We describe the spatial organization of the two NEAT1 noncoding (nc)RNAs required for the integrity of the paraspeckle nuclear bodies. The central sequences of the long transcript are internal when its extremities and the short isoform are peripheral, indicating how RNA can contribute to the architecture of nuclear bodies.
Abstract
Paraspeckles (PSPs) are nuclear bodies associated with the retention in the nucleus of specific mRNAs. Two isoforms of a long noncoding RNA (NEAT1_v1/Menε and NEAT1_v2/Menβ) are required for the integrity of PSPs. Here, we analyzed the molecular organization of PSPs by immuno- and in situ hybridization electron microscopy. Detection of the paraspeckle markers PSPC1 and P54NRB/NONO confirm the identity between PSPs and the previously described interchromatin granule-associated zones (IGAZs). High-resolution in situ hybridization of NEAT1 transcripts revealed a highly ordered organization of IGAZ/PSPs. Although the 3.7-kb NEAT1_v1 and the identical 5′ end of the 22.7-kb NEAT1_v2 transcripts are confined to the periphery, central sequences of NEAT1_v2 are found within the electron-dense core of the bodies. Moreover, the 3′ end of NEAT1_v2 also localize to the periphery, indicating possible architectures for IGAZ/PSPs. These results further suggest that the organization of NEAT1 transcripts constrains the geometry of these bodies. Accordingly, we observed in HeLa and NIH 3T3 cells that IGAZ/PSPs are elongated structures with a well-defined diameter. Our results provide new insight on the ability of noncoding RNAs to form subcellular structures.
INTRODUCTION
Nuclear compartmentalization is marked by the formation of various membraneless and dynamic nuclear bodies (NBs), including promyelocytic leukemia (PML) NBs, Cajal bodies (CBs), and paraspeckles (PSPs) (Spector, 2006). Studies on CBs and PML NBs suggest that core proteins have scaffolding roles in NB formation (Ishov et al., 1999; Lallemand-Breitenbach et al., 2001; Tucker et al., 2001; Shen et al., 2006; Kaiser et al., 2008). However, it was recently shown that several different components of NBs are competent to initiate NB formation, suggesting self-organization rather than an ordered assembly pathway (Kaiser et al., 2008). These models do not provide clues as to why there are several NBs of a limited size per nucleus rather than a single large one as a result of aggregation.
Insight into paraspeckle composition and formation may be informative within this context. PSPs are punctate ribonucleoprotein NBs (Fox et al., 2002), with a function in controlling gene expression by the nuclear retention of RNA (Bond and Fox, 2009; Chen and Carmichael, 2009). The core PSP proteins are the three members of the Drosophila Behavior Human Splicing (DBHS) family: PSPC1, P54NRB/NONO, and SFPQ/PSF. Several groups have recently shown that a long noncoding RNA (lncRNA), NEAT1(Menε/β), is a structural scaffold for PSPs and is essential for PSP formation (Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). This is the first example of an lncRNA with a role in NB formation and adds to the burgeoning list of functions for many of the thousands of newly described lncRNAs being discovered with transcriptomic approaches (Mercer et al., 2009). PSPs form close to the NEAT1 gene and their prevalence correlates with levels of NEAT1 expression, suggesting that NEAT1 is the limiting factor in PSP formation (Chen and Carmichael, 2009; Clemson et al., 2009; Sunwoo et al., 2009). In contrast, although the DBHS proteins are also required for PSP formation, unlike NEAT1, high levels of their expression do not result in PSP formation (Chen and Carmichael, 2009).
The human NEAT1 gene encodes two lncRNA isoforms of 3.7 kb (NEAT1_v1) and 22.7 kb (NEAT1_v2) that overlap completely at the 5′ end (see Figure 2A). These two transcripts were shown to be expressed to similar levels in HeLa cells (Sasaki et al., 2009). Orthologue mouse MENε/β transcripts are slightly shorter at 3.17 and 20.7 kb, respectively, and they share functionality in PSP localization (Sunwoo et al., 2009). The relative roles of the two isoforms in PSP formation are yet to be fully characterized. Stable overexpression of NEAT1_v1 increases PSP number, but only residual PSPs containing NEAT1_v1 are observed when NEAT1_v2 is knocked down (Clemson et al., 2009; Sunwoo et al., 2009), suggesting that NEAT1_v1 may not be sufficient for PSP formation. Moreover, transient NEAT1_v1 overexpression cannot rescue a NEAT1 knockdown, and NEAT1_v2 is preferentially associated with DBHS proteins in vivo, suggesting a model whereby NEAT1_v2 forms the PSP core, whereas NEAT1_v1 is recruited as a subsidiary factor (Sasaki et al., 2009).
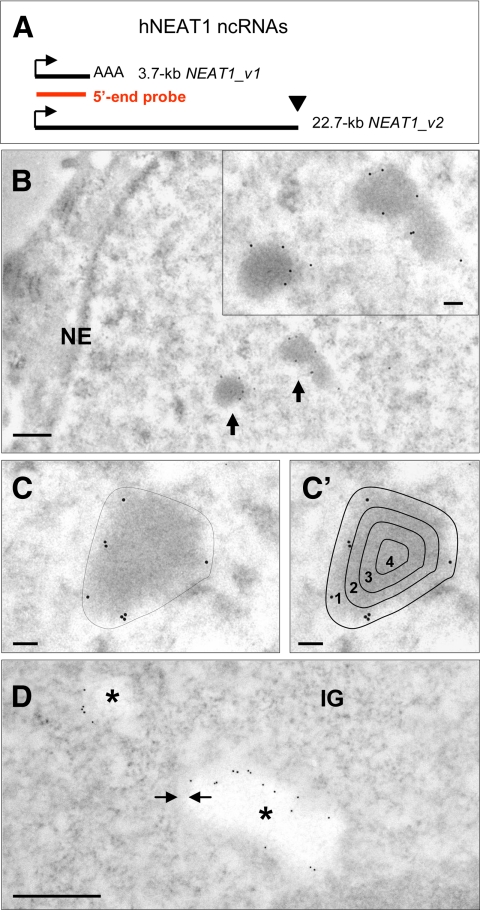
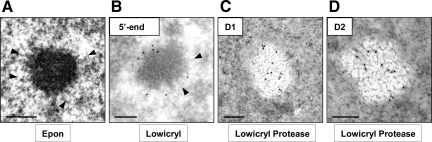
Figure 2.
Localization of NEAT1 ncRNAs by EM-ISH in HeLa cells. (A) Schematic representation of hNEAT1 ncRNAs. The black lines indicate different isoforms as labeled, and the red line indicates the position of the 5′-end biotinylated DNA probe hybridizing to NEAT1_v1 and to the 5′ end of NEAT1_v2. Identical start sites of the two transcripts are shown by arrows. Unconventional nonpolyadenylated 3′ end of the 22.7-kb transcript is illustrated by a black triangle. (B) Detection of DNA–RNA hybrids on Lowicryl-embedded thin section of HeLa cells with an anti-biotin antibody coupled to 10-nm gold particles reveals high specificity of labeling at the periphery of two IGAZ/PSPs (arrows and inset). (C and C′) Density of labeling (gold particles per surface unit) over the IGAZ/PSPs was measured as described in C, and peripheral and internal labelings were quantified as percentage of gold particles in four areas obtained by 71, 50, and 25% down-scaling of the external contour as described in (C′). (D) Protease treatment of the thin sections before EM-ISH–induced bleaching of the IGAZ/PSPs but no modification of peripheral labeling. Gold particles are located in a thin gray border (arrows) separating the bleached IGAZ/PSPs (stars) from the nucleoplasm. NE, nuclear envelope; IG, interchromatin granules. Bars, 500 nm (B and D) and 100 nm (C, C′, and inset of B).
PSPs were first described using fluorescence microscopy and were often observed adjacent to nuclear speckles in the interchromatin space. By electron microscopy (EM), PSPs are speculated to overlap the interchromatin granule-associated zones (IGAZs) (Cardinale et al., 2007; Bond and Fox, 2009) that were characterized by EM-in situ hybridization (ISH) on the basis of their high and low contents in U1 and U2 small nuclear (sn)RNA, respectively (Visa et al., 1993).
To study the distribution of the NEAT1 lncRNAs with respect to the PSP nuclear domains, we first characterized the PSPs ultrastructurally by immuno-EM. By high-resolution EM-ISH, we then studied the spatial distribution of the structural NEAT1 short and long isoforms in PSP subcompartments that we identified both in human and mouse cells.
MATERIALS AND METHODS
Cell Culture
Human HeLa and retinal pigment epithelial RPE-1 cells were maintained in DMEM and DMEM/F-12, respectively, supplemented with 10% fetal calf serum. Mouse NIH3T3 cells were maintained in DMEM supplemented with 10% calf serum.
Antibodies and Plasmids
Rabbit anti-PSPC1 was described previously (Fox et al., 2005). P54NRB/NONO monoclonal antibody was made against a peptide at the C terminus of the PSPC1 protein with 50% identity to P54NRB/NONO (Absolutions Monoclonal Antibody Service, Perth, Australia). This monoclonal recognizes P54NRB/NONO in mouse and human cells (data not shown). An SC35 monoclonal was obtained from Sigma-Aldrich (St. Louis, MO). pCRII-TOPO-hNEAT1 (1-3729 nt) was made by RTPCR of hNEAT1 from HeLa cDNA and TOPO cloning (Invitrogen, Paisley, United Kingdom). pCDNA3-mNEAT1 (1-3177 nt) was a kind gift of John Hutchinson, Harvard Medical School, Boston, MA (Hutchinson et al., 2007).
DNA Probes for EM-ISH
NEAT1 probes were biotin-labeled polymerase chain reaction (PCR)-amplified DNA fragments. We mixed 0.5 μg each of two adjacent or slightly overlapping 1- to 1.5-kb DNA fragments and then biotinylated them by nick-translation, except for human D1 probe in which 1 μg of a single 1.49-kb amplicon was used.
The human 5′-NEAT1 probe consisted of 1492-base pair and 1494-base pair DNA fragments corresponding to nt 230-1721 and 1751-3244 of the 22743 base pairs NEAT 1/MENε/β sequence (GQ859162) amplified with pCRII-TOPO-hNEAT1 DNA as a template. D1, D2 and 3′-end human NEAT1 probes were amplified from 100 ng of human genomic DNA, as follows: a 1491-base pair fragment (nt 7257-8748) for the D1 probe, fragments of 1299 base pairs and 1162 base pairs (nt 12841-14160 and 14735-15897) for D2 and fragments of 1087 base pairs (nt 20260-21346) and 1036 base pairs (nt 21647-22682) for the 3′-end NEAT1 probe. The mouse 5′-end NEAT1 probe was with 1511 and 1623 base pairs DNA fragments (nt 224-1734 and to nt 1481-3082 of NR_003513.2) amplified from 10 ng of pCDNA3-mNEAT1.
A 324-base pair fragment (nt 303-616 of HU1-1 sequence J00318.1) amplified from human genomic DNA was used as U1 snRNA probe. Amplicons were biotinylated by nick-translation for 3 h at 15°C in reactions containing 1 μg of DNA; 0.02 mM dATP, dCTP, and dGTP; and 0.05 mM biotin-16-dUTP.
Fixation and Embedding for Electron Microscopy
Cells fixed in 1.6% glutaraldehyde were dehydrated in ethanol and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate.
For embedding in Lowicryl K4M, cells were fixed 1 h either in 4% paraformaldehyde or in 1.6% glutaraldehyde at 4°C and dehydrated in methanol. Polymerization was at −30°C for 5 d under UV light. Ultrathin sections were stained with 4% uranyl acetate.
Antibodies and DNA probes used in this study were routinely tested on thin sections obtained with both fixatives. Because results obtained with both fixatives were comparable and to preserve the structures, the data shown and quantified in this study were from glutaraldehyde-fixed thin sections.
Localization of Proteins by Immunoelectron and Fluorescence Microscopy
For immuno-EM localization of the P54NRB/NONO and PSPC1 proteins, Lowicryl-embedded thin sections were incubated at room temperature for 1 h with the primary antibodies diluted 1/25 in phosphate-buffered saline (PBS) and for 30 min with anti-mouse or anti-rabbit antibodies coupled to 10-nm gold particles (BBInternational, Cardiff, United Kingdom). For immunofluorescence (IF), HeLa cells were fixed in 4% paraformaldehyde/PBS for 5 min at 4°C, permeabilized in 1% Triton-X-100/PBS for 15 min, and incubated with primary (rabbit anti-PSPC1, 1:500; mouse anti-P54NRB/NONO, 1:500; or mouse anti-SC35, 1:1000) and secondary anti-mouse-fluorescein isothiocyanate and/or anti-rabbit tetramethylrhodamine B isothiocyanate antibodies diluted at 1:250 (Jackson ImmunoResearch Laboratories, West Grove, PA). Cells were 4,6-diamidino-2-phenylindole (DAPI) stained, mounted in Vectashield (Vector Laboratories, Burlingame, CA), and imaged with an inverted Ti-E microscope (Nikon, Tokyo, Japan).
In Situ Hybridization at the Ultrastructural Level
Hybridization was performed as described previously (Visa et al., 1993; Souquere et al., 2009), for 3 h at 37°C or at 65°C for U1 snRNA and NEAT1 probes, respectively. Hybrids were detected with a goat anti-biotin antibody conjugated to 10-nm gold particles (BBInternational). Detection of DNA requires NaOH treatment of the thin sections before hybridization (Puvion-Dutilleul and Puvion, 1991; Puvion-Dutilleul and Pierron, 1992). This treatment was omitted in all our experiments. Specificity of RNA detection was controlled by a 89 or a 97% decrease of the U1 snRNA nuclear labeling when sections were pretreated either with RNase A (from 9.6 ± 3.4 to 1.1 ± 0.8 gold particles/μm2; n = 18 and 14, respectively) or were pretreated with protease and subsequently with RNase (0.3 ± 0.3 gold particles/μm2; n = 15) as in Supplemental Figure 1C.
Protease digestion was performed before hybridization with 0.2 mg/ml protease (from Streptomyces griseus, P6911, Sigma-Aldrich) in distilled water for 15 min at 37°C. RNase treatment was with 1 mg/ml RNase A (BDH, Poole, Dorset, United Kingdom) in 10 mM Tris-HCl buffer, pH 7.3, for 1 h at 37°C.
Thin sections were analyzed with a Tecnai Spirit (FEI, Hillsboro, OR) transmission electron microscope. Digital images were taken with a SIS MegaviewIII charge-coupled device camera (Olympus, Tokyo, Japan).
Quantifications
Gold particle density after EM-ISH was established on 17–28 cell profiles containing at least one IGAZ/PSP of a size ≥0.05 μm2. Approximately 125 μm2 of surrounding nucleoplasm were considered for each sample. Surface areas were determined with analySIS (Olympus Soft Imaging Solutions, Münster, Germany). Gold particles were counted by eye. Calculations and standard deviations were obtained with Excel (Microsoft, Redmond, WA).
To analyze gold particles distribution within IGAZ/PSP, four layers were delineated by progressive down-scaling of the external contour to 71, 50, and 25%. This generated four regions from the periphery to the interior with relative surface areas of 50, 25, 18.75, and 6.25%, respectively. Gold particles were counted and expressed as percentage of gold particles per area as shown in Figure 3, with no corrections for differences in surface areas.
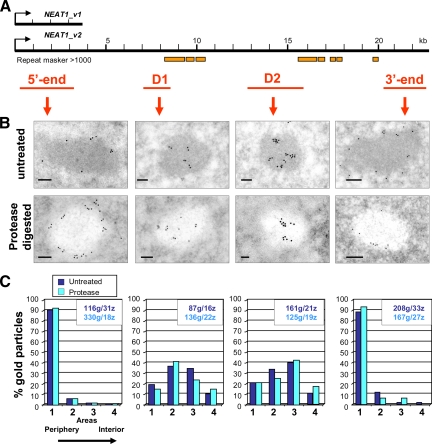
Figure 3.
Localization of various regions of NEAT1_v2 in IGAZ/PSPs of HeLa cells by EM-ISH. (A) Schematic representation of the NEAT1 transcripts and of the probes used for EM-ISH (in red). Location of repeats is indicated by orange boxes (repeat masker scores >1000). The 5′-end probe is as in Figure 2. (B) Representative labeling patterns are shown in micrographs of untreated and protease-digested thin sections in top and bottom row, respectively. Bars, 100 nm. (C) Percentage of gold particles from periphery to the interior was determined in areas 1–4 as described in Figure 2. The number of gold particles (g) and the number of IGAZ/PSPs (z) analyzed are indicated in the box of each histogram. Histograms exemplify peripheral labeling with the 5′- and 3′-end probes and internal labeling with D1 and D2 probes, irrespective of presence (dark blue) or absence (light blue) of proteins on the thin sections.
IGAZ/PSP dimensions were determined on glutaraldehyde/Lowicryl thin sections after immunolabeling with an antibody or by EM-ISH with the human or mouse 5′-NEAT1 probes. Long and short axis of the fibrillar region of the IGAZ/PSP was measured in HeLa and NIH3T3 cells by using analySIS. A two-tailed Student's test was used to compare human and murine short axis (Sx).
RESULTS
PSPs Are Identical to IGAZs
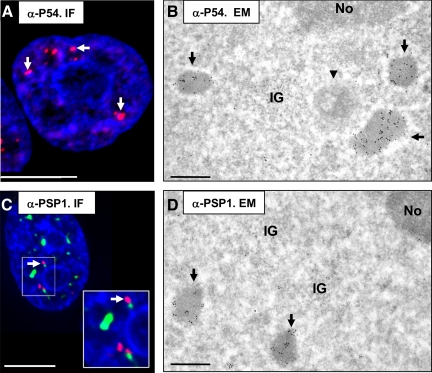
In HeLa cells, IGAZs are roundish or oblong, moderately electron-dense fibrillar zones, often found in clusters (Visa et al., 1993). We used antibodies against PSP marker proteins P54NRB/NONO and PSPC1 to determine the relationship between PSP and IGAZ. As shown in Figure 1, both antibodies specifically labeled the PSPs by IF (Figure 1, A and C) and the IGAZs, identified by their finely fibrillar structure, by immunogold labeling in the EM (Figure 1, B and D). Similar results were obtained in NIH3T3 mouse cells (see below) and in a human cell line (RPE-1) (data not shown), confirming that IGAZs correspond to PSPs (thereafter named IGAZ/PSPs).
Figure 1.
PSPs are identical to IGAZs. (A) Immunofluorescence staining of a HeLa cell with anti-P54NRB/NONO (red) to highlight paraspeckles (arrows) and chromatin (DAPI, blue). Bar, 5 μm. (B) Immunogold labeling of glutaraldehyde-fixed, Lowicryl-embedded HeLa cells with anti-P54NRB/NONO and an anti-mouse antibody coupled to 10-nm gold particles. Partial view of a nucleus showing three IGAZs (arrows), in which the P54NRB/NONO protein is highly concentrated. Notice scarce labeling of other nuclear structures, including a classical nuclear body (arrowhead), the cluster of interchromatin granules (IG) and the nucleolus (No). The cytoplasm was unlabeled. Bar, 0.5 μm. (C) Fluorescence micrograph of a HeLa nucleus stained to highlight the relationship among PSPs (anti-PSPC1, red, arrows), nuclear speckles (anti-SC35, green), and chromatin (DAPI, blue). Bar, 5 μm. Inset shows an increased magnification of the panel as indicated. (D) Immunogold labeling of cells treated as described in B with an anti-PSPC1-rabbit antibody coupled to 10-nm gold particles. IGAZs (arrows), in which the PSPC1 protein is highly concentrated, are shown. The cytoplasm was unlabeled. Bar, 0.5 μm.
EM-ISH Localization of the NEAT1 Noncoding (nc)RNAs
Relying on the ultrastructural characterization of the IGAZ/PSPs, we studied the localization at high resolution of the NEAT1 lncRNAs. A biotin-labeled DNA probe targeting the 0.23–3.2-kb region of both NEAT1 transcripts (Figure 2A) was hybridized to ultrathin sections of glutaraldehyde-fixed, Lowicryl-embedded HeLa cells. The probe does not hybridize to genomic DNA in these conditions (Puvion-Dutilleul and Pierron, 1992). DNA–RNA hybrids were revealed with an anti-biotin antibody coupled to 10-nm gold particles. Significant labeling was only found in the nucleus (Figure 2B), at a low density in the nucleoplasm (0.52 ± 0.49 gold particles/μm2; n = 24) and at a 100 times higher density (55.5 ± 32 gold particles/μm2) over the IGAZ/PSPs (Figure 2B and Table 1). Moreover, gold particles were concentrated at the periphery of the IGAZ/PSPs (Figure 2, B and C). To quantify this peripheral labeling, gold particles were counted in four areas delineated by down-scaling of the external contour (Figure 2C′). Analysis of 31 labeled structures (detailed results in Figure 3C, left) indicated that ∼90% of the gold particles were located in the outer ring that represents 50% of the surface of the IGAZ/PSPs. Thus, intensity of peripheral labeling was close to 110 particles/μm2, exceeding 220 times the labeling of the surrounding nucleoplasm. This confirms the strong enrichment of NEAT1 in PSPs as described previously (Hutchinson et al., 2007; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009), but it also highlights a preferential localization at the periphery, which was not apparent by fluorescence in situ hybridization (FISH). We repeated EM-ISH on protease-treated thin sections to determine whether proteins prevent access of the probe to more centrally located transcripts. As shown previously (Visa et al., 1993), the IGAZs are the most protease-sensitive nuclear structures and look bleached in these conditions (Figure 2D). Despite higher labeling of the IGAZ/PSPs (129 ± 31 gold particles/μm2; n = 19) and of the surrounding nucleoplasm (2.1 ± 1.55 particles/μm2; n = 19) the distribution of the gold particles was unchanged. Again, ∼90% of the particles were found in the outer ring when counted in the four areas mentioned above (Figure 3C, left). They overlap a narrow gray zone, which limits the bleached center of the IGAZ/PSPs from the more contrasted nucleoplasm (Figure 2D, arrows). Finally, we determined that U1 snRNA molecules are readily detected within the interior of the IGAZ/PSPs in our experimental conditions (Supplemental Figure 1A), even after protease treatment (Supplemental Figure 1B) but not, as expected, after protease and RNase treatments before hybridization (Supplemental Figure 1C). We concluded that the 3.7-kb NEAT1_v1 lncRNA and the 5′ end of the 22.7-kb NEAT1_v2 isoform are precisely delineating the frontier between the IGAZ/PSPs and the nucleoplasm.
Table 1.
Comparing efficiency of the various biotinylated human NEAT1 DNA probes
| NEAT1 probe | Gold particles (μm2 ± SD) |
||||
|---|---|---|---|---|---|
| Thin sections | Nucleoplasm | IGAZ/PSP | Enrichment | Frames | |
| 5′ end | Untreated | 0.52 ± 0.5 | 55.5 ± 32 | 106× | 24 |
| Protease | 2.10 ± 1.5 | 29.0 ± 31 | 61× | 19 | |
| D1 | Untreated | 0.57 ± 0.5 | 47.1 ± 29 | 82× | 18 |
| Protease | 0.70 ± 0.3 | 56.9 ± 36 | 81× | 17 | |
| D2 | Untreated | 0.65 ± 0.5 | 59.7 ± 28 | 92× | 21 |
| Protease | 0.43 ± 0.3 | 46.2 ± 19 | 107× | 17 | |
| 3′ end | Untreated | 0.53 ± 0.5 | 48.4 ± 32 | 91× | 28 |
| Protease | 0.62 ± 0.4 | 56.1 ± 31 | 90× | 22 | |
After EM-ISH with the human NEAT1 probes, density of labelling (gold particles per square micrometer of surface) was determined in the nucleoplasm and on IGAZ/PSPs of HeLa cells. For each probe, EM-ISH was carried out on untreated and on protease-digested thin sections. Mean values and SDs were calculated on number of frames indicated in the right column. Notice similar labelling densities of the IGAZ/PSP and of the surrounding nucleoplasm with the various probes. Ratio of labelling densities in the IGAZ/PSPs versus nucleoplasm in each experiment is defined as × fold enrichment.
Localization of Various Domains of the 22.7-kb NEAT1_v2 ncRNA
To specifically determine the localization of the long NEAT1_v2 ncRNA, we derived three DNA probes from sequences downstream of the first 3.7 kb, avoiding significant overlap with repeats contained within the NEAT_v2 sequence (Figure 3A). Labeling intensities after EM-ISH on HeLa cells (Supplemental Figure S2) were quantified (Table 1), and gold particles distribution was compared with results obtained with the 5′-end probe (Figure 3). As judged from absence of cytoplasmic labeling (data not shown) and from low nucleoplasmic hybridization signals (Supplemental Figure S2), the three NEAT1_v2 probes seem to be specific to the NEAT1_v2 transcript. Furthermore, with all three probes, the density of gold particles was ∼100-fold higher over IGAZ/PSPs than in the surrounding nucleoplasm as observed with the 5′-end probe (Table 1) and in agreement with FISH data (Sasaki et al., 2009; Sunwoo et al., 2009).
Remarkably, as shown in Figure 3B, EM-ISH with probe D1, corresponding to nt 7259-8729, revealed that this region of NEAT1_v2 is preferentially located in the interior of the IGAZ/PSPs, with little peripheral labeling. This sequence-specific detection of RNA segments was confirmed with probe D2, targeting nt 12843-15945, which also resulted in an internal labeling of the IGAZ/PSPs. Spectacularly, however, the labeling obtained with the 3′-end probe (nt 20260-22682) was peripheral and at first sight superimposable to the one obtained with the 5′-end probe (Figure 3B). Gold particles were counted from the periphery to the interior of the IGAZ/PSPs as illustrated in Figure 2. Histograms of the percentage of gold particles (Figure 3C), from the external to the central areas 1–4, demonstrate the highly specific localization of the 5′ and 3′ extremities of the 22.7-kb NEAT1 RNA to the outer area. In contrast, the D1 and D2 probe signals peak in areas 2 and 3, respectively. Importantly, these sequence-specific localizations were confirmed by independent EM-ISH carried out on protease digested thin sections (Figure 3, B and C).
These results demonstrate NEAT1_v2 RNA sequences internally located within the IGAZ/PSPs and reveal a highly ordered spatial organization of the NEAT1_v2 transcripts. In turn, this provides further support for the exclusive localization of NEAT1_v1 and the 5′ and 3′ extremities of NEAT1_v2 at the periphery of the IGAZ/PSPs.
Size Limit of the IGAZ/PSP
Our results suggest two extreme models for the positioning of NEAT1_v2 within the IGAZ/PSPs (Figure 4A). In one, the 5′ and 3′ ends of a single transcript would be on opposite sides (the cross-wise hypothesis). Alternatively, our results are compatible with both extremities of a transcript adjacent on the same side of an IGAZ/PSP (the looped hypothesis). Although we cannot discriminate between these two hypotheses with the data presented in Figure 3, they both suggest that one dimension of the IGAZ/PSPs is constrained by the size of the NEAT1_v2 transcript.
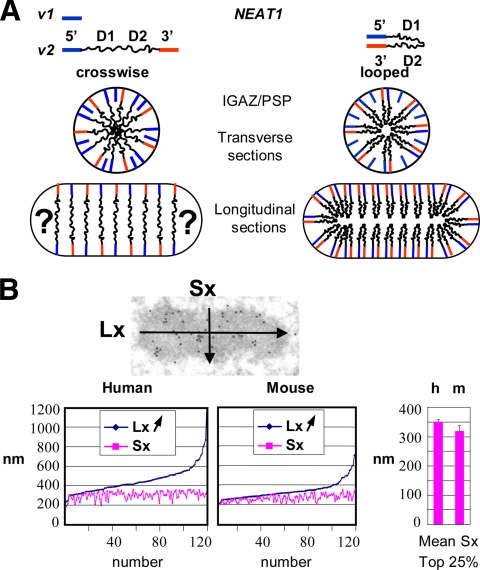
Figure 4.
Spatial organization of NEAT1_v2 and limit in size of IGAZ/PSPs. (A) Schematic representation of NEAT1_v1 and NEAT1_v2. Alternative cross-wise and looped structure models are drawn in transverse and longitudinal sections of an IGAZ/PSP. Question marks underline difficulties in drawing extremities of oblong structures with the cross-wise hypothesis. For clarity, peripheral location of the 3.7-kb NEAT1_v1 isoform was omitted in longitudinal sections. (B) Lx and Sx of 120 IGAZ/PSPs were measured on thin sections of Lowicryl-embedded human (HeLa) and mouse (NIH3T3) cells and plotted according to increasing Lx values. A size limit of Sx is obvious in both cell types. Right, comparison of Sx limit values by comparing top 25% Sx values in human and mouse cells.
We measured the short (Sx) and the long axis (Lx) of 120 IGAZ/PSPs, all validated by P54NRB/NONO immunolabeling or by EM-ISH on thin sections in HeLa cells. When dimensions are plotted by increasing Lx values as in Figure 4B, it is immediately apparent that although Lx is readily variable, Sx is constrained, reaching a limit at ∼360 nm. This distribution fits with what would be expected from the cross-sectioning of cylindrical objects with a fixed diameter.
To determine whether the constraint in size is restricted to HeLa cells, we measured, in the same conditions, the dimensions of 120 IGAZ/PSPs in mouse NIH3T3 cells. Murine IGAZ/PSPs were first characterized by EM-ISH (with a mouse-specific 5′-end NEAT1 DNA probe) or by immunolabeling with the P54NRB/NONO antibody. As with their human counterparts, murine IGAZ/PSPs were highly enriched in the mouse P54NRB/NONO homologue and coated with the 5′ ends of the NEAT1 lncRNAs (Supplemental Figure 3). Plotting their Lx and Sx values showed a distribution similar to that observed in human cells, albeit with slightly smaller dimensions (Figure 4B). Once again, the Sx values were strongly constrained. To limit underestimation of the size by partial longitudinal or transversal thin sectioning, we arbitrarily took the last quarter centile of the Sx values for comparison and found a significant difference (p < 0.001) for their mean values in human and mouse cells at 351 ± 8 and 319 ± 18 nm, respectively.
Architecture of the IGAZ/PSP Nuclear Bodies
Conventional EM in Epon-embedded HeLa or NIH3T3 cells revealed two compartments within the IGAZ/PSPs in the form of a crown of relatively large fibers (20–25 nm in diameter) emanating from a dense central core (Figure 5A). The crown of peripheral fibers is contained within a faint, white halo visible around the dense central core. This halo, that is also seen after Lowicryl embedding (Figure 5A), corresponds to the peripheral region of the IGAZ/PSPs in which the identical 5′ ends of the NEAT1 isoforms (Figure 5A) and the 3′ extremity of the long NEAT1_v2 transcript (Figure 3 and Supplemental S2) are localized.
Figure 5.
Evidencing two compartments in IGAZ/PSPs that coincide with NEAT1 isoform localization. (A) Ultrastructure of IGAZ/PSPs in conventional EM (Epon embedding), revealing a crown of fibers (arrowheads) radiating out of an electron-dense core. (B–D) Ultrastructural observations after EM-ISH on Lowicryl thin sections. (B) The crown of peripheral fibers (arrowheads) is specifically labeled by the 5′-end NEAT1 probe. (C and D) Contrast was adjusted to reveal network of fibrils after bleaching of the electron-dense core of the IGAZ/PSP by protease digestion. These fibrils are decorated by gold particles after EM-ISH with the NEAT1_v2 D1 and D2 probes, revealing internal sequences of the long NEAT1 isoform. The small and uniform diameter of these fibrils is evidenced by comparison with 10-nm gold particles. Bars, 200 nm.
The electron density of the central core is indicative of high protein content. This is consistent with its hypersensitivity to protease treatment on Lowicryl thin sections. When the central core is bleached by protease treatment, a regular network of underlying fibrils (barely visible with the contrast used in Figures 2 and 3 but seen in Figure 5, C and D) is unmasked. As expected, these fibrils contain NEAT1_v2 internal sequences as shown by EM-ISH with D1 and D2 probes in Figure 5, C and D, respectively. Therefore, our results illustrate a bipartite architecture of the IGAZ/PSPs, which reflects the differential localization of the long and short NEAT1 isoforms.
DISCUSSION
Specific Localization of the NEAT1_v1 and NEAT1_v2 lncRNAs within the IGAZ/PSP
Using a high-resolution EM-ISH protocol, we have been able to determine the specific localization of the NEAT1 transcripts in HeLa cells. Collectively, our data illustrate the high enrichment of the architectural NEAT1/Menε/β lncRNAs in PSPs in agreement with RNA-FISH experiments (Hutchinson et al., 2007; Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). We further demonstrate that the two NEAT1 isoforms are differentially located in subcompartments of IGAZ/PSPs.
NEAT1_v1, the identical 5′ end of NEAT_v2 and the 3′ end of NEAT1_v2 showed a remarkably constant peripheral labeling of the IGAZ/PSPs (Figures 2 and 3 and Supplemental Figures S2–S4). Thus there is a high density of specific ncRNA sequences coating the IGAZ/PSPs and delineating the frontier of these bodies within the nucleoplasm. This conclusion was reinforced by identification of central sequences of NEAT1_v2 that are spreading through the IGAZ/PSPs (Figure 3). The different localization of the structural NEAT1 isoforms provides a rationale for transcription of two evolutionarily conserved RNA from a single promoter. The additional supply of peripheral RNA molecules provided by the 3.7-kb NEAT1_v1 transcript may correspond to a need for more building blocks at the periphery than the interior of the IGAZ/PSP.
Size Limit of Nuclear Bodies?
NBs were originally described as nuclear inclusions as seen in the EM (de The et al., 1960). Models of assembly have been proposed for the PML bodies and the CBs. PML is essential for PML body assembly (Ishov et al., 1999). A SUMO-interacting motif (SIM) of PML and PML SUMOylation were shown to promote PML multimerization and subsequent recruitment of PML bodies components through noncovalent SUMO–SIM interactions (Shen et al., 2006). From tethering experiments in which many fusion proteins were shown to share the capacity to nucleate a new body, a nonsequential model of self-organization has been derived for CBs assembly (Kaiser et al., 2008). These models, based on protein–protein interactions, do not explain multiplicity of NBs of similar size in many cell types. In the same vein, IGAZ/PSPs are often found in clusters of closely apposed structures with a limited size.
The overlap of the 5′ and 3′ ends of NEAT1_v2 at the periphery of the IGAZ/PSPs (Figure 3) suggested that the size of these bodies might be limited by the length of the RNA. When the Lx and Sx axis of IGAZ/PSPs were measured, a size limit of the Sx was revealed (Figure 4B), whereas the Lx was more variable. Thus, considering IGAZ/PSPs as three-dimensional objects, dimensions along two of their three axes are strictly limited. Analysis of mouse NIH3T3 cells (Supplemental Figure S3) indicated that the constraint on Sx is an evolutionary conserved feature. We also noticed a 9% reduction of the size limit that mirrors the reduction in size of the mNEAT1_v2/Menβ transcript from 22.7 to 20.7 kb, strengthening the hypothesis that this transcript plays a central role in the organization of IGAZ/PSPs and determines their diameter (Figure 4B).
Sequence analysis of the human and mouse NEAT1 genes indicates short stretches of conservation, with the highest conservation occurring within NEAT1_v1 and the 3′ end of NEAT1_v2 (Sunwoo et al., 2009), raising the possibility that these regions may be critical for establishing the architecture of NEAT1_v2 within IGAZ/PSP. Of interest is the observation that transient overexpression of the first 15 kb of NEAT1_v2 was unable to rescue the NEAT1 knockdown phenotype and reform PSPs, suggesting that a complete NEAT1_v2 isoform and therefore its 3′ end is crucial for PSP formation (Sasaki et al., 2009).
Ultrastructure of the IGAZ/PSP
The size limit of the IGAZ/PSPs determines the level of folding of the architectural NEAT1 lncRNAs. The 22.7-kb human NEAT1_v2 transcript, in a linear extended form, would be 7 μm long. Superposition of its 5′ and 3′ ends at the periphery of a 360-nm-diameter structure implies a 20-fold level of compaction in the cross-wise hypothesis in Figure 4A and at least a 40-fold level of compaction in the looped hypothesis. These high levels of compaction are well within the range of folding of other structural RNAs, such as the human 18 and 28S rRNAs (6.9 kb altogether), that are ∼75-fold compacted in a 30-nm ribosome. However, the high level of compaction of the NEAT1_v2 transcript is taking place in structures that are at least 1000 times bigger than a ribosome, demonstrating an ordered molecular organization on a much higher scale.
The cross-wise-hypothesis would easily apply to spherical structures but is less suited for oblong structures, particularly at their extremities as stressed in Figure 4A. Furthermore, it implies a high concentration of RNA along the central axis. This is not seen in our observations after a short protease treatment on thin sections. In these conditions, IGAZ/PSPs are selectively bleached, whereas CBs and other NBs are much less affected (Supplemental Figure S4). Within the bleached center of the IGAZ/PSP, homogeneous thin fibers are unmasked (see Figure 5, C and D, and Supplemental Figure S4). These fibers, which are labeled by EM-ISH with the NEAT1_v2 D1, D2 and U1 snRNA probes (Figure 5, C and D, and Supplemental Figure S1B), disappear after sequential protease and RNase A treatments (Supplemental Figure S1C; data not shown), indicating that they correspond to a lattice made, at least in part, of the internal sequences of the NEAT1_v2 transcript. These RNA fibers form a regular network and never showed the central accumulation of RNA inherent to the cross-wise model. We therefore favor the looped hypothesis for NEAT1_v2 organization, a structural model that introduces the flexibility necessary for assembling bodies that, like the IGAZ/PSPs, are often elongated. Along these lines, it has been shown that after bromo-(Br)-UTP injection, IGAZs do not contain rapidly labeled Br-RNA but that their center starts to be labeled after a 12- to 13-min lag, a time that would be required for the synthesis of the 22.7-kb NEAT1_v2 (Cmarko et al., 1999).
Protease treatment also revealed a second compartment in the IGAZ/PSP in the form of a peripheral gray layer of 50 nm in width that contains NEAT1_v1 and the extremities of NEAT1_v2 (Figures 2 and 3). The bipartite structure of the IGAZ/PSP was confirmed in conventional EM as a crown of large fibers (20–25 nm) radiating out of an electron-dense network (Figure 5A).
Molecular Composition of IGAZ/PSP
Our immuno-EM and EM-ISH studies demonstrate a complete overlap between IGAZs, as originally defined in the EM, and the PSP nuclear domain originally defined by IF, in both human and mouse cells. This corroborates a published report showing by immuno-EM an enrichment of SFPQ/PSF, one of the PSP marker proteins, in IGAZ (Cardinale et al., 2007). PSPs were defined as the sites of accumulation of mammalian DBHS proteins (Fox et al., 2002). Recently, it has been shown that PSPs are critical for the nuclear retention of inverted repeat-containing, adenosine-to-inosine–edited mRNAs, probably acting as a nuclear anchor for P54NRB/NONO complexes that were first shown to trap inosine RNA in the nucleus (Zhang and Carmichael, 2001; Chen and Carmichael, 2009). These observations follow a seminal study of a retained mouse specific transcript which is released under stress conditions after cleavage of its edited 3′-untranslated region (Prasanth et al., 2005). Interestingly, an mRNA 3′-end cleavage factor also has been shown to be present in PSPs (Cardinale et al., 2007). In contrast, IGAZs were shown to contain U1 snRNA, the Sm core protein (Visa et al., 1993; Malatesta et al., 1999), the transcription factor TFIIH, and RNA polymerase II (Cmarko et al., 1999) that, associated with P54NRB/NONO and SFPQ/PSF, were isolated as specific complexes from HeLa cell extracts (Kameoka et al., 2004). This raises the possibility that IGAZ/PSPs, although not associated with active transcription (because they do not contain rapidly labeled bromo-RNA after bromo-UTP injection, Cmarko et al., 1999), or splicing (because they do not contain U2 snRNA, Visa et al., 1993), are sites of maturation or recycling of complexes involved in transcription and splicing.
Other lncRNA NBs?
Of the many thousands of lncRNAs that are transcribed, only a handful has been functionally characterized. However, the functions of this small number have provided new clues to mammalian development and pathologies, including, for example, XIST and X-inactivation (Brown et al., 1992), HOTAIR and metastasis (Gupta et al., 2010), and Evf2 and formation of the nervous system (Bond et al., 2009). NEAT1 is the first lncRNA shown to function by forming an NB, in this case essential for altering gene expression by the nuclear retention of RNA (Chen and Carmichael, 2009). Could other lncRNAs mediate their function by forming NB's? Often, lncRNA expression is highly tissue restricted, generally much more so than protein-coding genes. Indeed, two other lncRNA that are found in subnuclear foci are both tissue restricted, and in one case, Bsr, is restricted to the genus Rattus (rodents) (Royo et al., 2007; Sone et al., 2007). It will be important to examine the ultrastructure of such foci to determine whether similar lncRNA architectures are observed.
We show that the ultrastructure of a nuclear body reflects an elaborate spatial arrangement of two architectural lncRNAs. Given the striking protein enrichment of the IGAZ/PSPs, and the importance of the DBHS proteins in IGAZ/PSP formation, it will now be interesting to understand how interactions between NEAT1 lncRNAs and proteins form the IGAZ/PSP.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Hutchinson for pCDNA3-mNEAT1 plasmid; and M. Bénard, C. Bond, F. Dautry, M Ernoult-Lange, and D. Weil for help and discussions. This work was supported by the Centre National de la Recherche Scientifique, Université Paris XI, Association pour la Recherche contre le Cancer, and the National Health and Medical Research Council of Australia (to A. F.).
Abbreviations used:
- EM-ISH
electron microscopy-in situ hybridization
- IGAZ
interchromatin granule-associated zone
- NEAT1_v1/Menε and NEAT1_v2/Menβ
3.7- and 22.7-kb isoforms of nuclear enriched abundant transcript 1/multiple endocrine neoplasia noncoding RNA
- PSP
paraspeckle nuclear body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0690) on September 29, 2010.
REFERENCES
- Bond A. M., Vangompel M. J., Sametsky E. A., Clark M. F., Savage J. C., Disterhoft J. F., Kohtz J. D. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C. S., Fox A. H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Hendrich B. D., Rupert J. L., Lafreniere R. G., Xing Y., Lawrence J., Willard H. F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Cardinale S., Cisterna B., Bonetti P., Aringhieri C., Biggiogera M., Barabino S. M. Subnuclear localization and dynamics of the Pre-mRNA 3′ end processing factor mammalian cleavage factor I 68-kDa subunit. Mol. Biol. Cell. 2007;18:1282–1292. doi: 10.1091/mbc.E06-09-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L., Carmichael G. G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C. M., Hutchinson J. N., Sara S. A., Ensminger A. W., Fox A. H., Chess A., Lawrence J. B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D., Verschure P. J., Martin T. E., Dahmus M. E., Krause S., Fu X. D., van Driel R., Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell. 1999;10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de The G., Riviere M., Bernhard W. Examination by electron microscope of the VX2 tumor of the domestic rabbit derived from the Shope papilloma. Bull. Assoc. Fr. Etud. Cancer. 1960;47:570–584. [PubMed] [Google Scholar]
- Fox A. H., Bond C. S., Lamond A. I. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A. H., Lam Y. W., Leung A. K., Lyon C. E., Andersen J., Mann M., Lamond A. I. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Gupta R. A., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. N., Ensminger A. W., Clemson C. M., Lynch C. R., Lawrence J. B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T. E., Intine R. V., Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kameoka S., Duque P., Konarska M. M. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta M., Fakan S., Fischer U. The Sm core domain mediates targeting of U1 snRNP to subnuclear compartments involved in transcription and splicing. Exp. Cell Res. 1999;249:189–198. doi: 10.1006/excr.1999.4468. [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mattick J. S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Prasanth K. V., Prasanth S. G., Xuan Z., Hearn S., Freier S. M., Bennett C. F., Zhang M. Q., Spector D. L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F., Pierron G. Localization by high resolution in situ hybridization of the ribosomal minichromosomes during the nucleolar cycle of Physarum polycephalum. Exp. Cell Res. 1992;203:354–364. doi: 10.1016/0014-4827(92)90009-w. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F., Puvion E. Ultrastructural localization of defined sequences of viral RNA and DNA by in situ hybridization of biotinylated DNA probes on sections of herpes simplex virus type 1 infected cells. J. Electron Microsc. Tech. 1991;18:336–353. doi: 10.1002/jemt.1060180403. [DOI] [PubMed] [Google Scholar]
- Royo H., Basyuk E., Marty V., Marques M., Bertrand E., Cavaille J. Bsr, a nuclear-retained RNA with monoallelic expression. Mol. Biol. Cell. 2007;18:2817–2827. doi: 10.1091/mbc.E06-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y. T., Ideue T., Sano M., Mituyama T., Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Hayashi T., Tarui H., Agata K., Takeichi M., Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Souquere S., Mollet S., Kress M., Dautry F., Pierron G., Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 2009;122:3619–3626. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- Spector D. L. SnapShot: cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Sunwoo H., Dinger M. E., Wilusz J. E., Amaral P. P., Mattick J. S., Spector D. L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K. E., Berciano M. T., Jacobs E. Y., LePage D. F., Shpargel K. B., Rossire J. J., Chan E. K., Lafarga M., Conlon R. A., Matera A. G. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Puvion-Dutilleul F., Bachellerie J. P., Puvion E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur. J. Cell Biol. 1993;60:308–321. [PubMed] [Google Scholar]
- Zhang Z., Carmichael G. G. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.