Abstract
Many RNA viruses remodel intracellular membranes to generate specialized sites for RNA replication. How membranes are remodeled and what properties make them conducive for replication are unknown. Here we show how RNA viruses can manipulate multiple components of the cellular secretory pathway to generate organelles specialized for replication that are distinct in protein and lipid composition from the host cell. Specific viral proteins modulate effector recruitment by Arf1 GTPase and its guanine nucleotide exchange factor GBF1, promoting preferential recruitment of phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ) to membranes over coat proteins, yielding uncoated phosphatidylinositol-4-phosphate (PI4P) lipid-enriched organelles. The PI4P-rich lipid microenvironment is essential for both enteroviral and flaviviral RNA replication; PI4KIIIβ inhibition interferes with this process; and enteroviral RNA polymerases specifically bind PI4P. These findings reveal how RNA viruses can selectively exploit specific elements of the host to form specialized organelles where cellular phosphoinositide lipids are key to regulating viral RNA replication.
PaperFlick
Keywords: CELLBIO, MICROBIO
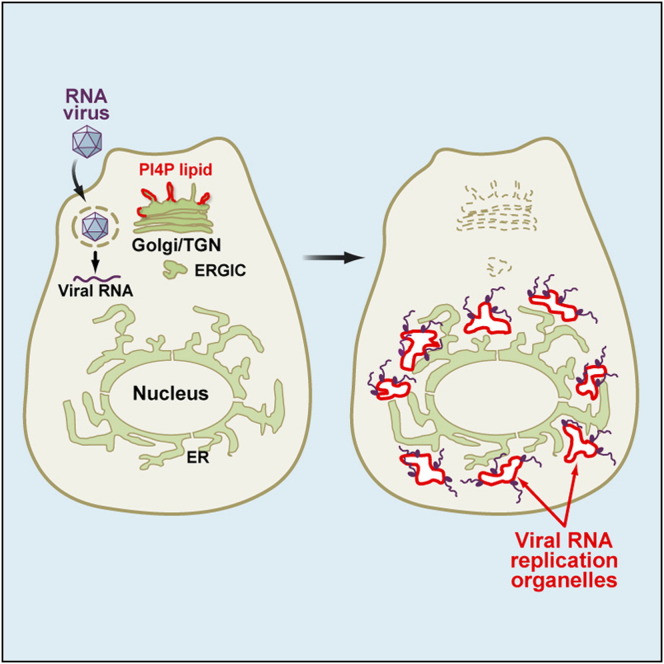
Graphical Abstract
Highlights
► RNA viruses generate specialized RNA replication organelles enriched in PI4P lipids ► Host PI4KIIIβ enzyme is co-opted by the virus to generate PI4P lipid-rich organelles ► Enteroviral RNA polymerases specifically and preferentially bind PI4P lipids ► PI4P microenvironment is essential for enteroviral and flaviviral RNA synthesis
Introduction
Viruses rely on their host for viability and replication. During infection, the virus and host become engaged in a dynamic duet, lasting from several hours to potentially years (in persistent infections), in which the virus initiates spatio-temporally ordered sequences of subcellular events, along the way dramatically altering cellular architecture and physiology. The host cell not only provides building blocks such as nucleotides and amino acids for viral metabolism but also can provide a structural platform for replication and viral assembly (Uetz et al., 2006). Many RNA viruses and even some DNA viruses such as the poxviruses rely on host intracellular membranes for replication (Miller and Krijnse-Locker, 2008, Salonen et al., 2005). In particular plus-strand RNA virus families, so called because upon infection their RNA can be directly translated into protein by host machinery, replicate and assemble on modified intracellular membranes (Miller and Krijnse-Locker, 2008, Salonen et al., 2005). The group of plus-strand RNA viruses includes many important human pathogens like picornaviruses (such as the enteroviral genus members poliovirus [PV] and Coxsackievirus B3 [CVB3], rhinovirus, and hepatitis A), coronaviruses (SARS), and flaviviruses (hepatitis C virus [HCV], Yellow Fever virus, Dengue Fever virus, West Nile virus).
Cells infected with plus-strand RNA viruses undergo a dramatic remodeling of their intracellular membranes, and RNA replication frequently takes place on the cytosolic leaflet of these remodeled membranes (Dales et al., 1965, Miller and Krijnse-Locker, 2008, Salonen et al., 2005). Replication membranes for picornaviruses, flaviviruses, and coronaviruses appear to originate from the endoplasmic reticulum (ER) (Schlegel et al., 1996), whereas for togaviruses and nodaviruses the endosomes/lysosomes and mitochondria are thought to be the membrane source (Magliano et al., 1998). Some viral replication enzymes have sequences that integrate into the host membrane bilayer, such as NS4B, a polytopic membrane protein of HCV (Lundin et al., 2003), and the 2B and 3A proteins of picornaviruses (Richards and Ehrenfeld, 1990). Many, however, are soluble proteins, whose mechanism of membrane association is unknown.
Plus-strand RNA viruses are critically dependent on intracellular membranes (Miller and Krijnse-Locker, 2008, Richards and Ehrenfeld, 1990, Salonen et al., 2005), but the properties of the replication membranes that are required to support viral RNA replication have not been defined. It has been speculated that membranes may limit diffusion of viral/host proteins and viral RNA, thereby increasing the local concentration of reaction elements; or that membranes may provide specific lipids that participate in the replication reactions themselves (Miller and Krijnse-Locker, 2008). A number of cellular factors have been implicated in viral RNA replication; e.g., several high-throughput siRNA screens have identified potential cellular factors whose knockdown reduces flavivirus replication (including components of the endosomal machinery, actin modulators, and phospholipid-modifying enzymes such as phosphatidylinositol-4 kinases), although the mechanism by which any of these proteins might regulate replication is not known (Berger et al., 2009, Borawski et al., 2009, Tai et al., 2009, Trotard et al., 2009, Vaillancourt et al., 2009). For PV and CVB3, both members of the enterovirus genus of the picornavirus family, we have shown that GBF1, a guanine nucleotide exchange factor (GEF) of the small Ras-family GTPase Arf1, was required for enteroviral RNA replication (Belov et al., 2007, Lanke et al., 2009). GBF1 catalyzes GDP/GTP exchange on Arf1, stabilizing membrane association, which in turn recruits various effectors to these membranes (Altan-Bonnet et al., 2004, Niu et al., 2005). In uninfected mammalian cells, GBF1 and Arf1 are both localized to the ER, ER-Golgi intermediate compartment (ERGIC), and the Golgi apparatus. Arf1's known major effectors at these sites include coat proteins such as COPI complex and clathrin, which regulate membrane budding, and phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ), which catalyzes the production of phosphatidylinositol-4-phosphate (PI4P) lipids at the membrane bilayer (Godi et al., 1999, Altan-Bonnet et al., 2004, Lee et al., 2004).
Here we focus on the in situ properties of the viral RNA replication membranes in cells infected with plus-strand RNA viruses. We demonstrate how remodeling of the host secretory pathway by enteroviral replication proteins generates organelles with unique protein and lipid composition geared for replicating viral RNA. We show that a specific enteroviral protein modulates effector recruitment by GBF1 and Arf1, promoting PI4KIIIβ recruitment to secretory organelle membranes. This leads to disassembly of conventional secretory organelles and assembly of “replication organelles” that are juxtaposed to ER exit sites. PI4KIIIβ at these organelle membranes produces a PI4P lipid microenvironment, which facilitates membrane binding of enteroviral RNA polymerase and viral RNA synthesis. Finally, we find that flaviviruses, specifically HCV, also induce and depend on the PI4P lipid microenvironments for RNA replication. Thus PI4KIIIβ is a key cellular protein exploited by several plus-strand RNA viruses for replication.
Results
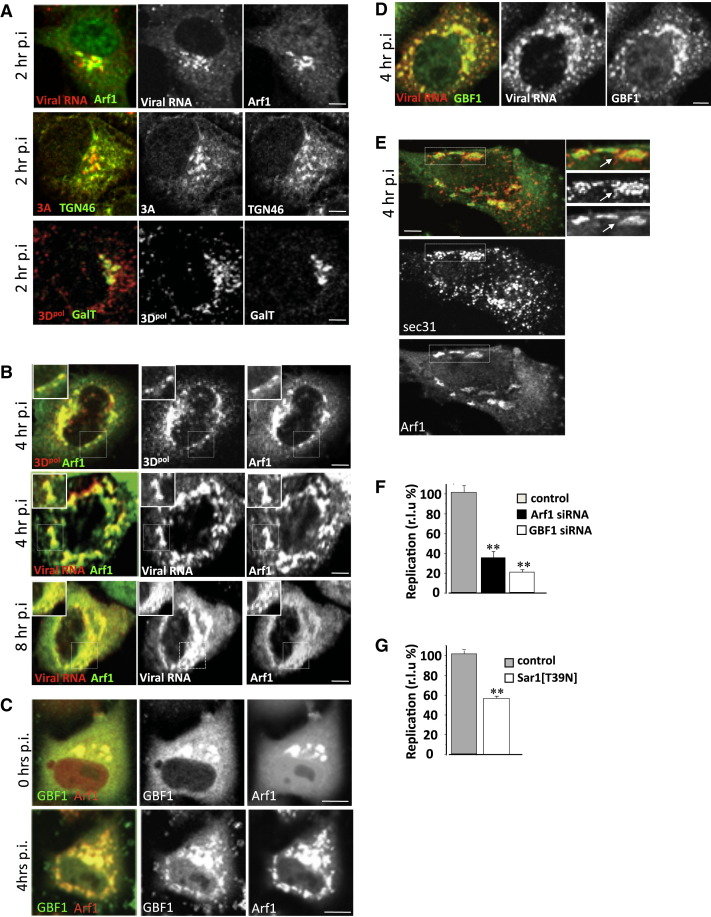
We first investigated the in situ properties of RNA virus replication sites during enteroviral infections. Two hours after infection with CVB3, we were able to detect newly synthesized viral plus-strand RNA molecules (by fluorescence in situ hybridization) localized to the host secretory Golgi/trans-Golgi network (TGN) compartments by colabeling with Arf1-GFP (Figure 1A), a fluorescent protein-tagged version of the host small GTPase Arf1, which functionally mimics the native Arf1 GTPase in both uninfected and virally infected cells (Niu et al., 2005, Presley et al., 2002; Figure S1 A available online). Viral replication protein 3A, a small tail-anchored membrane protein (Towner et al., 1996), and 3Dpol, the RNA-dependent RNA polymerase (RdRp), both components of the viral replication enzyme complex (Richards and Ehrenfeld, 1990), were also localized to the Golgi apparatus and TGN compartments (Figure 1A), signifying that these compartments were sites of initial viral RNA synthesis.
Figure 1.
Enteroviral RNA and Replication Machinery Are Localized to the Host Secretory Pathway Organelles
(A) Viral RNA and viral replication protein (3A, 3Dpol) subcellular distribution in early stages of CVB3 RNA replication.
(B) Viral RNA and viral replication protein (3A, 3Dpol) subcellular distribution at peak stages of CVB3 RNA replication. See also Figure S1.
(C) Arf1-RFP and GBF1-YFP dynamics in CVB3-infected HeLa cell. Confocal time-lapse images of single cell are presented. See also Movie S1.
(D) GBF1 and viral RNA are colocalized in HeLa cells during CVB3 infection.
(E) Replication organelles, at peak replication, form adjacent to ER exit sites.
(F) Arf1 and GBF1 facilitate viral RNA replication. CVB3 replicon assays in HeLa cells pretreated with siRNA against Arf1 or GBF1. Bar graph presents maximum replication values for each condition, normalized to control samples transfected with nontargeting siRNA. Error bars are SEM from eight replicates for each condition (∗∗p < 0.01). See also Figures S2A–S2C and Table S1.
(G) Functional ER exit sites facilitate viral RNA replication. PV replicon assays in HeLa cells transiently expressing Sar1[T39N] plasmid. Bar graph presents maximum replication values, normalized to control samples transfected with GFP. Error bars are SEM from eight replicates for each condition (∗∗p < 0.01). See also Table S1. r.l.u% = relative light unit %. All fluorescence images were confocal images of optical slice thickness ∼1 μm. Scale bar, 10 μm.
Figure S1.
Arf1 and Enteroviral Replication Protein Colocalization, Related to Figure 1
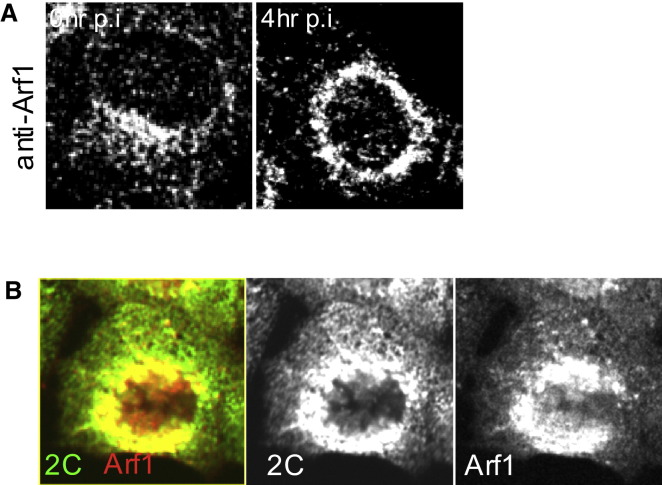
(A) Native Arf1 distribution in uninfected and CVB3-infected cells mimics that of Arf1-GFP.
(B) Confocal image (optical slice 2 μm) of a single HeLa cell expressing Arf1-CFP immunostained with antibodies to viral 2C protein and CFP, at 4 hr infection with PV.
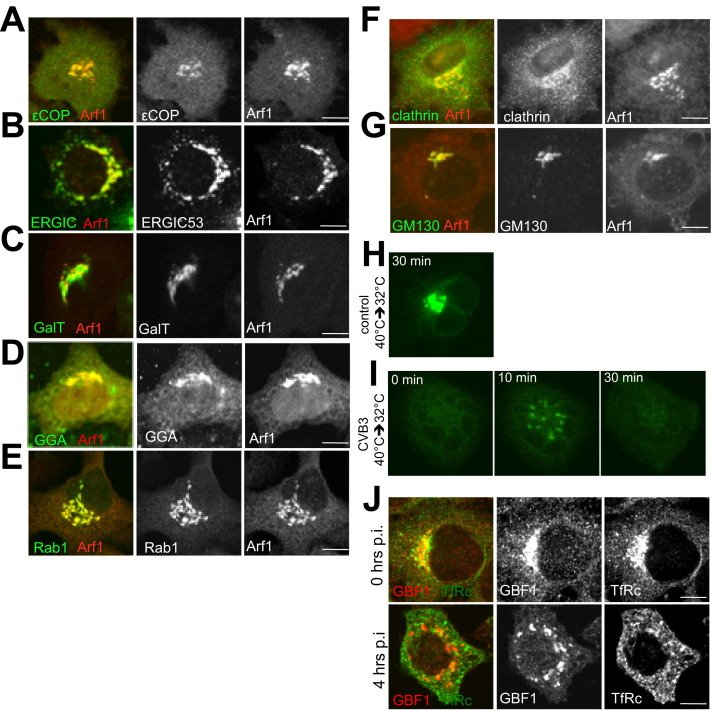
By 4 hr post-infection, at the peak of enteroviral RNA replication, both viral RNA and viral replication enzyme levels rapidly increase. At this stage, Arf1 GTPase, viral RNA, 3Dpol (Figure 1B, 4 hr p.i.), and other viral replication proteins (Figure S1B) were all redistributed to discrete cytoplasmic structures, which we term RNA replication organelles. These organelles persisted until the death of the cell, ∼10 hr after start of infection, during which time both viral RNA and viral protein levels continued to increase at these structures (Figure 1B, 8 hr p.i.). Cellular GBF1, the GEF for Arf1, which we have previously shown to be required for enteroviral RNA replication, was colocalized with Arf1 (Figure 1C, Movie S1) and viral RNA and replication enzymes throughout infection (Figure 1D). The replication organelles at 4 hr post-infection were formed adjacent to ER exit sites as determined by antibody labeling of sec31, a component of COPII coats, which are recruited by activated Sar1 GTPases to the ER (Figure 1E). ER exit site number and distribution were largely unaffected throughout infection (not shown). Furthermore, reduction in levels of GBF1 (>70% depletion) and Arf1 (>50% depletion) proteins with siRNA (Figure 1F; Figures S2A–S2C) or transient expression of a dominant-negative, GTPase-inactive Sar1 protein (Sar1 [T39N]) (Figure 1F), which is known to block the formation of ER exit sites (Kuge et al., 1994), inhibited by ∼70% (for Arf1), ∼80% (for GBF1), and ∼50% (for Sar1T39N) the replication of enteroviral RNA molecules (Figures 1F and 1G), whereas none of these treatments alone had any effect on cell viability (Table S1). Thus secretory pathway machinery GBF1, Arf1, and Sar1 proteins were all required for or facilitated viral RNA replication. Note that in all siRNA treatment or transient plasmid expression experiments, viral RNA replicons, where the capsid protein-encoding sequences have been replaced by Renilla luciferase, were transfected into cells and assayed for bioluminescence as an indicator of viral RNA replication, thereby avoiding complications in the interpretation of data due to potential impact of any treatment on viral entry or virus assembly steps.
Figure S2.
siRNA-Mediated Reduction of Host Factors and Impact on Enterovirus Replication, Related to Figure 1 and Figure 5
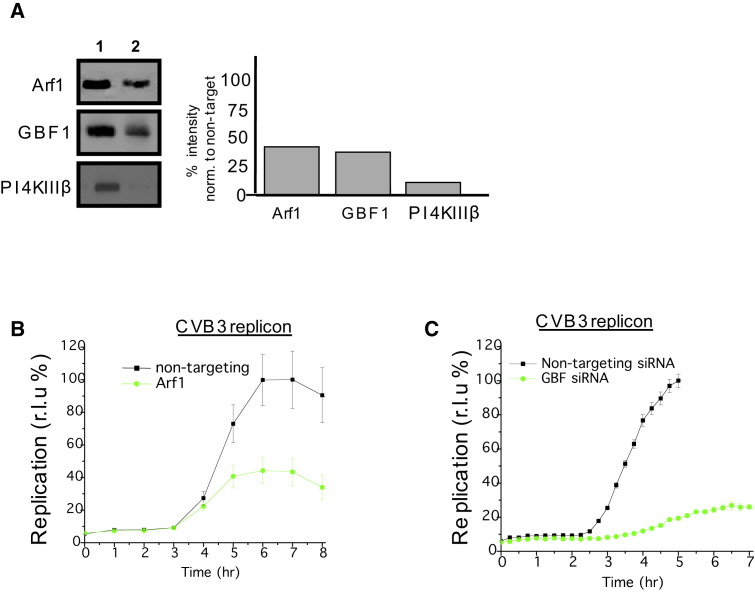
(A) Western blots of HeLa cells treated with nontargeting (lane 1) or Arf1, GBF1, PI4KIIIβ siRNAs (lane 2); equal amounts of protein (confirmed by actin blotting) for nontarget/Arf1, nontarget/GBF1, and nontarget/PI4KIIIβ conditions were loaded on SDS-PAGE gels. Bar graph shows quantification of representative blot, where the extent of siRNA depletion has been normalized to nontargeting siRNA condition.
(B and C) CVB3 replicon assays in HeLa cells treated with nontargeting siRNA and Arf1 siRNA (B) or GBF1 siRNA (C). Eight replicate samples were assayed for nontargeting, Arf1, and GBF1 siRNA treatment conditions.
Atypical Replication Organelles Formed at ER Exit Sites for Viral RNA Replication
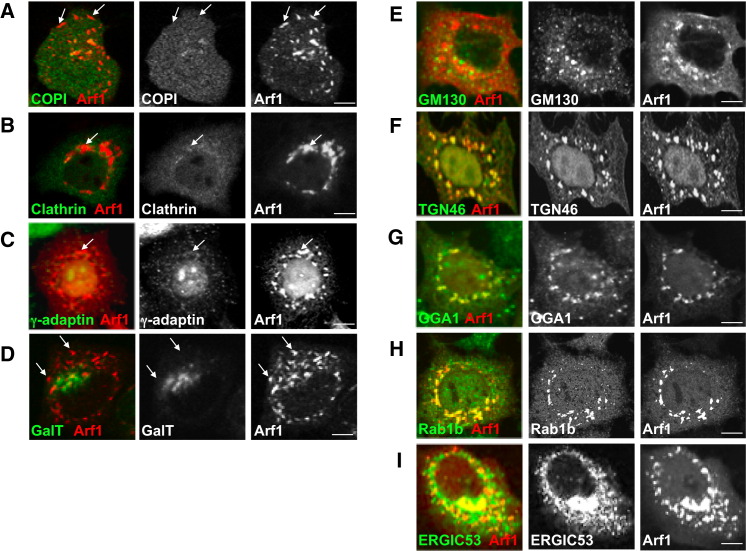
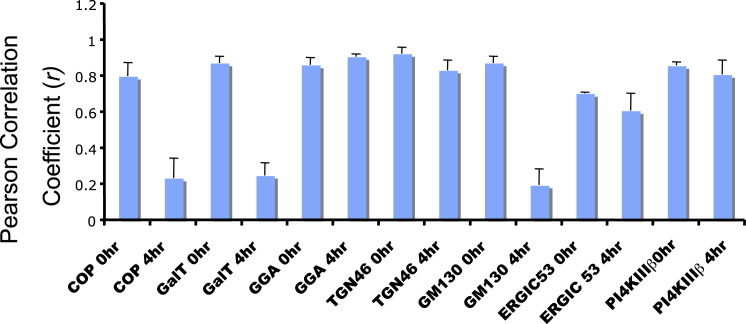
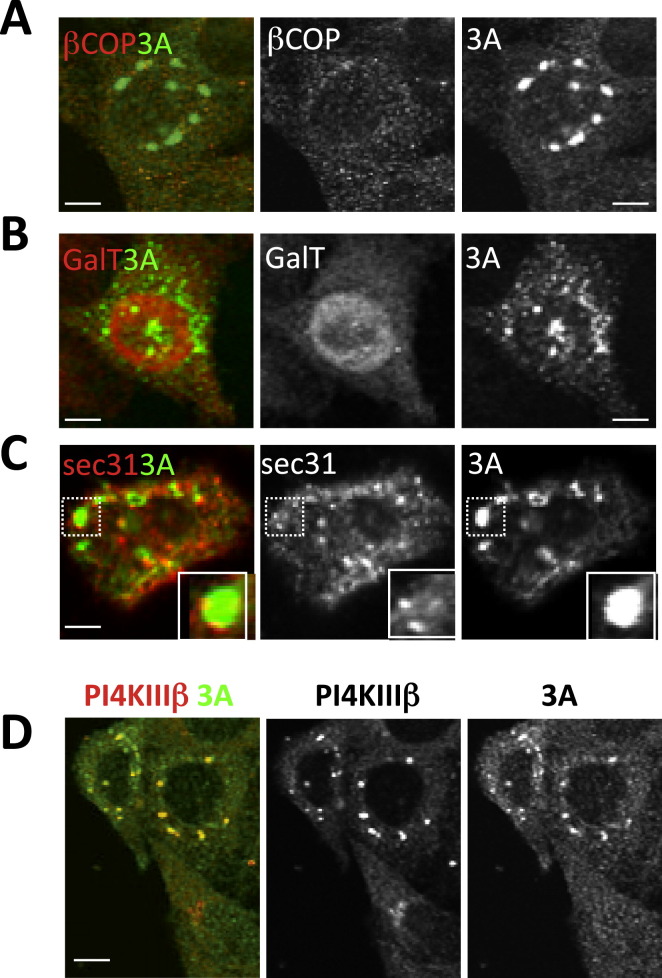
When we examined by high-resolution confocal imaging the replication membranes formed at 4 hr, we found that neither the coat proteins ɛCOPI (a component of the COPI coat complex) and clathrin nor the clathrin adaptor γ-adaptin, which are all known Arf1 effectors required for sorting/budding of cargo, including Golgi enzyme Galactosyltransferase (GalT), were colocalized with Arf1 at these organelles (Figures 2A–2C). Note that in uninfected cells Arf1 is colocalized at the Golgi/TGN/ERGIC with each of these components (Figures S3A–S3G). We assessed the colocalization between Arf1 and these components, at the start (0 hr) and at 4 hr post-infection, by calculating the Pearson correlation coefficients (Figure S4 ). In infected cells COPI, clathrin, and γ-adaptin were dispersed across the cytoplasm while their total cellular levels remained unchanged as determined by western blotting (not shown). The lack of localization of these proteins to replication membranes was surprising given that Arf1 is able to bind and hydrolyze GTP at these sites (Belov et al., 2007) and hence a priori capable of recruiting these effectors. Consistent with the absence of coats, these organelles were not labeled with Golgi enzymes (Figure 2D; Movie S2), which typically sort into GBF1/Arf1-GTP/COPI membranes at the ERGIC and Golgi apparatus of uninfected cells (Figure S3C) (Lanoix et al., 1999). COPI-dependent membrane budding mediates anterograde transport from the ERGIC and hence is required for the maintenance of the Golgi apparatus (Lee et al., 2004). The absence of Golgi enzyme GalT localization to these organelles suggested a disruption of anterograde transport, and indeed at 4 hr post-infection the Golgi apparatus was completely disassembled (Movie S2); consistent with this, trafficking of secretory cargo to the cell surface was blocked (Figures S3H and S3I). In addition, ERGIC/Golgi matrix protein GM130 (Figure 2E) and endosomal components such as transferrin receptor (Figure S3J) were also absent from these membranes. Nevertheless, these organelles did contain a combination of other TGN, Golgi, and ERGIC components including TGN46, GGA1, Rab1b proteins as well as some ERGIC 53 (Figures 2F–2I; Figure S4). Recent studies have found that depletion of COPI proteins from cells can result in a loss of secretory pathway compartmentalization and the formation instead of membrane-bound structures that contain components of TGN, Golgi, and ERGIC proteins (Styers et al., 2008). Similarly, our findings here suggest that decoupling GBF1/Arf1 activity from COPI recruitment to membranes also results in a complete reorganization of the secretory pathway away from distinct separate conventional organelles.
Figure 2.
Reorganization of Secretory Pathway Organelles after Enteroviral Infection
HeLa cells (A) coexpressing Arf1-RFP/ɛCOP-YFP; expressing Arf1-GFP (B, C, E–I); or coexpressing Arf1-RFP/GalT-YFP (see Movie S2) (D) were infected with CVB3 for 4 hr. Cells in (B), (C), (E)–(I) were fixed and coimmunostained with anti-GFP and (B) anti-clathrin heavy chain; (C) anti-γ-adaptin; (E) anti-GM130; (F) anti-TGN46; (G) anti-GGA1; (H) anti-Rab1b; (I) anti-ERGIC53 antibodies. Arrows in (A), (B), (C), and (D) indicate Arf1-labeled membranes that do not label with ɛCOP-YFP, clathrin, γ-adaptin, and GalT-YFP, respectively. See also Figures S3A–S3H and Figure S4.
All fluorescence images were confocal images of optical slice thickness ∼1 μm. Scale bar, 10 μm.
Figure S3.
Host Cell Secretory Machinery Distribution and Dynamics in Uninfected and Enterovius-Infected Cells, Related to Figure 2
(A–G) Colocalization in HeLa cells of Arf1-GFP and secretory pathway components in uninfected cells. Antibodies to GFP were utilized to localize Arf1.
(H and I) Disruption of secretory trafficking in CVB3 infected cells. HeLa cells expressing ts045VSVG-GFP were infected with CVB3 or mock for 2 hr prior to 32°C switch. Image showing Golgi accumulation of ts045VSVG-GFP 30 min after switch from non-permissive 40°C to permissive 32°C in mock-infected cells (H). Note the lack of ts045VSVG-GFP accumulation at the Golgi after 30 min in CVB3-infected cells (I).
(J) Transferrin Receptor is not localized to replication organelles. HeLa cells at 0 hr and 4 hr post CVB3 infection were fixed and coimmunostained with antibodies to native GBF1 to label the replication organelles and Transferrin Receptor. All images unless indicated are confocal sections 1 μm optical slice thickness. Scale bar, 10 μm.
Figure S4.
Pearson Correlation Coefficients of Arf1 and Host Secretory Components Pre- and Post-Enterovirus Infection, Related to Figure 2 and Figure 3
Pearson Correlation Coefficients were calculated as a measure of the degree of colocalization between Arf1-labeled membranes and ɛCOP, GalT, GGA, TGN46, GM130, ERGIC53, PI4KIIIβ proteins. The coefficients were calculated at 0 hr post-infection when Arf1 is localized to the Golgi apparatus and at 4 hr post-infection when Arf1 is localized at the replication organelles. In general small Pearson coefficients (<0.5) indicate low or no colocalization.
PI4KIIIβ Is a Component of Replication Organelles during Enteroviral Infection
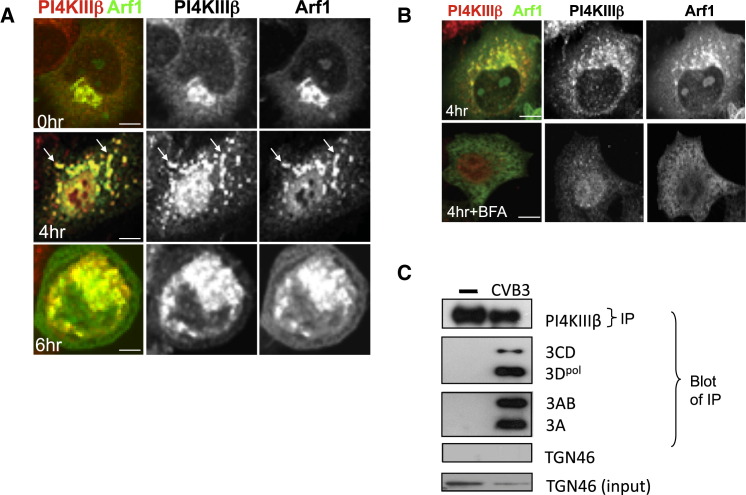
Phospholipid-modifying enzyme PI4KIIIβ, which catalyzes the production of PI4P lipids from PI (D'Angelo et al., 2008), is one of the critical downstream effectors of Arf1, recruited to and activated by Arf1 at the TGN and Golgi apparatus membranes and required for membrane trafficking (Balla and Balla, 2006, Godi et al., 1999). Given the absence of COPI or clathrin effectors at GBF1/Arf1-labeled replication organelles, we tracked the fate of PI4KIIIβ during enteroviral infection. Before infection, Arf1 and PI4KIIIβ were colocalized at the Golgi apparatus (Figure 3A, 0 hr), but at 4 hr post-infection, in striking contrast to COPI and clathrin, PI4KIIIβ remained colocalized with Arf1 (Figure 3A, 4 hr). This was confirmed by calculation of its Pearson coefficient, which showed no change between these two time points (Figure S4). This localization remained for the rest of the infection (Figure 3A, 6hr) and was correlated with GBF1/Arf1 localization and activity, as Brefeldin A (BFA) treatment, which inactivates GBF1/Arf1 (by stabilizing the GBF1/Arf1-GDP complex) (Niu et al., 2005), dispersed Arf1 along with PI4KIIIβ (Figure 3B). Furthermore, PI4KIIIβ was not just spatio-temporally correlated with replication organelles but was in a physical association with the viral replication enzyme complex. Upon immunoprecipitation of PI4KIIIβ from infected cells at 4 hr post-infection, the enteroviral 3A, 3AB, 3CD, and 3Dpol proteins, which are all components of the replication complex, coprecipitated with it (Figure 3C). Importantly, TGN46, which was spatio-temporally localized with Arf1 (and hence PI4KIIIβ) (Figure 2F), was not coprecipitated, indicating the specificity of the complex between PI4KIIIβ and the viral replication proteins. In addition, immunoprecipitation with IgG alone did not precipitate PI4KIIIβ or any of the viral proteins (not shown).
Figure 3.
PI4KIIIβ Is Localized to Enteroviral RNA Replication Organelles
(A) PI4KIIIβ and Arf1 are colocalized throughout CVB3 infection. HeLa cells expressing Arf1-GFP infected with CVB3 were immunostained with anti-PI4KIIIβ and anti-GFP antibodies.
(B) GBF1/Arf1 inactivation leads to PI4KIIIβ dispersal. HeLa cells were infected with CVB3 for 4 hr and treated with 10 μg/ml of BFA for 30 min. 75% ± 5% (n = 10 cells) of PI4KIIIβ-associated fluorescence was dispersed upon BFA treatment.
(C) PI4KIIIβ is in physical complex with viral replication enzymes. HeLa cell lysates from cells infected with CVB3 or mock (−) for 4 hr were immunoprecipitated with anti-PI4KIIIβ antibodies. Input samples verifying the presence of TGN46 in the lysates prior to immunoprecipitation are presented in the bottom panel.
All fluorescence images were confocal images of optical slice thickness ∼1 μm. Scale bar, 10 μm.
Enteroviral 3A Protein Can Selectively Recruit PI4KIIIβ to Membranes
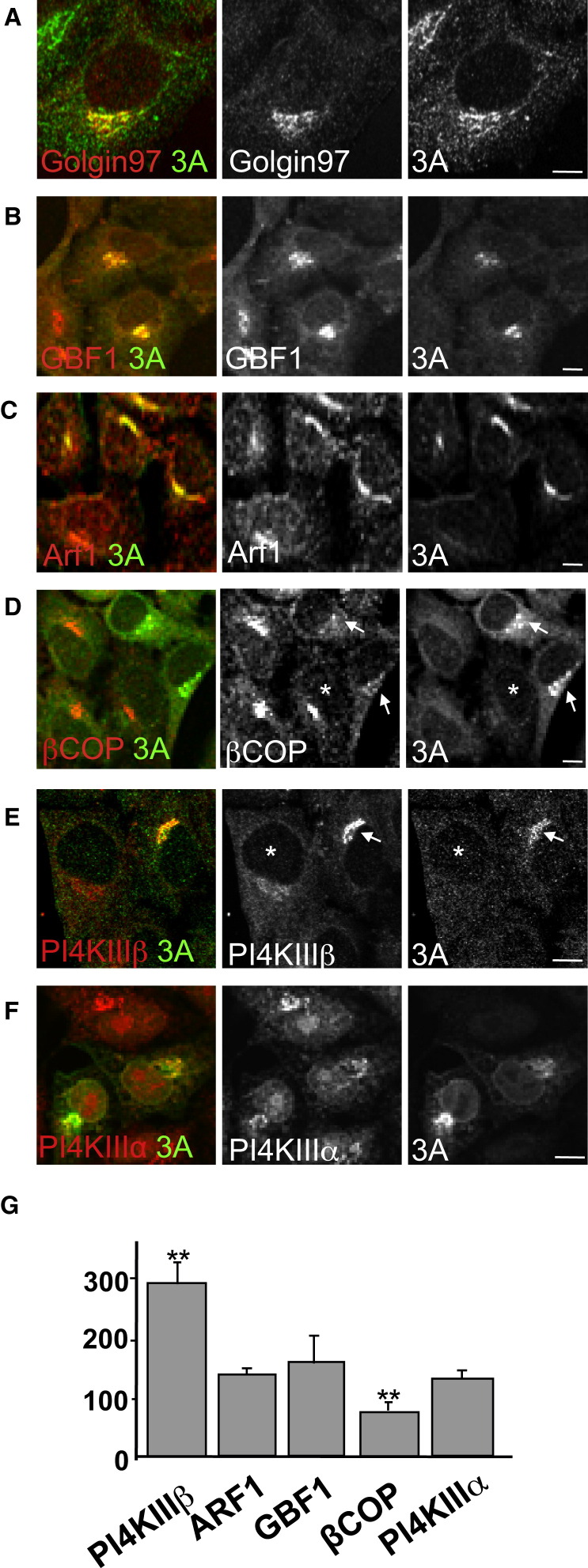
We next tested whether the CVB3 replication protein 3A could induce the selective recruitment of PI4KIIIβ to membranes observed during enteroviral infection. Membrane-anchored 3A proteins from CVB3 and PV have been documented to activate Arf1 by binding GBF1 and are required for viral RNA replication (Belov et al., 2007, Wessels et al., 2006, Lanke et al., 2009). At low levels of transient expression, 3A-myc was localized to the Golgi apparatus as determined by colocalization with Golgin97 (Figure 4A), along with native GBF1 and Arf1 proteins (Figures 4B and 4C). Although GBF1/Arf1 was present at these structures, the levels of βCOP, a component of the COPI coat complex, were decreased at these sites 2-fold compared to cells not expressing 3A-myc (Figure 4D, arrow; Figure 4G) whereas the total cellular βCOP levels were unchanged (data not shown). Significantly, despite a decrease in βCOP levels at 3A-labeled membranes, we found that PI4KIIIβ was redistributed with an ∼3-fold increase to these membranes relative to cells where 3A was absent (Figure 4E, arrow; Figure 4G). In comparison, recruitment to the Golgi of structurally homologous enzyme PI4KIIIα was unaffected by 3A-myc expression (Figures 4F and 4G). In cells expressing high levels of 3A-myc, the Golgi apparatus was disassembled and 3A was localized to discrete membranes juxtaposed to ER exit sites that were labeled with PI4KIIIβ but lacked βCOP and Golgi enzymes (Figures S5A–S5D)—a phenotype previously observed in CVB3-infected cells (Figure 2A, Figure 3A). Hence 3A expression alone enhanced the membrane recruitment of one Arf1 effector, PI4KIIIβ, over another, COPI, and caused disassembly of the Golgi apparatus, thus mimicking specific aspects of the virally infected phenotype.
Figure 4.
Enteroviral 3A Proteins Can Promote Selective PI4KIIIβ Recruitment over Coat Proteins
(A–F) Impact of 3A-myc ectopic expression on host secretory machinery. HeLa cells were immunostained with antibodies to myc-tag and native: (A) Golgin97, a Golgi resident protein; (B) GBF1; (C) Arf1; (D) βCOP; (E) PI4KIIIβ; (F) PI4KIIIα. Arrows and asterisk in (D) and (E) indicate cells where 3A is either expressed (arrow) or not (∗). See also Figures S5A–S5D.
(G) Quantification of GBF1 (n = 15 cells); Arf1 (n = 9 cells); βCOP (n = 10 cells); and PI4KIIIβ (n = 13 cells) antibody fluorescence associated with 3A-labeled membranes as (%) of their respective values at the Golgi apparatus of cells not expressing 3A. Error bars are SEM (∗∗p < 0.001).
All fluorescence images were confocal images of optical slice thickness ∼1 μm. Scale bar, 10 μm.
Figure S5.
High-Level Transient Ectopic Enteroviral 3A Expression, Related to Figure 4
Secretory machinery distribution in cells expressing high levels of 3A-myc. All cells were transfected with 3A-myc and then coimmunostained with antibodies against –myc and (A) βCOP, (B) GalT, (C) sec31, (D) PI4KIIIβ. Scale bar, 5 μm.
PI4KIIIβ Activity Regulates Viral RNA Synthesis
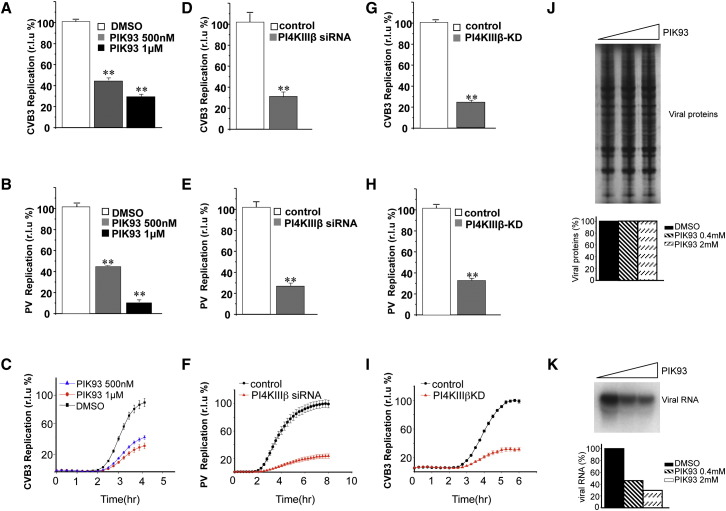
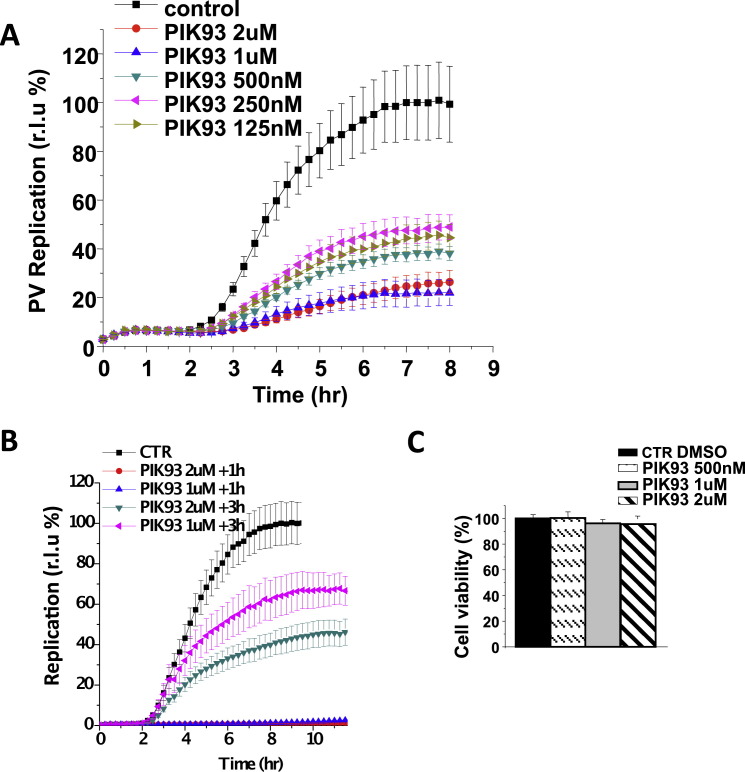
To determine whether PI4KIIIβ kinase activity was required for enteroviral RNA replication, we measured the effect of the small molecule PIK93, which selectively inhibits PI4KIIIβ (Knight et al., 2006), and found that enteroviral RNA replication was significantly reduced (Figures 5A–5C). RNA replication could be inhibited at concentrations as low as 125 nM and when PIK93 was added at time points after start of replication, indicating a requirement for PI4KIIIβ activity throughout the replication process; and PIK93, up to 2 μM, had little effect on cell viability (Figures S6A–S6C). When we reduced PI4KIIIβ levels with siRNA (Figures 5D–5F; Figure S2A) or transiently expressed a kinase-dead PI4KIIIβ (PI4KIIIβ-KD) (Tóth et al., 2006) (Figures 5G–5I), both PV and CVB3 viral RNA replication were considerably delayed (Figures 5F and 5I) and accumulated reduced viral RNA levels compared to nontargeting siRNA or GFP-expressing control cells. PI4 kinase activity and PI4P lipids have been implicated in ER exit site maintenance (Blumental-Perry et al., 2006), but we found no change in distribution or number of ER exit sites in cells treated with PI4KIIIβ siRNA whereas cells treated with PI4KIIIα siRNA had ∼50% reduction (Figure S7 ). Hence downregulating PI4KIIIβ activity—a kinase critical for PI4P lipid synthesis—inhibited enteroviral RNA replication.
Figure 5.
PI4KIIIβ Activity Is Required for Enteroviral RNA Replication
(A and B) PIK93 block of PI4KIIIβ activity inhibits CVB3 and PV RNA replication. CVB3 and PV replicon assays in cells treated with 500 nM and 1 μM PIK93 are shown. Bar graphs present maximum replication values for CVB3 (A) and PV (B) with PIK93 treatment, normalized to control (DMSO) treatment. See also Figures S6A–S6C.
(C) Kinetics of inhibition by PIK93 presented for the CVB3 replicon.
(D and E) Reduction of PI4KIIIβ levels with siRNA inhibits CVB3 and PV RNA replication. CVB3 and PV replicon assays in HeLa cells; bar graphs present maximum replication values for each condition, normalized to control (nontargeting) siRNA. See also Figure S2A.
(F) Kinetics of inhibition by PI4KIIIβ siRNA presented for PV replicon.
(G and H) Expression of ectopic kinase-dead PI4KIIIβ (PI4KIIIβ-KD) inhibits CVB3 and PV replication. CVB3 and PV replicon assays in HeLa cells; bar graphs present maximum replication values for each condition, normalized to control (GFP) plasmid ectopic expression.
(I) Kinetics of inhibition by PI4KIIIβ-KD presented for the CVB3 replicon. Error bars in each CVB3 assay are SEM of six CVB3 samples and each PV assay is SEM for eight PV samples. (∗∗p < 0.01).
(J and K) PI4KIIIβ activity regulates viral RNA synthesis. Cell-free PV RNA translation (J) and synthesis (K) assays performed in the presence of PIK93.
Figure S6.
PIK93 Impact on Enterovirus RNA Replication and Cell Viability, Related to Figure 5
(A) Impact on PV replication of different concentrations of PIK93 (added at time 0 hr).
(B) Impact on replication of PIK93 added at 1 hr and 3 hr post replicon transfection.
(C) PIK93 effects on cell viability. Cells were treated with the indicated PIK93 concentrations for 24 hr.
Figure S7.
Impact of PI4KIIIα and PI4KIIIβ Reduction on ER Exit Sites in HeLa Cells, Related to Figure 5
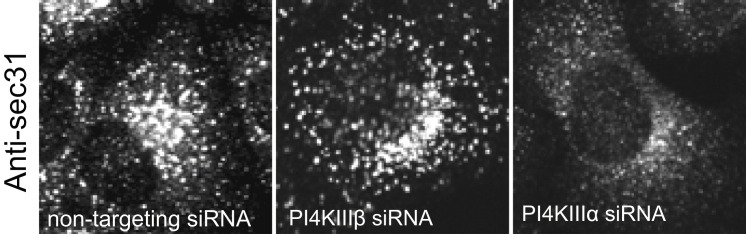
HeLa cells were treated with nontargeting, PI4KIIIβ, or PI4KIIIα siRNA's for 72 hr; fixed and immunostained with antibodies to sec31 to label ER exit sites. Note the decrease in the number of ER exit sites in PI4KIIIα siRNA-treated cells whereas it is unaffected in PI4KIIIβ siRNA-treated cells.
The inhibition of replication observed after reduction of PI4KIIIβ activity could be due to decreased viral RNA translation or proteolytic processing of the viral polyprotein into viral replication enzymes; alternatively inhibition could result from a direct effect on viral RNA synthesis. A block in any one or more of these stages would result in the inhibition of viral RNA replication observed (Figures 5A–5I). To distinguish among these we utilized a cell-free assay where viral RNA translation and processing can be uncoupled from viral RNA synthesis (Barton and Flanegan, 1993, Fogg et al., 2003). We found that inhibition of PI4KIIIβ kinase activity had no impact on viral RNA translation or its subsequent proteolytic processing of the polytopic protein into the individual viral replication proteins (Figure 5J). There was, however, a >70% inhibition of viral RNA synthesis (Figure 5K). Thus viral RNA synthesis is regulated by PI4KIIIβ activity.
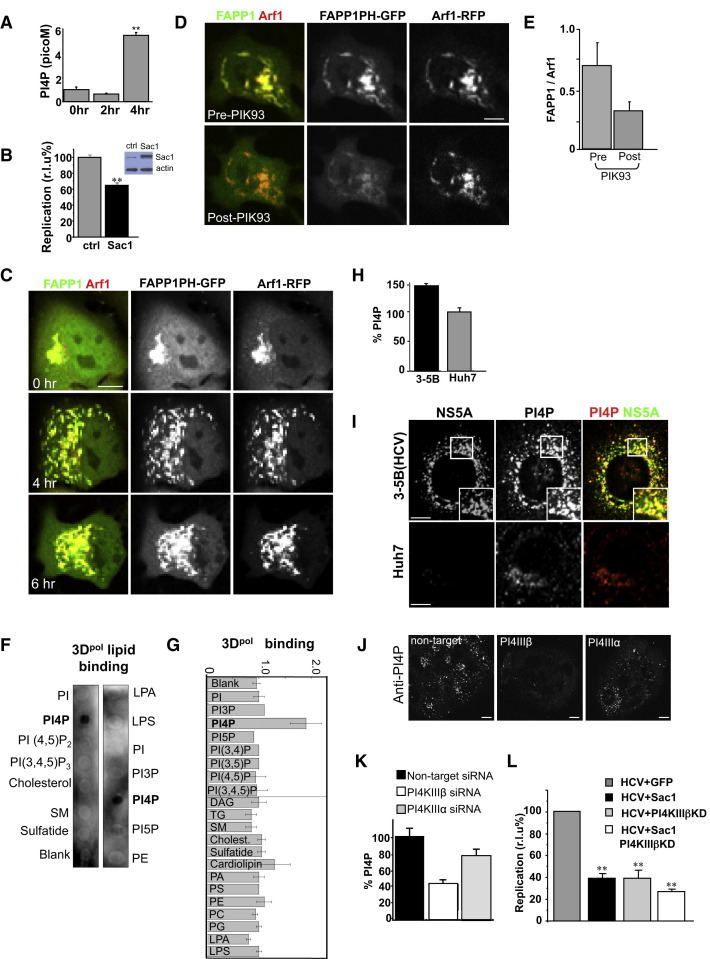
PI4P Lipid Microenvironment at Replication Organelles Regulates Enteroviral RNA Replication
Given the localization of PI4KIIIβ to replication organelles, and the inhibition of replication after knockdown of PI4KIIIβ activity, we monitored the levels of cellular PI4P, a known lipid product of PI4KIIIβ activity, during infection with CVB3. PI4P levels increased ∼5-fold within the first 4 hr of CVB3 infection (Figure 6A). Notably levels of PIP2, which can be derived from PI4P lipids, were not changed and even decreased during this time (data not shown). To test if PI4P lipids themselves, independent of PI4KIIIβ, regulated viral RNA replication, we ectopically expressed in cells Sac1 phosphatase, which specifically converts PI4P lipids back to PI (Blagoveshchenskaya et al., 2008). The inhibition was only ∼40% (Figure 6B) likely due to continued production of PI4P by endogenous PI4 kinase activity, or because not all cells that were transfected with enterovirus RNA were transiently expressing the Sac1 phosphatase.
Figure 6.
PI4P Lipid Microenvironment within Replication Organelles Regulates both Enteroviral and Flaviviral RNA Replication
(A) Cellular PI4P lipid levels rise in CVB3 infection. Total cellular PI4P lipids were quantified over time. Error bars are SEM from duplicate samples (∗∗p < 0.01).
(B) Reduction in PI4P lipid levels inhibits enteroviral RNA replication. PV replicon assays in HeLa cells ectopically expressing Sac1 are shown. Inset: western blot showing increase in Sac1 levels after 16 hr of ectopic expression. Bar graph presents maximum replication values, normalized to control (GFP) plasmid ectopic expression. Error bars are SEM from eight replicates cells (∗∗p < 0.01).
(C) PI4P lipids localize to enteroviral replication organelles. Time-lapse confocal images of single HeLa cell infected with CVB3, coexpressing FAPP1PH-GFP/Arf1-RFP (see also Movie S3).
(D) PI4KIIIβ activity is responsible for PI4P lipids at enteroviral replication organelles. Time-lapse confocal image of single HeLa cell coexpressing FAPP1-PH-GFP/ARF1-RFP pre- and post-PIK93 treatment at 4 hr post-infection with CVB3.
(E) Quantification of FAPP1PH-GFP to Arf1-RFP fluorescence from (D) expressed as a ratio. Error bars are SEM from ten cells for each condition.
(F and G) PV RNA polymerase specifically and preferentially binds PI4P lipids. Purified recombinant 3Dpol enzyme was incubated with membrane strips that were previously spotted with different lipids. Antibodies detected 3Dpol binding. Two representative blots are shown. Binding was quantified and plotted in bar graph as a ratio over background non-lipid spotted membrane from three different experiments for each lipid type.
(H) Cellular PI4P lipid levels rise in HCV replicating cells. Quantification of PI4P with anti-PI4P primary antibodies of 3-5B(HCV) cells, normalized as % of PI4P lipid quantification within Huh7 cells. Error bars are SEM from ten cells for each cell type.
(I) PI4P lipids localize to HCV replication membranes. PI4P lipid and NS5A protein distribution in Huh7 and 3-5B(HCV) cells were determined by immunostaining with anti-PI4P and anti-NS5A antibodies.
(J and K) PI4KIIIβ is responsible for a significant fraction of PI4P lipids at HCV replication membranes. 3-5B(HCV) cells were treated with nontargeting, PI4KIIIβ, or PI4KIIIα siRNA. Cells were immunostained and quantified for PI4P lipids. Representative images of groups of siRNA-treated cells are shown (J). Quantification was done on 20 cells for each siRNA treatment condition.
(L) Reduction of PI4P lipids inhibits HCV replication. HCV replicon assays conducted with J6/JFH (p7-Rluc2A) replicons in Huh7 cells ectopically expressing Sac1, PI4KIIIβ-KD, or both plasmids are shown. Bar graph presents maximum replication value normalized to control (GFP) plasmid ectopic expression. Error bars are SEM from eight replicates of cells for each treatment condition (∗∗p < 0.01).
All fluorescence images were confocal images of optical slice thickness ∼1 μm. Scale bar, 10 μm.
We next determined if the PI4P lipids were localized to replication membranes. We utilized GFP-tagged FAPP1-PH proteins, which are known to bind PI4P lipid-containing membranes in the presence of Arf1 (Godi et al., 2004, Tóth et al., 2006). We coexpressed Arf1-RFP and FAPP1-PH-GFP and acquired time-lapse confocal images of individual cells during infection with CVB3 to monitor the distribution of both Arf1 and PI4P lipids (Figure 6C; Movie S3). At the start of infection the major PI4P pool in the cell was found at the Golgi/TGN compartment. By 4 hr post-infection, all the newly formed Arf1-labeled organelles contained PI4P lipids (Figure 6C, 4 hr). The PI4P labeling was independently confirmed by staining with antibodies to PI4P lipids (data not shown).
PI4KIIIβ Activity Responsible for PI4P Lipid Microenvironment at Replication Organelles
By acutely treating cells coexpressing FAPP1-PH-GFP and Arf1-RFP with PIK93 inhibitor at 4 hr post-infection, we demonstrated that the PI4P lipids at replication organelles were the products of PI4KIIIβ activity. Using time-lapse imaging, we followed the fate of FAPP1-PH-GFP and Arf1-RFP within individual cells pre- and post-PIK93 treatment. We found that within ∼30 min of PIK93 addition, ∼60% of the FAPP1-PH-GFP associated with replication organelles was lost whereas the Arf1-RFP pattern was largely unaffected (Figures 6D and 6E). This demonstrated that a significant fraction of the PI4P lipids at replication sites were a product of PI4KIIIβ activity and that Arf1 binding to membranes was not dependent on PI4P lipids. Furthermore it indicated that the PI4P lipid microenvironment at these membranes was not static but turning over, as in the absence of PI4 kinase activity, the lipids were quickly lost from these sites. This could be due to conversion back to PI, potentially as a result of endogenous Sac1 phosphatase activity or transport out of replication membranes. The lack of complete FAPP1-PH-GFP dispersal upon PIK93 treatment also suggested that other PI4 kinase family members might be harnessed for PI4P lipid synthesis as well during infection at these sites.
RNA Polymerase 3Dpol Binds PI4P Lipids
Enteroviral RNA is synthesized by viral replication complex enzymes, which include the RdRp 3Dpol. The mechanism by which the soluble 3Dpol protein associates with membranes and catalyzes the synthesis of viral RNA on these membranes is unknown. Given the in vivo localization of the viral replication complex proteins including 3Dpol to membranes enriched in PI4P lipids (Figures 1A and 1B, Figure 6C), we tested whether the 3Dpol protein had itself any affinity for PI4P lipids. We assayed for binding of 3Dpol to PI4P lipids by incubating purified recombinant 3Dpol with membrane strips spotted with different cellular lipids including PI4P. We found that 3Dpol specifically and preferentially bound to PI4P lipids over all other phosphatidylinositides and phospholipids (Figures 6F and 6G). Thus 3Dpol alone, independent of any of the other components of the viral replication complex, has a high and specific affinity for PI4P lipids that potentially can regulate both its binding to cellular membranes and its subsequent RNA synthesis activities.
PI4P Lipid Microenvironment Regulates HCV RNA Replication
Given that the PI4P lipid microenvironment was required for enteroviral RNA replication, we next tested if any other plus-strand RNA virus was similarly dependent. HCV, an enveloped virus that is a member of the flaviviruses, assembles its replication enzymes and replicates its RNA on remodeled ER membranes (Wölk et al., 2008). We coimmunostained the Huh7-derived liver cell line 3-5B(HCV), which contains autonomously replicating, subgenomic, dicistronic, selectable HCV RNAs from the infectious HCV-N1b strain as well as expressing HCV nonstructural proteins (Ikeda et al., 2002), with antibodies against PI4P lipids and the HCV protein NS5A. NS5A, a membrane-associated protein that is part of the HCV replication complex of enzymes and is colocalized with HCV RNA molecules (Wölk et al., 2008), allowed us to identify the cellular viral RNA replication sites. First by quantification of PI4P-associated antibody fluorescence we found that 3-5B(HCV) cells, compared to parental Huh7, had an ∼45% increase in the total cellular levels of PI4P (Figure 6H). Second, whereas in the parental Huh7 cells the PI4P lipid pool was localized to the perinuclear Golgi apparatus/TGN region, in 3-5B(HCV) cells the abundant PI4P lipids were found colocalized with NS5A at discrete punctate sites all across the cytoplasm (Figure 6I, inset). Notably PI4KIIIβ reduction by siRNA knockdown (>90% by immunofluorescence staining, not shown) had greater effect on lowering PI4P levels within 3-5B(HCV) cells compared to PI4KIIIα reduction, though it is likely that both enzymes contribute to the increased cellular PI4P levels (Figures 6J and 6K). Finally, to test whether PI4P lipids were also required for HCV RNA replication, we measured the replication of HCV J6/JFH (p7-Rluc2A) replicons (Jones et al., 2007) in Huh7 cells where we had depleted the PI4P lipid pool by transiently expressing the Sac1 phosphatase either alone or in combination with catalytically inactive PI4KIIIβ-KD. HCV RNA replication was inhibited by >50% when Sac1 was expressed alone, and the effect was greater (∼70%) in the presence of PI4KIIIβ-KD, as would be predicted (Figure 6L, Sac+PI4KIIIβKD). Thus the PI4P lipid microenvironment in the membrane is an important regulator of both enteroviral and flaviviral RNA replication.
Discussion
Here we demonstrate that both enteroviruses and flaviviruses exploit host PI4KIIIβ enzymes and replicate their respective viral RNA on PI4P lipid-enriched membranes. Furthermore we show that the enteroviral membrane protein 3A can reorganize the host secretory trafficking pathway to enhance the recruitment of PI4KIIIβ to host membranes in order to generate a PI4P lipid-enriched membrane microenvironment to which the soluble viral RdRp 3Dpol can bind.
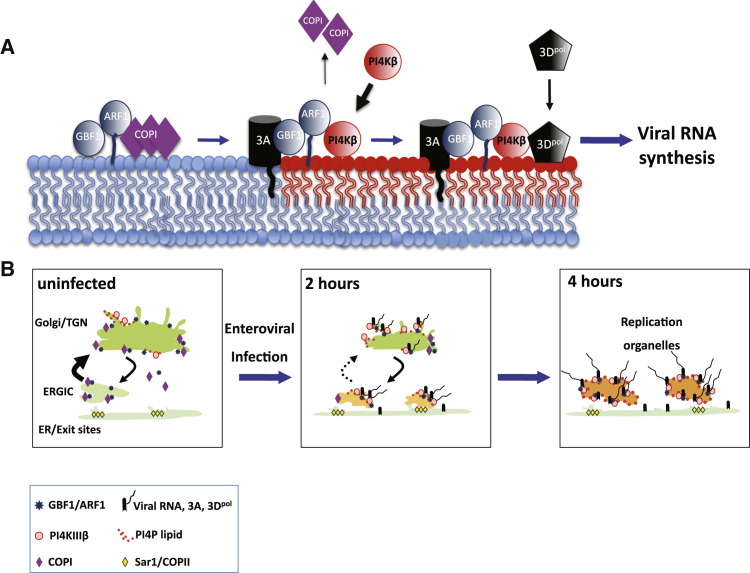
Based on our findings we propose a model for the reorganization of the secretory pathway in enteroviral infections, which generates PI4P lipid-enriched replication organelles (Figure 7 ). Here membrane-bound enteroviral 3A proteins bind and modulate host GBF1/Arf1 to enhance recruitment of PI4KIIIβ to membranes, over COPI and other coat proteins, where it will catalyze the production of PI4P lipids, leading to the biogenesis of a PI4P lipid-enriched membrane microenvironment that is distinct from that in uninfected cells. This PI4P lipid-rich microenvironment will in turn promote the recruitment and stabilization on the membrane of the RdRp 3Dpol from the cytosolic pool. 3Dpol as part of a replication complex of 3A and several other viral proteins will then initiate RNA synthesis at these membranes (Figure 7A).
Figure 7.
Model for Secretory Pathway Reorganization in Enteroviral Infections
(A) Tail-anchored membrane protein 3A by binding and modulating GBF1/Arf1 promotes PI4KIIIβ recruitment to the membrane bilayer at the expense of coat protein COPI. Recruited PI4KIIIβ will catalyze the production of a PI4P lipid microenvironment (red lipids) that will in turn facilitate the recruitment of 3Dpol from the cytosolic pool to the membrane and promote the synthesis of viral RNA.
(B) Uninfected (0 hr): Steady-state exchange of membranes, Golgi enzymes, and cargo through bidirectional trafficking between the ERGIC and Golgi/TGN compartments. Golgi enzymes and cargo are sorted out of Sar1/COPII-labeled ER exit sites into ERGIC compartments whereupon GBF1/Arf1 and COPI coats mediate the trafficking to and from the Golgi/TGN compartments. PI4KIIIβ enzymes are recruited to Golgi/TGN membranes by Arf1 and catalyze the production of PI4P lipids at these membranes.
2 hr: Upon infection, newly synthesized viral replication enzymes such as the membrane-bound 3A target to and assemble on secretory organelle membranes but concentrate and initiate viral RNA synthesis on the Golgi/TGN membranes, where the pre-existing (i.e., prior to infection) steady-state pool of PI4P lipids facilitates viral replication protein assembly and RNA replication. Rising levels of 3A combined with its modulation of effector recruitment by GBF1/Arf1 will enhance the recruitment of PI4KIIIβ over COPI, leading to a decreased rate of anterograde transport out of the ERGIC and subsequent disassembly of the Golgi/TGN organelles.
4 hr: Enhanced recruitment of PI4KIIIβ over COPI results in the formation of uncoated PI4P lipid-enriched organelles adjacent to ER exit sites. The PI4P lipid-enriched microenvironment of these organelles facilitates the ongoing assembly of newly synthesized viral replication proteins such as RdRp 3Dpol and viral RNA replication.
Enteroviral RNA replication begins on existing PI4P lipid-containing organelles including the Golgi and TGN, which have the highest steady-state levels of this lipid in uninfected cells (Godi et al., 1999) (Figure 7B, uninfected). Viral RNA replication is a positive feedback loop, where newly synthesized viral RNA molecules are translated into increasing amounts of viral replication proteins including 3A, which then further replicate viral RNA. Since 3A modulates GBF1/Arf1 effector recruitment, its impact on GBF1/Arf1/coat/PI4KIIIβ interactions will become inescapable as its levels rise. The selective recruitment of PI4KIIIβ to membranes over COPI will eventually disrupt secretory membrane trafficking and lead to Golgi disassembly by decreasing anterograde transport from the ERGIC and intra-Golgi trafficking, both COPI-dependent processes (Lee et al., 2004) (Figure 7B, 2 hr). Furthermore 3A-bound membranes emerging de novo from ER exit sites will develop into PI4P lipid-enriched uncoated membranes (Figure 7B, 4 hr). In addition to forming replication organelles, disruption of secretory pathway integrity has been implicated in the suppression of cytokine secretion and MHC-dependent antigen presentation (Deitz et al., 2000), thus serving multiple functions for efficient enterovirus replication and propagation.
Newly formed replication organelles have an atypical combination of host protein and lipids, including being enriched in PI4P lipids and containing PI4KIIIβ, TGN46, GGA1, Rab1b, and ERGIC53, which in uninfected cells would be segregated among different secretory compartments. Modulation of host secretory machinery by viral proteins and its consequences are highly complex. The localization of some components can be explained through affinity for either PI4P lipids (e.g., GGA1) or GBF1 (e.g., GGA1, Rab1b) (Monetta et al., 2007, Wang et al., 2007). However GGA1 can bind clathrin and γ-adaptin (Bonifacino, 2004, Lefrançois and McCormick, 2007) and GBF1 can bind COPI (Deng et al., 2009), but neither COPI, clathrin, nor γ-adaptin are present at replication sites. Potentially the high PI4P lipid microenvironment in conjunction with viral proteins may modulate interactions among GBF1, Arf1, and specific effectors, promoting the sorting/partitioning of some components while deterring others.
Arf1 recruits a variety of effectors to membranes, yet little is known on the mechanism of the selection process. ArfGEFs not only facilitate Arf recruitment and activation but also regulate effector selection: e.g., GBF1 can bind COPI and GGA proteins. Which ArfGEF recruits PI4KIIIβ is not known in uninfected cells, but GBF1 in the context of 3A proteins may facilitate the recruitment of PI4KIIIβ in infected cells, given that PI4KIIIβ is colocalized with GBF1 and coimmunoprecipitated with 3A (Figures 3A and 3C), where the latter is known to bind GBF1 (Wessels et al., 2006). It also remains to be explored whether GBF1, Arf1, or any other viral or host proteins may be stimulating PI4KIIIβ activity to reach the high PI4P lipid levels observed during infection.
HCV, an enveloped flavivirus whose replication enzymes are sequence- and structure-wise distinct from enteroviral enzymes (Dubuisson et al., 2002), nevertheless depends on PI4P lipid-enriched membranes and PI4KIIIβ for replication (Figures 6H–6L). Whereas most enteroviral infections disrupt secretory trafficking, flaviviruses utilize the secretory pathway to mature into virions and exit the cell (Mackenzie and Westaway, 2007). HCV RNA is replicated on remodeled ER membranes whereas structural proteins are located to lipid droplets (Miyanari et al., 2007). Through a complex assembly process not yet understood virions bud out of the ER and are released from the cell through exocytosis. The presence of high levels of PI4P lipids at ER membranes, in addition to regulating HCV RNA replication, could impact the organization and kinetics of secretory trafficking and budding/export of HCV. Indeed secretory trafficking is attenuated in HCV-infected cells (Konan et al., 2003).
In mammalian cells, PI4P lipids, the most abundant monophosphorylated inositol phospholipids, were previously viewed only as PIP2 precursors (D'Angelo et al., 2008). However independent functions have recently emerged: several host proteins including CERT, OSBP, and FAPP1/2 specifically bind PI4P lipids (Lemmon, 2008); and PI4P lipids regulate selective autophagy and ER exit site biogenesis (Blumental-Perry et al., 2006, Yamashita et al., 2006). PI4P lipids can locally change membrane curvature (Ishiyama et al., 2002, McMahon and Gallop, 2005). PI4P lipid-enriched membranes during viral infection may generate high-curvature membrane pockets to shield viral components from host defense. Little is known about how soluble viral RNA polymerases are recruited to membranes. PI4P lipids may provide docking sites to concentrate viral proteins for efficient RNA synthesis. Enteroviral RdRp 3Dpol preferentially binds PI4P lipids over all other cellular lipid components, and PI4P depletion specifically perturbs viral RNA synthesis (Figure 5 and Figure 6). This raises the possibility that enteroviruses rewire host secretory machinery to generate PI4P lipid-enriched membranes to recruit to and concentrate on membrane RNA polymerases. Furthermore, PI4P lipid binding may also induce conformational changes and modulate RdRp enzymatic activity. The phosphoinositide-binding domain on 3Dpol is unknown and studies to identify the lipid-binding site and investigate its occurrence across different RdRps are underway.
In summary, we show that the PI4P lipid microenvironment is an important facilitator of plus-strand viral RNA replication. Enteroviruses reorganize the cellular secretory trafficking machinery away from building conventional secretory organelles to generate organelles whose membranes are enriched in PI4P lipids. Cellular PI4 kinases are key players in this process. The findings with both picornavirus and flavivirus family members highlight the importance of PI4P lipids and PI4 kinases for viral RNA replication and will instigate studies to determine how widespread this dependence on PI4P lipids is among RNA viruses and the mechanism by which these lipids modulate viral RNA synthesis machinery. Furthermore small molecules targeting PI4 kinases, such as PIK93, may provide a basis for the design of new classes of therapeutics against viral RNA replication.
Experimental Procedures
Live-Cell Imaging
All imaging was performed on a Zeiss LSM510META confocal laser scanning confocal microscope (Carl Zeiss, USA) using high-magnification, high numerical aperture objectives. Live cells were maintained on the microscope stage in a temperature, CO2, and humidity-controlled environmental chamber. Time-lapse images were acquired every 5 min for the duration of infection.
Immunofluorescence and Analysis
Cells were plated on coverslips, fixed with 4% formaldehyde PBS solution, permeabilized with 0.2% saponin, incubated with primary, and fluorophore-tagged secondary antibodies, and mounted. Confocal images were obtained and analyzed with Zeiss LSM or Image J software.
Fluorescence in Situ Hybridization
Alexa555-labeled CVB3 plus-strand RNA-specific probes were synthesized using FISH Tag RNA kit (Invitrogen Corp., CA). For colocalization of CVB3 plus-strand RNA with Arf1 or GBF1, infected cells were fixed with 4% formaldehyde followed by overnight permeabilization with 70% ethanol. Cells were rehydrated in SSC buffer and hybridized with RNA probes overnight in hybridization buffer. GBF1 or Arf1 was immunostained with primary and Alexa488-labeled secondary antibodies in the absence of detergents.
PI4P Lipid Extraction and Quantification
Cells were harvested, PI4P extracted, spotted on nitrocellulose membrane strips, and detected by FAPP1 protein-derived PI4P detectors followed by secondary and tertiary antibodies as described in Dowler et al. (2002).
siRNA Transfection
Cells were seeded in 96-well plates (for the replicon assay) or 12-well plates for western blot analysis to verify knockdown efficiency of proteins, 1 day before siRNA transfection. Typically 50 nM of each siRNA was transfected via Dharmafect1 (Dharmacon, CO) and incubated for 48 hr.
Replicon Assays
pRib-Rluc (CVB3), pXpA-RenR(PV), and J6/JFH (p7-Rluc2A) (HCV) plasmids with Renilla luciferase gene as reporters in place of structural genes were utilized to measure viral RNA replication. Plasmids were in vitro-transcribed and RNAs transfected into HeLa or Huh7 cells grown in 96-well plates. Cells were incubated with live-cell Renilla substrate and light signal was recorded with multiwell plate reader at 15 min intervals up to 16 hr at 37°C.
RNA Polymerase Lipid-Binding Assay
Recombinant PV polymerase (3Dpol) in pET26Ub-3D was purified as described (Gohara et al., 1999). Lipid dot-blot strips were purchased (Echelon Biosciences, UT). The strips were incubated in blocking buffer for 1 hr at room temperature (RT) and then incubated in the same buffer with purified 3Dpol overnight at 4°C. The blots were then washed in TBST-50 buffer. To detect lipid protein interactions, strips were incubated with anti-3Dpol antibody for 1 hr at RT. Blots were washed as before and incubated with anti-rabbit horseradish peroxidase for 1 hr at RT. 3Dpol bound to the lipids immobilized on the membrane were visualized by incubating with chemiluminescent substrate.
Cell-free Translation and Replication Assays
HeLa S10 extracts for translation-replication reactions were prepared and utilized for in vitro RNA translation and replication reactions as described (Barton and Flanegan, 1993, Fogg et al., 2003). Translation reaction mixtures included in vitro-synthesized PV RNA transcripts in the presence of 2 mM GuHCl to block replication. An aliquot from each translation reaction mixture was mixed with Redivue [35S] methionine (Amersham GE, NJ), incubated, and then resolved by SDS-PAGE for visualization of translation products. Total membrane pellets from translation reactions were suspended in GuHCl free-medium containing cyclohexamide and P32-CTP and replication reactions were performed in the absence of any protein synthesis. Replicated PV RNA was resolved on agarose gels, which were then exposed to film for detection and quantification of PV RNA. PIK93 dissolved in DMSO was added to a final concentration of 0.4 mM or 2 mM in both the translation and replication assay stages; control reaction mixtures contained DMSO only.
Statistical Analysis
Data were expressed and plotted as means ± standard error of the mean (SEM). Unpaired student's t tests were used to compare the mean of control and experimental groups. The actual p value and sample size of each experimental group were provided in the respective figure legends.
Extended Experimental Procedures
Cell and Virus Propagation
HeLa cells (ATCC, MD) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplement with 10% fetal bovine serum (FBS), 25mM HEPES buffer, 2mM glutamine, 100units/ml of penicillin and 100 μg/ml of streptomycin. NNeo/3-5B(RG) cells were provided by Dr. Stanley Lemon (Univ. Texas-Galveston). This Huh7–derived cell line contains autonomously replicating, subgenomic, dicistronic, selectable HCV RNAs from an infectious molecular clone HCV-N of genotype 1b virus and expresses the HCV non-structural proteins NS3-NS5B.The replicon-bearing cell line was cultured in DMEM, supplemented with 2 mM L-glutamine, 5% heat-inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 300 μg/mL G418. Huh7 was maintained in the same media described above in the absence of G418. To generate CVB3 viral stock, confluent HeLa cells were infected by CVB3 for 24 hr. The infected cells underwent freeze-and-thaw cycle 3 times, supernatant was collected and stored at −80°C. Virus titer was determined by plaque assays.
Plasmids, Antibodies, and Chemicals
Arf1-GFP, Arf1-RFP, ɛCOP-GFP, GalT-YFP, Sar1T39N plasmids and anti-GBF1 antibodies were gifts of Jennifer Lippincott-Schwartz, NIH; Sac1 plasmid was gift of Peter Mayinger, OHSU;ERGIC53-GFP was gift of Theresa Ward, London School of Tropical Medicine; anti- CVB3_3A was gift of J.Lindsay Whitton, Scripps Research Institute; anti-GalT was gift of EG Berger, University of Zurich); anti-Arf1 was gift of Julie Donaldson, NIH; anti-HCV NS5A and HCV replicon J6/JFH (p7-Rluc2A)/delta E1 E2 were gifts of Charles Rice, The Rockefeller University; and PIK93 was gift of Kevin Shokat, UC Berkeley. Following reagents were commercially obtained: Anti-PI4KIIIβ and anti-myc (Cell Signaling, MA); anti-βCOP (Sigma, MO); anti-TGN46 (Abd Serotec, NC); anti-ERGIC53 (Sigma); anti-GFP (Millipore Corp., MA), anti- sec31 (BD Biosciences) and all secondary antibodies (Invitrogen Corp. CA; Jackson ImmunoResearch Labs Inc, PA). Brefeldin A, saponin (Sigma, MO), Live cell Renilla substrate Endu-Ren (Promega Corp. WI), Bovine serum albumin (BSA) (Fisher Scientific, PA), DMEM (Mediatech, VA), FBS (Atlanta biologicals, GA).
Live-Cell Imaging
All imaging was performed on a Zeiss LSM510META confocal laser scanning confocal microscope (Carl Zeiss, USA) equipped with lasers emitting 458nm, 488nm, 514nm, 565nm and 633nm laser lines. For all live-cell imaging cells were grown in coverslip bottomed Lab-Tek chambers (ThermoFisher, NY), and selected plasmids were transfected using FuGENE6 (Roche Applied science, IN). After 16 hr, incubation medium were replaced with imaging medium (phenol red free complete media buffered with 25mM HEPES pH7.3). Cells were maintained on the microscope stage in an environmental chamber where temperature (37°C) and CO2 (5%) and humidity were controlled for the duration of experiment. For high-resolution light-level imaging only 40X and 63X oil immersion objectives with 1.4 numerical aperture were utilized with pinhole set at 1.2 Airy units. Time lapsed images were acquired every 5 min. For fluorescence quantification pinhole was opened up to collect fluorescence from the entire depth of fluorescent structure of interest. Fluorescence on selected regions of interest within images was quantified using either Zeiss LSM image analysis (Carl Zeiss, USA) or Image J (NIH, MD) software.
Immunofluorescence and Analysis
Cells were plated on glass coverslips, fixed with 4% formaldehyde PBS solution (10 mM sodium phosphate, 150 mM sodium chloride, pH 7.4) at room temperature. Coverslips were permeabilized with either 0.2% Triton or 0.2% saponin and sequentially incubated with primary and fluorophore-tagged secondary antibodies. To quantify the relative fluorescence of Arf1, GBF1, PI4KIIIβ, PI4KIIIα or βCOP in 3A-myc labeled structures, images were analyzed using LSM-FCS software (Carl Zeiss, USA). The Golgi and cytosol regions of interest were selected for both 3A and neighboring non-3A expressing cells, and mean fluorescence intensity of Arf1, GBF1, PI4KIIIβ, PI4KIIIα and βCOP in those regions were obtained. The relative fluorescence was determined by following formula: [(IGolgi-ICytosol) 3A cell /(IGolgi-ICytosol)non-3A expressing cell ] ∗100, where IGolgi is the mean fluorescence intensity per pixel at the Golgi and ICytosol is the mean fluorescence intensity in the cytosol. PI4P staining on 3-5B cells was done as described previously (Jovic et al., 2009). Cells were fixed with 3.7% formaldehyde PBS solution for 10 min and incubated with anti-PI4P (Echelon Biosciences, UT) and HCV NS5A antibodies in 0.2% Saponin, 0.5%BSA PBS solution for 1 hr followed by fluorophore –tagged secondary antibodies for detection.
Fluorescence In Situ Hybridization
Codetection of protein (Arf1, GBF1) and RNA (CVB3 plus-strand RNA) in cells was adapted from http://www.singerlab.org/protocols. Alexa555 labeled CVB3 plus strand RNA specific ∼500nt length probes were made using FISH Tag RNA kit (Invitrogen, CA). HeLa cells were infected with CVB3 for 4 hr, fixed with 4% formaldehyde followed by overnight permeabilization with 70% ethanol at 4°C. Cells were re-hydrated in 2XSSC buffer (300 mM sodium chloride, 30 mM sodium citrate, pH 7.0), 50% formamide and hybridized with Alexa555 labeled CVB3 plus strand RNA specific probe at 37°C overnight in hybridization buffer (10%dextran sulfate, 2 mM vanadyl-ribonucleoside complex, 0.02% RNase-free BSA, 40 μg E. coli tRNA, 2xSSC, 50%formamide, 30ng of probe). The excess probes were washed, and Arf1 or GBF1 was subsequently immunostained with primary and Alexa488-secondary antibodies in PBS/BSA in the absence of detergents.
siRNA Transfection
Arf1 siRNA was part of the human on-target plus membrane trafficking and remodeling siRNA library (Dharmacon, CO); GBF1, PI4KIIIβ and nontargeting siRNA were purchased separately (QIAGEN,CA). HeLa cells were seeded at 10000/well in a 96-well plate (for the replicon assay) or in 200000/well in a 12-well plate for Western blot analysis to verify knockdown efficiency of proteins one day before siRNA transfection. 50 nM of siRNA were transfected via Dharmafect1 transfection reagent (Dharmacon, CO) and incubated for 48 hr.
Cell Viability Quantification
Optimal plasmid expression times, siRNA concentrations, siRNA incubation times and PIK93 concentration and incubation times that maximize cell viability were assessed by both quantifying cell number and by CellTiter-Glo cell viability assays (Promega Corp, WI). Plasmid concentration range tested was 0.1 μg/μl-1 μg/μl; and siRNA concentration range tested was 25 nM-100 nM.
Replicon Assays
The CVB3 pRib-Rluc, PV pXpA-RenR, and J6/JFH (p7-Rluc2A) plasmids with Renilla luciferase gene as reporters in place of structural genes were utilized to measure viral RNA replication. Plasmids were linearized and in vitro-transcribed and RNA purified with MEGAscript system and MEGAclear Kit respectively (Applied BioSystems/Ambion, TX). RNA was quantified by spectrophotometry and quality of runoff transcripts were estimated by agarose gels. Replicon RNAs were transfected into HeLa cells grown in 96-well white plates with flat bottom (Corning Corp., NY) at 15 ng/well with Trans-it mRNA transfection reagent (Mirus Bio LLC, WI). Incubation media contained 80 μM live-cell Renilla substrate Endu-Ren (Promega Corp., WI). Renilla light signal was recorded with Biotek HT multi-well plate reader (Biotek, VT) at 15 min intervals up to 16 hr at 37°C.
PI4P Lipid Extraction and Quantification
Cells were harvested, and PI4P lipids were extracted in cold 0.5M of Trichloroacetic acid (TCA) for 5 min. Cell pellets were washed twice with 3 ml of 5% TCA/1 mM EDTA solution and pellets were suspended in 3 ml of methanol:chloroform (2:1) solution, incubated at room temperature for ten minutes and subjected to centrifugation. To obtain phosphatidylinositol lipids, the 2.25 ml of methanol: chloroform: 12N hydrochloric acid solution (80:40:1) was added to pellets followed by fifteen minutes incubation at room temperature. Supernatant was mixed with 0.75 ml of chloroform before adding 1.35 ml of 0.1N hydrochloric acid to generate organic and aqueous phases. The organic phase containing the phosphatidylinositide was dried in a vacuum dryer, and subjected to PI4P quantification by protein lipid overlay assay (Echelon Biosciences, UT). PI4P samples from different post-infection time points were spotted on nitrocellulose membrane strips and blocked with 3% BSA 0.1% tween-20 PBS solution for 1 hr to remove non-specific binding. PI4P lipids were detected by PI4P detectors derived from FAPP1 proteins (Dowler et al., 2002) followed by secondary and tertiary antibody incubation. The signal was visualized by SuperSignal chemiluminescent substrate (Pierce, IL). The concentration of PI4P was determined by correlating the intensity of samples to a standard curve generated from known concentrations of PI4P.
RNA Polymerase Lipid-Binding Assay
Recombinant PV polymerase (3Dpol) in pET26Ub-3D was purified as described (Gohara et al., 1999). Lipid dot-blot strips (PIP micro Strips and membrane lipid Strips) were purchased (Echelon Biosciences, UT). The nitrocellulose lipid blots were incubated in blocking buffer [3% (w/v) fatty acid-free BSA fraction V (EMD, NJ) in TBST-50 (50 mM Tris, 150 mM NaCl, 0.1% Tween20 at pH 7.4)] for 1 hr at RT and then incubated in the same buffer with purified PV 3Dpol overnight at 4°C. The lipid blots were then washed in TBST-50 (5 times, 10 min each). To detect lipid protein interactions, strips were then incubated with an anti-3Dpol antibody (1:5000 in blocking buffer) for 1 hr at RT. Blots were washed as before and incubated with anti-rabbit horseradish peroxidase (1:10000 in blocking buffer) (Pierce, IL) for 1 hr at RT. 3Dpol bound to the lipids immobilized on the membrane were visualized by incubating with chemiluminescent substrate (Pierce, IL). To ensure lipid-binding specificity, several different concentrations of a protein in range from 1 to 4 μg/ml were tested. The lowest concentration giving specific binding and a detectable signal on a western blot was chosen for the lipid dot-blot experiments. Control experiments included parallel blots using no 3Dpol protein and no lipid spotted membranes.
Immunoprecipitation
Infected (CVB3; 4 hr) and noninfected HeLa cells were scraped and suspended in hypotonic buffer (50 mM KCl, 25 mM HEPES, 1 mM DTT, 1 mM EDTA). Suspended sells were broken up using cell homogenizer (Isobiotec, Germany) to preserve the replication organelle membranes. The nucleus and unbroken cells were removed by centrifugation, and the supernatant cell lysates were incubated with 1ug of monoclonal PI4KIIIβ antibody (Millipore Corp., MA) at 4°C overnight. Protein A/G magnetic beads (New England Biolabs, MA) were added and incubated at 4°C for additional 2 hr. The pull down fractions were resolved by SDS-PAGE and transferred to nitrocellulose membranes which are then blocked with 5% Nonfat Milk (3A, 3AB, 3C, 3CD blot) or 5%FCS 1%BSA (PI4KIIIβ blot) in TBST solution (10 mM Tris pH = 7.4, 150 mM NaCl, 0.1%Tween20), incubated anti-3A, anti-3Dpol or anti-PI4KIIIβ antibody and washed 5 times with TBST before incubating with horseradish peroxidase-conjugated secondary antibodies. Bound antibody complexes were detected by chemiluminescence.
Pearson Correlation Coefficient Calculations
We statistically assessed the degree of colocalization between two components using a classical Pearson correlation coefficient for the image intensity in the red and green channels using the following formulae (Bolte and Cordelieres 2006):
Acknowledgments
We thank J. Lippincott-Schwartz, S.M. Simon, K. Hirschberg, T. Ward, R. Hegde, R. Collins, J. Arnold, and G. Altan-Bonnet for insightful discussions. E.E. and G.B. were supported by the Intramural Research Program of the NIH (NIAID); T.B. was supported by the Intramural Research Program of the NIH(NICHD); C.E.C. was supported by grant AI053531 (NIAID,NIH); F.J.M.v.K. was supported by Netherlands Organisation for Scientific Research (NWO-VIDI-917.46.306, NWO-ECHO-700.57.001); N.K.B. was supported by grant DK06687 (NIDDK,NIH); and N.A.-B. was supported with grant MCB0822058 from the National Science Foundation and grant 649117 from the Busch Foundation.
Published: May 27, 2010
Footnotes
Supplemental Information includes Extended Experimental Procedures, seven figures, one table, and three movies and can be found with this article online at doi:10.1016/j.cell.2010.03.050.
Supplemental Information
Cell viability measured by Celltiter-glo viability assays.
Arf1-RFP (red) and GFB1-YFP (green) were coexpressed in a HeLa cell, infected with CVB3 and immediately imaged. Time-lapse confocal images were taken 5 min apart for a total of 8 hr.
Arf1-RFP (red) and GalT-YFP (green) were coexpressed in a HeLa cell, infected with CVB3 and immediately imaged. Time-lapse confocal images were taken 5 min apart for a total of 6 hr.
Arf1-RFP (red) and FAPP1-PH-GFP (green) were coexpressed in a HeLa cell, infected with CVB3, and immediately imaged. Time-lapse confocal images were taken 5 min apart for a total of 6 hr.
References
- Altan-Bonnet N., Sougrat R., Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Balla A., Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Barton D.J., Flanegan J.B. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J. Virol. 1993;67:822–831. doi: 10.1128/jvi.67.2.822-831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., Altan-Bonnet N., Kovtunovych G., Jackson C.L., Lippincott-Schwartz J., Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger K.L., Cooper J.D., Heaton N.S., Yoon R., Oakland T.E., Jordan T.X., Mateu G., Grakoui A., Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A., Cheong F.Y., Rohde H.M., Glover G., Knödler A., Nicolson T., Boehmelt G., Mayinger P. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J. Cell Biol. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumental-Perry A., Haney C.J., Weixel K.M., Watkins S.C., Weisz O.A., Aridor M. Phosphatidylinositol 4-phosphate formation at ER exit sites regulates ER export. Dev. Cell. 2006;11:671–682. doi: 10.1016/j.devcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Borawski J., Troke P., Puyang X., Gibaja V., Zhao S., Mickanin C., Leighton-Davies J., Wilson C.J., Myer V., Cornellataracido I., et al. Class III phosphatidylinositol-4 kinase alpha & beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dales S., Eggers H.J., Tamm I., Palade G.S. Electron microscopic study of the formation of Poliovirus. Virology. 1965;26:379–389. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- D'Angelo G., Vicinanza M., Di Campli A., De Matteis M.A. The multiple roles of PtdIns(4)P–not just the precursor of PtdIns(4,5)P2. J. Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- Deng Y., Golinelli-Cohen M.P., Smirnova E., Jackson C.L. A COPI coat subunit interacts directly with an early-Golgi localized Arf exchange factor. EMBO Rep. 2009;10:58–64. doi: 10.1038/embor.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitz S.B., Dodd D.A., Cooper S., Parham P., Kirkegaard K. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA. 2000;97:13790–13795. doi: 10.1073/pnas.250483097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S., Kular G., Alessi D.R. Protein lipid overlay assay. Sci. STKE. 2002;129:PI6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]
- Dubuisson J., Penin F., Moradpour D. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 2002;12:517–523. doi: 10.1016/s0962-8924(02)02383-8. [DOI] [PubMed] [Google Scholar]
- Fogg M.H., Teterina N.L., Ehrenfeld E. Membrane requirements for uridylylation of the poliovirus VPg protein and viral RNA synthesis in vitro. J. Virol. 2003;77:11408–11416. doi: 10.1128/JVI.77.21.11408-11416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A., Pertile P., Meyers R., Marra P., Di Tullio G., Iurisci C., Luini A., Corda D., De Matteis M.A. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D.R., Kular G.S., Daniele T., Marra P., Lucocq J.M., De Matteis M.A. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Gohara D.W., Ha C.S., Kumar S., Ghosh B., Arnold J.J., Wisniewski T.J., Cameron C.E. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 1999;17:128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
- Ikeda M., Yi M., Li K., Lemon S.M. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 2002;76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N., Hill C.M., Bates I.R., Harauz G. The formation of helical tubular vesicles by binary monolayers containing a nickel-chelating lipid and phosphoinositides in the presence of basic polypeptides. Chem. Phys. Lipids. 2002;114:103–111. doi: 10.1016/s0009-3084(02)00002-6. [DOI] [PubMed] [Google Scholar]
- Jones C.T., Murray C.L., Eastman D.K., Tassello J., Rice C.M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight Z.A., Knight Z.A., Gonzalez B., Feldman M.E., Zunder E.R., Goldenberg D.D., Williams O., Loewith R., Stokoe D., Balla A., et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konan K.V., Giddings T.H., Jr., Ikeda M., Li K., Lemon S.M., Kirkegaard K. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J. Virol. 2003;77:7843–7855. doi: 10.1128/JVI.77.14.7843-7855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge O., Dascher C., Orci L., Rowe T., Amherdt M., Plutner H., Ravazzola M., Tanigawa G., Rothman J.E., Balch W.E. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J. Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanke K.H., van der Schaar H.M., Belov G.A., Feng Q., Duijsings D., Jackson C.L., Ehrenfeld E., van Kuppeveld F.J. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J. Virol. 2009;83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix J., Ouwendijk J., Lin C.C., Stark A., Love H.D., Ostermann J., Nilsson T. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COP I vesicles. EMBO J. 1999;18:4935–4948. doi: 10.1093/emboj/18.18.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.C., Miller E.A., Goldberg J., Orci L., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Lefrançois S., McCormick P.J. The Arf GEF GBF1 is required for GGA recruitment to Golgi membranes. Traffic. 2007;8:1440–1451. doi: 10.1111/j.1600-0854.2007.00623.x. [DOI] [PubMed] [Google Scholar]
- Lemmon M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lundin M., Monné M., Widell A., Von Heijne G., Persson M.A. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 2003;77:5428–5438. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.M., Westaway E.G. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 2007;75:10787–10799. doi: 10.1128/JVI.75.22.10787-10799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliano D., Marshall J.A., Bowden D.S., Vardaxis N., Meanger J., Lee J.Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Monetta P., Slavin I., Romero N., Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol. Biol. Cell. 2007;18:2400–2410. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu T.K., Pfeifer A.C., Lippincott-Schwartz J., Jackson C.L. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J.F., Ward T.H., Pfeifer A.C., Siggia E.D., Phair R.D., Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- Richards O.C., Ehrenfeld E. Poliovirus RNA replication. Curr. Top. Microbiol. Immunol. 1990;161:89–119. doi: 10.1007/978-3-642-75602-3_4. [DOI] [PubMed] [Google Scholar]
- Salonen A., Ahola T., Kääriäinen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A., Giddings T.H., Jr., Ladinsky M.S., Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styers M.L., O'Connor A.K., Grabski R., Cormet-Boyaka E., Sztul E. Depletion of beta-COP reveals a role for COP-I in compartmentalization of secretory compartments and in biosynthetic transport of caveolin-1. Am. J. Physiol. Cell Physiol. 2008;294:C1485–C1498. doi: 10.1152/ajpcell.00010.2008. [DOI] [PubMed] [Google Scholar]
- Tai A.W., Benita Y., Peng L.F., Kim S.S., Sakamoto N., Xavier R.J., Chung R.T. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth B., Balla A., Ma H., Knight Z.A., Shokat K.M., Balla T. Phosphatidylinositol 4-kinase IIIbeta regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem. 2006;281:36369–36377. doi: 10.1074/jbc.M604935200. [DOI] [PubMed] [Google Scholar]
- Towner J.S., Ho T.V., Semler B.L. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- Trotard M., Lepère-Douard C., Régeard M., Piquet-Pellorce C., Lavillette D., Cosset F.L., Gripon P., Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780–3789. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- Uetz P., Dong Y.A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S.V., Roupelieva M., Rose D., Fossum E., Haas J. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- Vaillancourt F.H., Pilote L., Cartier M., Lippens J., Liuzzi M., Bethell R.C., Cordingley M.G., Kukolj G. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Wang J., Sun H.Q., Macia E., Kirchhausen T., Watson H., Bonifacino J.S., Yin H.L. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol. Biol. Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels E., Duijsings D., Niu T.K., Neumann S., Oorschot V.M., de Lange F., Lanke K.H., Klumperman J., Henke A., Jackson C.L., et al. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell. 2006;11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Wölk B., Büchele B., Moradpour D., Rice C.M. A dynamic view of hepatitis C virus replication complexes. J. Virol. 2008;82:10519–10531. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S., Oku M., Wasada Y., Ano Y., Sakai Y. PI4P-signaling pathway for the synthesis of a nascent membrane structure in selective autophagy. J. Cell Biol. 2006;173:709–717. doi: 10.1083/jcb.200512142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Bolte, S., and Cordelières, F.P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232. [DOI] [PubMed]
- Dowler, S., Kular, G., and Alessi, D.R. (2002). Protein lipid overlay assay. Sci. STKE 129, PL6. [DOI] [PubMed]
- Gohara, D.W., Ha, C.S., Kumar, S., Ghosh, B., Arnold, J.J., Wisniewski, T.J., and Cameron, C.E. (1999). Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17, 128–138. [DOI] [PubMed]
- Jović, M., Kieken, F., Naslavsky, N., Sorgen, P.L., and Caplan, S. (2009). Eps15 homology domain1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol- (4,5)-bisphosphate and are required for efficient recycling. Mol. Biol. Cell 20, 2731–2743. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell viability measured by Celltiter-glo viability assays.
Arf1-RFP (red) and GFB1-YFP (green) were coexpressed in a HeLa cell, infected with CVB3 and immediately imaged. Time-lapse confocal images were taken 5 min apart for a total of 8 hr.
Arf1-RFP (red) and GalT-YFP (green) were coexpressed in a HeLa cell, infected with CVB3 and immediately imaged. Time-lapse confocal images were taken 5 min apart for a total of 6 hr.
Arf1-RFP (red) and FAPP1-PH-GFP (green) were coexpressed in a HeLa cell, infected with CVB3, and immediately imaged. Time-lapse confocal images were taken 5 min apart for a total of 6 hr.