C expression is detected in E11.5 and E12.5 lungs. NM II-C is detected in E13.5 lung epithelial cells (C, green) that are surrounded by smooth muscle cells (C, red). NM II-C is not detected in C−/C− lungs (D). Green spots in C and D are autofluorescence from red blood cells. (E–G) Immunofluorescence confocal images of E16.5 mouse lung stained for NMHC II-A (E, green), II-B (F, green), and II-C (G, green) and for E-cadherin (red). NMHC II-A (E) and II-B (F) are ubiquitously expressed in developing mouse lung. NMHC II-A is enriched in epithelial cells, whereas II-B is enriched in parenchymal cells. In contrast, NMHC II-C (green) is only detected in epithelial cells (G), where both NMHC II-A and II-B also are expressed.
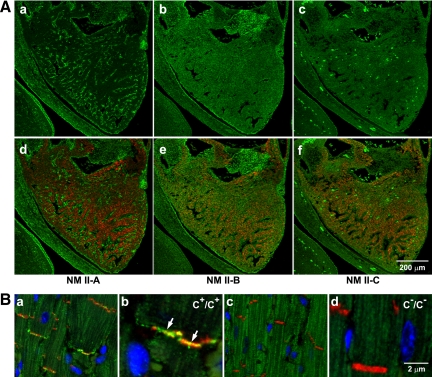
Unlike the lung epithelial cells, ventricular cardiac myocytes lack NM II-A after E7.5 and only contain NM II-B and II-C (Figure 2A, green). Coexpression of desmin (red), a marker for cardiac myocytes, with NM II-B and II-C can be seen in the E13.5 heart shown in Figure 2A, e and f (green; also see Ma et al., 2009). Moreover, as Figure 2B shows, NM II-C, similarly to II-B (Takeda et al., 2000), localizes to the intercalated discs in the adult heart (Figure 2Ba, green; enlarged in b). This colocalization of NM II-B and II-C to the intercalated disc allowed us to make a direct comparison of possible compensatory interactions between NM II-B and II-C. First we examined the hearts of NM II-C ablated mice for pathological changes. Supplemental Figure S1B, b and c, shows that no obvious abnormalities are observed in C−/C− mouse hearts compared with C+/C+ hearts. A magnetic resonance imaging analysis confirmed a normal ejection fraction at 6 mo for C−/C− mice (69 ± 10%, n = 3, compared with wild-type 80 ± 12%, n = 4). An ECG analysis by telemetry also shows no abnormalities for C−/C− mice. These results indicate that ablation of NM II-C alone has no obvious effects on mouse heart development. We therefore decided to study the effect of ablating NM II-C in mice with decreased amount of NM II-B (NM II-B hypomorphic mice).
Figure 2.
Expression of NM II-C in embryonic and adult mouse hearts. (A) Immunofluorescence confocal images of an E13.5 mouse heart stained for NMHC II-A (a, green), II-B (b, green), and II-C (c, green) together with desmin (d–f, red), a marker for (cardiac) myocytes. NM II-A is only expressed in nonmyocytes (a, green). Note the lack of colocalization with desmin-positive cardiac myocytes. NM II-B is detected in both myocytes (e, red and green colocalization) and nonmyocytes (e, green) in the heart. NM II-C is detected in myocytes (f, red and green colocalization) but not in nonmyocytes. The bright green spots are autofluorescence from red blood cells in c and f. (B) Immunofluorescence confocal images of adult heart sections from C+/C+ (a, magnified in b) and C−/C− (c, magnified in d) mice. N-Cadherin is a marker for the intercalated disc (red). Nuclei are stained with DAPI (blue). Arrows in b indicates the presence of NMHC II-C (green) in the intercalated disc. NMHC II-C is absent from C−/C− intercalated discs (c and d).
Ablation of NM II-C in NM II-B Hypomorphic Mice
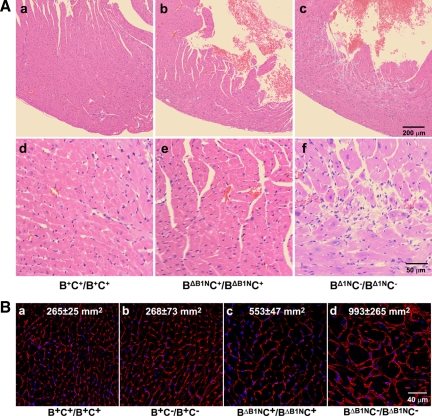
C−/C− mice were crossed with hypomorphic II-B (BΔB1N/BΔB1N) mice, which express only 12% of wild-type amounts of NMHC II-B in the heart to generate II-B-hypomorphic/II-C-ablated mice (BΔB1NC−/BΔB1NC−). Supplemental Figure S3 shows that ablation of NM II-C in BΔB1N/BΔB1N mice does not affect the expression level of NM II-B. Compared with the wild-type mouse (Supplemental Figure S3D), 13 ± 6 and 15 ± 5% of wild-type levels of NM II-B are expressed in BΔB1NC+/BΔB1NC+ (Supplemental Figure S3E) and BΔB1NC−/BΔB1NC− (Supplemental Figure S3F) mouse hearts, respectively (n = 3 mice) as determined by quantification of immunofluorescence staining of E13.5 mouse heart sections with specific antibodies to NMHC II-B. Previous studies showed that BΔB1N/BΔB1N mice develop cardiac myocyte hypertrophy by 11 mo (Uren et al., 2000). Figure 3A shows H&E staining of heart sections from B+C+/B+C+ (Figure 3A, a and d), BΔB1NC+/BΔB1NC+ (Figure 3A, b and e), and BΔB1NC−/BΔB1NC− (Figure 3A, c and f) mice at 6 mo. The sections from the BΔB1NC−/BΔB1NC− hearts show evidence for myocyte hypertrophy as well as cardiac myocyte death and fibrosis. Neither wild-type (Figure 3Ad) nor II-C ablated mice (Supplemental Figure S1Bc) show these defects. WGA staining of the heart at this age to outline the membrane of the myocytes illustrates more clearly the differences in the size of cardiac myocytes among these four genotypes (Figure 3B). Measuring the cross-sectional area of cardiac myocytes shows that the average area in the BΔB1NC−/BΔB1NC− mouse heart is almost 4 times greater than that in either B+C+/B+C+ or B+C−/B+C− mouse hearts (Figure 3B, a and d, p < 0.01, n = 2 mice; >100 myocytes/mouse), whereas no difference is found between B+C+/B+C+ and B+C−/B+C− myocytes. Note that the BΔB1NC+/BΔB1NC+ cardiac myocytes (Figure 3Bc) were twice as large as wild-type or II-C ablated myocytes (p < 0.05, n = 2 mice; >100 myocytes/mouse). These data indicate that the absence of NM II-C together with a decrease in NM II-B reinforces the cardiac phenotype.
Figure 3.
Ablation of NM II-C accelerates development of cardiomyopathy in NM II-B hypomorphic mice. (A) H&E-stained heart sections of B+C+/B+C+, BΔB1NC+/BΔB1NC+, and BΔB1NC−/BΔB1NC− mice at 6 mo. Compared with wild-type hearts (a, B+C+/B+C+; magnified in d), BΔB1NC+/BΔB1NC+ hearts show no obvious changes (b; magnified in e); however, BΔB1NC−/BΔB1NC− hearts show marked cardiac hypertrophy and interstitial fibrosis (c; magnified in f). (B) WGA staining shows plasma membranes in heart sections from B+C+/B+C+, B+C−/B+C−, BΔB1NC+/BΔB1NC+, and BΔB1NC−/BΔB1NC− mice. The average cross-sectional area of the cardiac myocytes for each genotype was measured and is shown in each panel. Compared with B+C+/B+C+ cardiac myocytes (a), the average cross-sectional area for B+C−/B+C− myocytes remains unchanged (b), but it is doubled in BΔB1NC+/BΔB1NC+ myocytes (c) and increased to 4 times in BΔB1NC−/BΔB1NC− myocytes (d). Nuclei are stained with DAPI (blue).
Because both NM II-B and NM II-C localize to the intercalated disc of the adult cardiac myocytes, and cardiac specific ablation of NM II-B disrupted the integrity of the intercalated discs in mouse hearts at 10 mo (Ma et al., 2009), we next examined the distribution of the intercalated disc associated proteins connexin43, β-catenin, and N-cadherin in these mice by using immunofluorescence confocal microscopy. As shown in Supplemental Figure S4, β-catenin and N-cadherin staining of the intercalated discs is very discrete in B+C+/B+C+, BΔB1NC+/BΔB1NC+, and B+C−/B+C− cardiac myocytes (Supplemental Figure S4A, a, e, c, and d, red); however, it is noticeably diffuse in BΔB1NC−/BΔB1NC− myocytes, indicating a disruption of the discs (Supplemental Figure S4A, b and f arrows, red). Quantification of β-catenin staining at the intercalated discs shows that the average width of staining in BΔB1NC−/BΔB1NC− hearts is 2.48 ± 0.67 μm compared with 1.41 ± 0.23 μm for wild-type hearts (Supplemental Figure S4B). In contrast, no difference in the staining of connexin43 (Supplemental Figure S4A, a–d, green), is seen in BΔB1NC−/BΔB1NC− myocytes compared with the wild-type myocytes. These results indicate that complete loss of NM II-C together with decreased amounts of NM II-B, but not the loss of NM II-C or decreased amounts of NM II-B alone, causes instability of adhesion junctions in the intercalated discs of the cardiac myocytes. This is consistent with the function of NM II-B and II-C, along with actin in maintaining the adhesion junctions of the intercalated discs.
Simultaneous Ablation of NM II-B and II-C in Cardiac Myocytes
To better understand the interplay between NM II-C and II-B, we next generated NM II-B and II-C double knockout mice (B−C−/B−C−) that survive only until E14.5, similarly to B−/B− mice. An immunofluorescence study confirms the loss of NM II-B and II-C in the hearts of the B−C−/B−C− mice (Supplemental Figure S5, D and F). By this age (E13.5) NMHC II-A is only present in the nonmyocyte cells and not in the myocytes (Supplemental Figure S5, A and B). Note the lack of colocalization between NM II-A (green) and MF20 (red), a marker for sarcomeric myosin. Ablation of both NM II-B and II-C by E13.5 therefore results in no NM II expression in B−C−/B−C− cardiac myocytes. Although severely hypoplastic, the B−C−/B−C− mouse hearts have sufficient numbers of cardiac myocytes to support life to E14.5 (Supplemental Figure S6Ab).
To understand how cardiac myocytes develop in the absence of NM II-B and II-C, we examined NM II-A expression in cardiac myocytes between E7.5 and E12.5 in wild-type mouse hearts. Immunofluorescence confocal microscopy shows that NM II-A (Supplemental Figure S6Ba, green) is expressed in cardiac myocytes (red, MF20) of the wild-type heart tube at E7.5. Note that NM II-B is enriched in cardiac myocytes at this time (Supplemental Figure S6Bd, green). By E8.5 and later however, NM II-A is only detected in cardiac myocytes at the arterial pole region where the latest addition of cardiac myocytes occurs, but it is not detected in the ventricular myocytes (Supplemental Figure S6Bb, green; enlarged in c, arrows) that continue to show NM II-B expression (Supplement Figure S6Be, green; enlarged in f, arrows). These results show that NM II-A is expressed in cardiac myocytes but only in the E7.5 heart tube or in myocytes newly added from their precursors at the arterial pole. In B−C−/B−C− mice, expression of NM II-A therefore ensures a continuous supply of cardiac myocytes from their precursors at the arterial pole that contributes to the development of B−C−/B−C− hearts. However, the absence of NM II (NM II-B and II-C) greatly impairs cell division of B−C−/B−C− ventricular myocytes and consequently leads to a marked reduction in the number of cardiac myocytes in B−C−/B−C− hearts (see below).
It has been proposed that NM II plays a role in the early stages of sarcomere formation in vertebrates during cardiac myocyte development (Sanger et al., 2005). We examined the sarcomeres in B−C−/B−C− and wild-type cardiac myocytes by immunostaining using antibodies to desmin, which stain the Z-lines, and antibodies to cardiac myosin II (MF20), which stain the thick filaments. Despite the loss of NM II-B and II-C in B−C−/B−C− cardiac myocytes and absence of NM II-A in E13.5 cardiac myocytes, there are no significant alterations in sarcomere formation in the knockout cardiac myocytes (Supplemental Figure S7, D–F) compared with wild-type myocytes (Supplemental Figure S7, A–C). This suggests that NM II is not necessary for sarcomere formation at least in vivo (see Discussion).
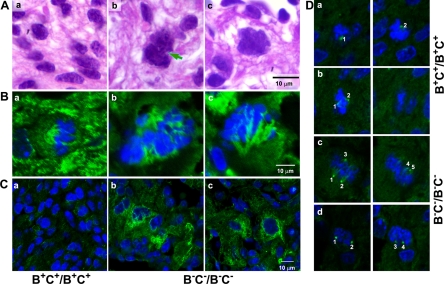
Abnormal Karyokinesis in NM II-B/II-C Doubly Ablated Cardiac Myocytes
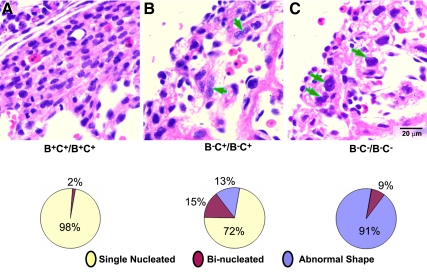
It was reported previously that NM II-B null mice have increased numbers of binucleated cells (23%) at E13.5, which was attributed to the loss of NM II-B with a resultant defect in cytokinesis (Takeda et al., 2003). In the B−C−/B−C− null mice, there is a marked increase in multilobed nuclei and in Figure 4, we show quantification of the number of abnormally shaped, double-nucleated, and single-nucleated cardiac myocytes present in the compact myocardium of wild-type, B−C+/B−C+, and B−C−/B−C− mice (Figure 4, bottom). Whereas only 13 ± 8% of the nuclei of B−C+/B−C+ myocytes were abnormal in shape (n = 3 mice, >1000 myocytes/mouse counted), 91 ± 8% of the nuclei of the B−C−/B−C− myocytes had major abnormalities in shape (n = 3 mice, >1000 myocytes/mouse counted). Figure 5A, b and c, shows examples of abnormally shaped nuclei in the cardiac myocytes of B−C−/B−C− hearts compared with normal nuclei in wild-type hearts (Figure 5Aa). The nuclei are irregular and significantly larger in B−C−/B−C− myocytes, sometimes with a connection between two unsuccessfully separated nuclei (Figure 5Ab, arrow), indicating a failure in karyokinesis. To further characterize karyokinesis in B−C−/B−C− cardiac myocytes, we stained heart sections from wild-type (Figure 5Ba) and B−C−/B−C− (Figure 5B, b and c) mice with β-tubulin (Figure 5B, green) to reveal the mitotic spindle of the dividing cells. The tissue sections were counterstained with DAPI (blue) to view chromosome alignment. In wild-type mouse hearts, the mitotic cardiac myocytes at metaphase have a bipolar mitotic spindle flanking the midaligned chromosomes (Figure 5Ba). In contrast, no bipolar spindles are found in B−C−/B−C− myocytes. Instead, only deformed spindles with irregularly aggregated chromosomes were observed (Figure 5B, b and c). More than 80 dividing cells from four wild-type and four B−C−/B−C− hearts were examined for these experiments. These results are consistent with a role for NM II in cardiac myocyte karyokinesis. To understand the possible cause of defective spindles in B−C−/B−C− cardiac myocytes, the effect of ablation of NM II on microtubule stability was examined by staining heart sections for acetylated tubulin. B−C−/B−C− cardiac myocytes show increased acetylated tubulin (Figure 5C, b and c, green) compared with B+C+B+C+ cardiac myocytes (Figure 5Ca, green). Quantification of acetylated tubulin staining shows that the fluorescence intensity for NM II-B/II-C doubly ablated myocytes is 17,354 ± 2520 pixels compared with 7855 ± 1937 pixels for wild-type myocytes (n = 3 mice, 20 myocytes counted for each mouse; p < 0.01). These results suggest that the microtubules in B−C−/B−C− cardiac myocytes become more stable after the loss of NM II. Furthermore, staining of the heart sections with γ-tubulin revealed abnormal formation of multiple centrosomes in mitotic B−C−/B−C− cardiac myocytes (Figure 5D, c and d). Ablation of NM II in the cardiac myocytes alters the dynamics of the microtubule system that subsequently contributes to a failure in karyokinesis.
Figure 4.
Abnormal nuclei in cardiac myocytes of B−C+/B−C+ and B−C−/B−C− mice. (A) H&E-stained images of heart sections from B+C+/B+C+ (A), B−C+/B−C+ (B), and B−C−/B−C− (C) mice at E13.5. Both B−C+/B−C+ (B) and B−C−/B−C− (C) hearts have significantly fewer cardiac myocytes than B+C+/B+C+ hearts (A). B−C+/B−C+ hearts show abnormal accumulation of binucleated myocytes (B, arrows), whereas B−C−/B−C− hearts contain many myocytes with multilobed nuclei (C, arrows). The percentage of abnormally shaped nuclei is shown in pie-charts (n = 3 mice for each genotype, >1000 myocytes are counted for each mouse; bottom).
Figure 5.
Impaired karyokinesis in B−C−/B−C− cardiac myocytes. (A) Images of H&E-stained cardiac myocytes from E13.5 B+C+/B+C+ (a) and B−C−/B−C− (b and c) mouse heart sections. The individual B+C+/B+C+ cardiac myocytes at this stage contain a single nucleus with regular and smooth surfaces (a). Most B−C−/B−C− nuclei are larger and seem multilobed (b and c), and sometimes have a connection between two unsuccessfully separated nuclei (arrow, b). (B) Immunofluorescence confocal images show the mitotic spindles of the cardiac myocytes from E13.5 B+C+/B+C+ (a) and B−C−/B−C− (b and c) mouse hearts stained for β-tubulin (green). Chromosomes are stained with DAPI (blue). The dividing B+C+/B+C+ myocytes at metaphase show bipolar mitotic spindles (a). In contrast, no regular bipolar spindles are identified in B−C−/B−C− myocytes (b and c). (C) Immunofluorescence confocal images of cardiac myocytes from E13.5 B+C+/B+C+ (a) and B−C−/B−C− (b and c) mouse hearts stained for acetylated-tubulin (green). B−C−/B−C− cardiac myocytes show an increase in acetylated-tubulin staining (b and c) compared with B+C+/B+C+ cardiac myocytes (a), suggesting an increased stability of the microtubules in these B−C−/B−C− cells. (D) Immunofluorescence confocal images of mitotic cardiac myocytes from E13.5 B+C+/B+C+ (a and b) and B−C−/B−C− (c and d) mouse hearts stained for γ-tubulin (green, centrosome marker). Although B+C+/B+C+ cells contains only two centrosomes (a and b), multiple centrosomes are seen in B−C−/B−C− cells (c and d). DAPI stains the nuclei (blue). The same cell is shown at different focal plains in both right and left panels. Numbers identify different centrosomes.
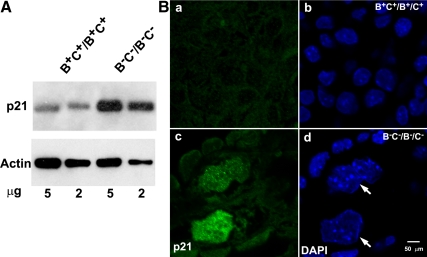
Immunoblot analysis (Figure 6A) and immunofluorescence confocal microscopy (Figure 6B) show a marked increase in expression of the cyclin-dependent kinase inhibitor p21(WAF1/CIP1) in the nuclei of B−C−/B−C− cardiac myocytes compared with the wild-type myocytes. This is also consistent with defects in karyokinesis of B−C−/B−C− cardiac myocytes. The increased p21 expression probably forces cardiac myocytes to exit from the cell cycle and thereby contributes to the development of a hypoplastic heart in B−C−/B−C− mice. Of note, cells with defects in karyokinesis are usually removed through apoptosis; however, terminal deoxynucleotidyl transferase dUTP nick-end labeling assay shows no increase in apoptosis in B−C−/B−C− cardiac myocytes. This is consistent with the increase in p21 expression in these cells.
Figure 6.
Increased p21 expression in B−C−/B−C− cardiac myocytes. (A) Immunoblot analysis of p21 expression from 5 and 2 μg of E13.5 mouse heart extracts shows increased expression of p21 in B−C−/B−C− hearts compared with B+C+/B+C+ hearts. Actin expression is used as a loading control. (B) Immunofluorescence confocal images of E13.5 heart sections from B+C+/B+C+ (a and b) and B−C−/B−C− (c and d) mice stained for p21 (green). DAPI stains the nuclei (blue). B−C−/B−C− cardiac myocytes (large nuclei) but not the nonmyocytes show increased p21 expression (c, green) compared with B+C+/B+C+ cardiac myocytes (a, green). Note the multilobed nuclei of B−C−/B−C− cardiac myocytes (d, arrows).
Defects in Karyokinesis in the HL-1 Cardiac Myocyte Cell Line
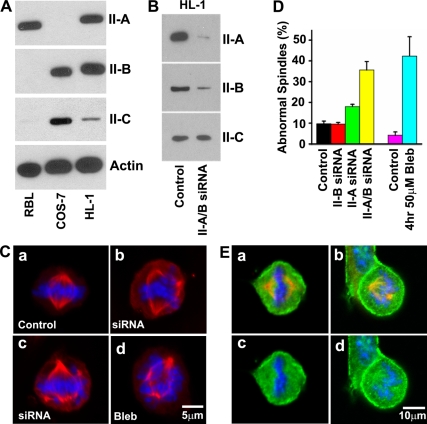
To further test the hypothesis that NM II plays a role in spindle formation in situ we made use of the HL-1 cell line, originally derived from a mouse atrial cardiac myocyte tumor (Claycomb et al., 1998). This cell line differs from primary cardiac myocytes in containing NM II-A, NM II-B, and NM II-C as detected by immunoblot analysis (Figure 7A). We first made use of mass spectroscopy to quantify the various peptides for each of the isoforms and found that 95 ± 3% were derived from NMHC II-A and 5 ± 3% of the peptides were derived from NMHC II-B. (NMHC II-C was not detected due to the very low levels). To evaluate the function of NM II in karyokinesis in HL-1 cells in culture, we either depleted NM II expression using siRNA (Figure 7B) or inhibited NM II activity using the small chemical inhibitor blebbistatin.
Figure 7.
NM II is required for mitotic spindle formation in HL-1 cells. (A) Immunoblot analysis of NMHC II isoform expression in HL-1 cells. All three isoforms of NMHC II are detected in HL-1 cells. RBL cells and COS-7 cells are used as positive controls for NMHC II-A or NMHC II-B and II-C, respectively. Actin is a loading control. (B) Immunoblot analysis shows double knockdown of NMHC II-A and II-B 72 h after siRNA II-A and II-B treatment. (C) Immunofluorescence confocal images show typical metaphase mitotic spindles (red, β-tubulin; blue, DAPI staining) from control (a), 72 h after NMHC II-A and II-B double siRNA treatment (b and c), and 4 h after 50 μM blebbistatin treatment (d) of HL-1 cells. Simultaneous siRNA NM II-A and II-B knockdown or blebbistatin-treated HL-1 cells develop abnormal spindles. (D) Quantification of abnormal mitotic spindles in HL-1 cells after 72 h of siRNA NMHC II knockdown or 4 h of blebbistatin treatment. Note that simultaneous siRNA knockdown of NM II-A and II-B markedly increases the percentage of HL-1 cells showing abnormal spindles compared with siRNA NM II-A or II-B knockdown alone. (E) Immunofluorescence confocal images of the mitotic spindle from control HL-1 cells stained for NMHC II-A (a and c, green) II-B (b and d, green), and β-tubulin (a and b, red). NMHC II-A and II-B costain with β-tubulin (top, yellow), indicating localization of both NMHC II-A and II-B in mitotic spindles of dividing HL-1 cells, in addition to their cortical localizations (green).
When HL-1 cells are treated simultaneously with siRNA to NM II-A and II-B for 72 h or with 50 μM blebbistatin for 4 h, they develop markedly abnormal spindles in the dividing cells (Figure 7C) similar to II-B and II-C ablated myocytes. Quantification (Figure 7D) shows that lowering both isoforms using siRNA results in 35.6 ± 4.1% of the nuclei having abnormalities in chromosome segregation. Inhibition of NM II motor activity by 50 μM blebbistatin results in 42.3 ± 9.3% of the nuclei having the same defect as siRNA-treated cells. These results indicate that siRNA knockdown of NM II expression or blebbistatin inhibition of NM II activity impairs karyokinesis in HL-1 cells in culture. These results also show an additive effect of siRNA induced reduction of NM II-A and II-B. As shown in Figure 7D, knockdown of NM II-B alone shows no difference in karyokinesis compared with the control (9.7 ± 1.3 vs. 9.3 ± 0.8%; p > 0.05, n = 4). Although knockdown of II-A alone significantly increases the number of cells with abnormal karyokinesis (18.0 ± 1.1%, p < 0.0001 compared with control, n = 4), knockdown of II-A and II-B together doubles the number of cells with this defect (35.6.±4.1%; p < 0.0001, n = 4).
Coimmunostaining of endogenous NM II-A (Figure 7E, a and c, green) or endogenous II-B (Figure 7E, b and d, green) and β-tubulin (red) in HL-1 cells shows the presence of both NM II-A and NM II-B in the mitotic spindle. Both NM II-A and II-B also are enriched in the cortical area of these cells.
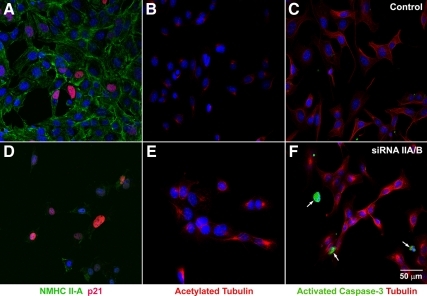
Staining of the HL-1 cells for p21 shows only a small increase in p21 expression in siRNA-treated HL-1 cells (Figure 8D, red) compared with control cells (Figure 8A, red). However a significant percentage (15 ± 5%) of control HL-1 cells are already positive for p21, and the number of p21-positive cells increases to 32 ± 8% after siRNA NM II-A/II-B treatment. In contrast to cardiac myocytes in vivo, staining for activated caspase-3 shows an increase in apoptotic cells after siRNA treatment (Figure 8F). This most likely reflects the differences between a cultured tumor cell line and intact cardiac tissue. Similar to NM II-B/II-C doubly ablated cardiac myocytes, NM II-A/II-B siRNA-treated HL-1 cells also show a marked increase in acetylated tubulin (4421 ± 1082 pixels; Figure 8E, red) compared with the control cells (574 ± 221 pixels; Figure 8B, red). The increase in acetylated tubulin is seen in both mitotic and nonmitotic HL-1 cells after siRNA NM II-A/B treatment.
Figure 8.
Increased acetylated tubulin in siRNA NM II-A/II-B–treated HL-1 cells. Immunofluorescence confocal images of B+C+/B+C+ HL-1 cells (A–C) and NM II-A/II-B siRNA-treated HL-1 cells (D–F) stained with antibodies to NMHC II-A (A and D, green), p21 (A and D, red), acetylated tubulin (B and E, red), activated caspase-3 (C and F, green), and β-tubulin (C and F, red). NMHC II-A is significantly lower in siRNA-treated cells (D, green) compared with controls (A, green). Although the number of p21-positive cells increases after siRNA treatment, the overall expression level of p21 is not obviously different in siRNA-treated individual cells (D) compared with control cells (A). siRNA-treated HL-1 cells show marked increase in acetylated tubulin (E, red) compared with control cells (B, red). siRNA treatment also increases the number of apoptotic cells detected by activated caspase-3 (F, green) compared with control HL-1 cells (C). DAPI (blue) stains the nuclei.
DISCUSSION
Although NM II-C–ablated mice seem healthy and have no difference in life span compared with the wild-type mice, we cannot exclude subtle changes we failed to detect or possible abnormalities that might develop when they are challenged by stress. This is in contrast to ablation of NM II-A or II-B that causes lethality by E6.5 and E14.5, respectively (Tullio et al., 1997; Conti et al., 2004). One reason may relate to both the temporal and spatial patterns of NM II-C expression in developing mice. Unlike NM II-A and NM II-B, NM II-C is not detected in mice before E10.5. In addition, except for epithelial cells the expression level of NM II-C in tissues is very low compared with NM II-A or II-B. In the heart and brain, for example, NM II-C is only detected by immunoblot or immunostaining analyses (Conti et al., 2004; Golomb et al., 2004). Relative to NM II-A and II-B, II-C levels are so low that mass spectroscopy analysis of lysates prepared from hearts and brains fails to detect peptides from NMHC II-C. Furthermore, NM II-C is always coexpressed along with II-A or II-B in cells. Therefore, expression of more abundant amounts of NM II-A or II-B may functionally compensate for the loss of NM II-C. Studies from cultured cells show putative functions for NM II-C in cytokinesis (Jana et al., 2006) and neurite outgrowth (Wylie and Chantler, 2008). However, ablation of NM II-C in mice failed to show these abnormalities. This may be due to the differences, including the content of NM II isoforms, between cultured cells and intact tissues in vivo. Cells in culture grow in a defined environment that may limit the compensatory effects from other isoforms of NM II.
B−C−/B−C− Mice Develop a Beating Heart with Normal Myofibril Formation
Studies from cultured cardiac myocytes proposed a role for NM II in myofibril formation (Sanger et al., 2005). Above, we present data on the absence of a requirement for NM II in myofibril formation in mice in vivo. Although NM II-A is detected in cardiac myocytes in the heart tube at E7.5 in the developing arterial pole area, it is not detected in the ventricular cardiac myocytes starting from E8.5, when they are still actively proliferating. During each myocyte division the sarcomeres disrupt and then reform (Engel et al., 2005). Ablation of NM II-B and II-C in mice results in ventricular cardiac myocytes that contain no NM II after E8.5. However, we find no difference in sarcomeric structures between B−C−/B−C− cardiac myocytes and B+C+/B+C+ cardiac myocytes. Our results suggest that there is no requirement for NM II in myofibril formation in developing cardiac myocytes in vivo, which differ from the findings in primary culture.
NM II Is Required for Karyokinesis in Cardiac Myocytes
When both NM II-B and II-C are ablated together in mouse hearts, most of the cardiac myocytes in the compact myocardium show multilobed nuclei, indicating abnormalities in karyokinesis in B−C−/B−C− cardiac myocytes. Although double ablation of both NM II-C and NM II-B markedly increases the incidence of karyokinesis defects in cardiac myocytes, ablation of II-C alone does not. Thus NM II-B alone is sufficient for normal cardiac myocyte karyokinesis during embryonic heart development. Conversely the presence of NM II-C in B−/B− mouse hearts also successfully supports normal karyokinesis for the majority of B−/B− cardiac myocytes. These results are consistent with the finding that cardiac myocytes contain larger quantities of NM II-B than II-C and both II-B and II-C play a similar role in karyokinesis. Studies from HL-1 cells demonstrate that NM II-A also plays the similar role in karyokinesis in this cell line. Previous work has demonstrated the presence of NM II-A in the mitotic spindle (Kelley et al., 1996), and the current study shows the presence of NM II-A and II-B in the mitotic spindle of HL-1 cells. These data provide strong evidence supporting the function of NM II in karyokinesis in the heart in vivo during mouse development and in cultured HL-1 cells.
It has been reported that NM II plays a role in mitotic spindle assembly by regulating centrosome separation and positioning after nuclear envelope breakdown in cultured cells (Rosenblatt et al., 2004). Inhibition of NM II activity by blebbistatin or siRNA depletion of NM II resulted in asymmetric spindles in PtK2, B6–8 hybridoma and Drosophila S2R+ cell lines that normally completed centrosome movement after nuclear envelope breakdown. However in a study analyzing the role of Rho GTP in mitosis, it was demonstrated that NM II was not required for spindle assembly in the Rat-2 cell line (Bakal et al., 2005). In addition, inhibition of NM II by blebbistatin in HeLa cells showed no spindle abnormalities except for blockage of cytokinesis (Straight et al., 2003). Thus, the requirement of NM II in spindle formation seems to be cell dependent in cultured cell lines. NM II is involved in asymmetric spindle positioning in Caenorhabditis elegans zygote mitosis, in mouse oocyte polar body extrusion and in Dictyostelium mitosis after mechanical disturbance (Effler et al., 2006; Deng et al., 2007; Goulding et al., 2007).
A requirement for NM II in karyokinesis in live organisms is not well documented in the literature. Genetic ablation or mutation of NM II in Drosophila or Dictyostelium showed no major defects in karyokinesis (Young et al., 1993; Zang et al., 1997). One possible explanation is that cells with defects in karyokinesis are eliminated by apoptosis that results in underscoring such abnormalities. Interestingly, in B−C−/B−C− mouse hearts, cardiac myocytes with defects in karyokinesis show enhanced p21 expression. It is well known that activation of p21 signaling can inhibit apoptosis (Gartel and Tyner, 2002). This may contribute to the accumulation of cardiac myocytes containing multilobed nuclei in B−C−/B−C− mouse hearts. However, cardiac myocytes are not the only cells that require NM II in karyokinesis. For example, germline ablation of NM II-A in mice results in a visceral endoderm that, in contrast to the other cells of the E6.5 developing embryo, is devoid of all NM IIs. The NM II-A null cells of the A−/A− visceral endoderm also show evidence for abnormalities of karyokinesis demonstrated by the presence of large and bizarre-shaped nuclei (Conti et al., 2004).
The defect in karyokinesis seems to be related to the abnormalities in mitotic spindle formation that consequently results in the failure of chromosome segregation during cell division. Increased microtubule stability in B−C−/B−C− cardiac myocytes also may contribute to abnormal spindle formation. It has been reported that the anticancer agent estramustine suppressed the dynamic instability of microtubules resulting in spindle abnormalities in MCF-7 cells in culture (Mohan and Panda, 2008). The spindle abnormalities seen in estramustine treated MCF-7 cells are similar to those observed in our NM II knockdown HL-1 cells as well as in B−C−/B−C− cardiac myocytes in vivo. An increase in microtubule stability was also observed in cultured human foreskin fibroblasts after siRNA depletion of NM II-A (Even-Ram et al., 2007). Alteration in microtubule dynamics also may contribute to the formation of multiple centrosomes in NM II-B/II-C doubly ablated cardiac myocytes. Formation of multiple centrisomes also could occur as a result of a failure in cytokinesis. This contribution in B−C−/B−C− mouse hearts is minimal, because the majority of B−C−/B−C− cardiac myocytes (>90%) have defects in karyokinesis. Our results propose a novel role for NM II in regulating mitotic spindle formation by altering microtubule dynamics.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Drs. Chengyu Liu, Yubin Du, and Wen Xie (Transgenic Core, National Heart, Lung, and Blood Institute, National Institutes of Health [NHLBI, NIH]) for providing outstanding service and advice. We acknowledge the professional skills and advice of Drs. Christian A. Combs and Daniela A. Malide (Light Microscopy Core Facility, NHLBI, NIH), Drs. Danielle Springer and Audrey Noguchi (Phenotype Core Facility, NHBLI, NIH), and Dr. Guanghui Wang (Proteomics Core Facility, NHLBI, NIH). We thank Dr. Kazuyo Takeda (NHLBI) for providing the NMHC II-C genomic clone. We thank the members of Laboratory of Molecular Cardiology for reagents and discussions. Antoine Smith's and Dalton Saunders's skillful technical assistance is gratefully acknowledged.
Abbreviations used:
- E
embryonic day
- ES
embryonic stem
- H&E
hematoxylin and eosin
- IB
immunoblot
- IF
immunofluorescence
- Neor
neomycin resistance cassette
- NM
nonmuscle myosin
- NMHC
nonmuscle myosin heavy chain
- WGA
wheat germ agglutinin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0293) on September 22, 2010.
REFERENCES
- Babbin B. A., Koch S., Bachar M., Conti M. A., Parkos C. A., Adelstein R. S., Nusrat A., Ivanov A. I. Non-muscle myosin IIA differentially regulates intestinal epithelial cell restitution and matrix invasion. Am. J. Pathol. 2009;174:436–448. doi: 10.2353/ajpath.2009.080171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C. J., Finan D., LaRose J., Wells C. D., Gish G., Kulkarni S., DeSepulveda P., Wilde A., Rottapel R. The Rho GTP exchange factor Lfc promotes spindle assembly in early mitosis. Proc. Natl. Acad. Sci. USA. 2005;102:9529–9534. doi: 10.1073/pnas.0504190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J., Jana S. S., Adelstein R. S. Vertebrate nonmuscle myosin II isoforms rescue small interfering RNA-induced defects in COS-7 cell cytokinesis. J. Biol. Chem. 2005;280:19594–19599. doi: 10.1074/jbc.M501573200. [DOI] [PubMed] [Google Scholar]
- Berg J. S., Powell B. C., Cheney R. E. A millennial myosin census. Mol. Biol. Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A., Broxson E., Witte D., Omoloja A. Thrombocytopenia and proteinuria. Nonmuscle myosin heavy-chain-9-related disease (MYH9 RD) or Epstein syndrome (ES) Pediatr. Nephrol. 2009;24:485–488. doi: 10.1007/s00467-008-0937-4. [DOI] [PubMed] [Google Scholar]
- Bresnick A. R. Molecular mechanisms of nonmuscle myosin-II regulation. Curr. Opin. Cell Biol. 1999;11:26–33. doi: 10.1016/s0955-0674(99)80004-0. [DOI] [PubMed] [Google Scholar]
- Claycomb W. C., Lanson N. A., Jr, Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., Izzo N. J., Jr HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti M. A., Even-Ram S., Liu C., Yamada K. M., Adelstein R. S. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J. Biol. Chem. 2004;279:41263–41266. doi: 10.1074/jbc.C400352200. [DOI] [PubMed] [Google Scholar]
- Deng M., Suraneni P., Schultz R. M., Li R. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev. Cell. 2007;12:301–308. doi: 10.1016/j.devcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Donaudy F., et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am. J. Hum. Genet. 2004;74:770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effler J. C., Kee Y. S., Berk J. M., Tran M. N., Iglesias P. A., Robinson D. N. Mitosis-specific mechanosensing and contractile-protein redistribution control cell shape. Curr. Biol. 2006;16:1962–1967. doi: 10.1016/j.cub.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel F. B., Schebesta M., Duong M. T., Lu G., Ren S., Madwed J. B., Jiang H., Wang Y., Keating M. T. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Ram S., Doyle A. D., Conti M. A., Matsumoto K., Adelstein R. S., Yamada K. M. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- Gartel A. L., Tyner A. L. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol. Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- Golomb E., Ma X., Jana S. S., Preston Y. A., Kawamoto S., Shoham N. G., Goldin E., Conti M. A., Sellers J. R., Adelstein R. S. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J. Biol. Chem. 2004;279:2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- Goulding M. B., Canman J. C., Senning E. N., Marcus A. H., Bowerman B. Control of nuclear centration in the C. elegans zygote by receptor-independent Galpha signaling and myosin II. J. Cell Biol. 2007;178:1177–1191. doi: 10.1083/jcb.200703159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Adelstein R. S. Neuronal cell expression of inserted isoforms of vertebrate nonmuscle myosin heavy-chain Ii-B. J. Biol. Chem. 1995;270:14533–14540. doi: 10.1074/jbc.270.24.14533. [DOI] [PubMed] [Google Scholar]
- Jana S. S., Kawamoto S., Adelstein R. S. A specific isoform of nonmuscle myosin II-C is required for cytokinesis in a tumor cell line. J. Biol. Chem. 2006;281:24662–24670. doi: 10.1074/jbc.M604606200. [DOI] [PubMed] [Google Scholar]
- Jana S. S., Kim K. Y., Mao J., Kawamoto S., Sellers J. R., Adelstein R. S. An alternatively spliced isoform of non-muscle myosin II-C is not regulated by myosin light chain phosphorylation. J. Biol. Chem. 2009;284:11563–11571. doi: 10.1074/jbc.M806574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley C. A., Sellers J. R., Gard D. L., Bui D., Adelstein R. S., Baines I. C. Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J. Cell Biol. 1996;134:675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Y., Kawamoto S., Bao J., Sellers J. R., Adelstein R. S. The B2 alternatively spliced isoform of nonmuscle myosin II-B lacks actin-activated MgATPase activity and in vitro motility. Biochem. Biophys. Res. Commun. 2008;369:124–134. doi: 10.1016/j.bbrc.2007.11.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Y., Kovacs M., Kawamoto S., Sellers J. R., Adelstein R. S. Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J. Biol. Chem. 2005;280:22769–22775. doi: 10.1074/jbc.M503488200. [DOI] [PubMed] [Google Scholar]
- Leal A., Endele S., Stengel C., Huehne K., Loetterle J., Barrantes R., Winterpacht A., Rautenstrauss B. A novel myosin heavy chain gene in human chromosome 19q13.3. Gene. 2003;312:165–171. doi: 10.1016/s0378-1119(03)00613-9. [DOI] [PubMed] [Google Scholar]
- Ma X., Bao J., Adelstein R. S. Loss of cell adhesion causes hydrocephalus in nonmuscle myosin II-B-ablated and mutated mice. Mol. Biol. Cell. 2007;18:2305–2312. doi: 10.1091/mbc.E07-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Kawamoto S., Hara Y., Adelstein R. S. A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Mol. Biol. Cell. 2004;15:2568–2579. doi: 10.1091/mbc.E03-11-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., et al. Conditional ablation of nonmuscle myosin II-B delineates heart defects in adult mice. Circ. Res. 2009;105:1102–1109. doi: 10.1161/CIRCRESAHA.109.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Mohan R., Panda D. Kinetic stabilization of microtubule dynamics by estramustine is associated with tubulin acetylation, spindle abnormalities, and mitotic arrest. Cancer Res. 2008;68:6181–6189. doi: 10.1158/0008-5472.CAN-08-0584. [DOI] [PubMed] [Google Scholar]
- Pecci A., et al. Position of nonmuscle myosin heavy chain IIA (NMMHC-IIA) mutations predicts the natural history of MYH9-related disease. Hum. Mutat. 2008;29:409–417. doi: 10.1002/humu.20661. [DOI] [PubMed] [Google Scholar]
- Phillips C. L., Yamakawa K., Adelstein R. S. Cloning of the cDNA encoding human nonmuscle myosin heavy chain-B and analysis of human tissues with isoform-specific antibodies. J. Muscle Res. Cell Motil. 1995;16:379–389. doi: 10.1007/BF00114503. [DOI] [PubMed] [Google Scholar]
- Robinson D. N., Spudich J. A. Mechanics and regulation of cytokinesis. Curr. Opin. Cell Biol. 2004;16:182–188. doi: 10.1016/j.ceb.2004.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J., Cramer L. P., Baum B., McGee K. M. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- Sanger J. W., Kang S., Siebrands C. C., Freeman N., Du A., Wang J., Stout A. L., Sanger J. M. How to build a myofibril. J. Muscle Res. Cell Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- Smutny M., Cox H. L., Leerberg J. M., Kovacs E. M., Conti M. A., Ferguson C., Hamilton N. A., Parton R. G., Adelstein R. S., Yap A. S. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat. Cell Biol. 2010;12:696–702. doi: 10.1038/ncb2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Cheung A., Limouze J., Chen I., Westwood N. J., Sellers J. R., Mitchison T. J. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Takeda K., Kishi H., Ma X., Yu Z. X., Adelstein R. S. Ablation and mutation of nonmuscle myosin heavy chain II-B results in a defect in cardiac myocyte cytokinesis. Circ. Res. 2003;93:330–337. doi: 10.1161/01.RES.0000089256.00309.CB. [DOI] [PubMed] [Google Scholar]
- Takeda K., Yu Z. X., Qian S., Chin T. K., Adelstein R. S., Ferrans V. J. Nonmuscle myosin II localizes to the Z-lines and intercalated discs of cardiac muscle and to the Z-lines of skeletal muscle. Cell Motil. Cytoskeleton. 2000;46:59–68. doi: 10.1002/(SICI)1097-0169(200005)46:1<59::AID-CM6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Tullio A. N., Accili D., Ferrans V. J., Yu Z. X., Takeda K., Grinberg A., Westphal H., Preston Y. A., Adelstein R. S. Nonmuscle myosin II-B is required for normal development of the mouse heart. Proc. Natl. Acad. Sci. USA. 1997;94:12407–12412. doi: 10.1073/pnas.94.23.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren D., et al. Gene dosage affects the cardiac and brain phenotype in nonmuscle myosin II-B-depleted mice. J. Clin. Invest. 2000;105:663–671. doi: 10.1172/JCI8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X. F., Adelstein R. S., Horwitz A. R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nature Reviews Mol. Cell. Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. M., Constantin P. E., Claycomb W. C. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am. J. Physiol Heart Circ. Physiol. 2004;286:H823–H829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- Wylie S. R., Chantler P. D. Myosin IIC: a third molecular motor driving neuronal dynamics. Mol. Biol. Cell. 2008;19:3956–3968. doi: 10.1091/mbc.E07-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P. E., Richman A. M., Ketchum A. S., Kiehart D. P. Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 1993;7:29–41. doi: 10.1101/gad.7.1.29. [DOI] [PubMed] [Google Scholar]
- Zang J. H., Cavet G., Sabry J. H., Wagner P., Moores S. L., Spudich J. A. On the role of myosin-II in cytokinesis: division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol. Biol. Cell. 1997;8:2617–2629. doi: 10.1091/mbc.8.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.