Abstract
Summary: Recent advances in single-cell manipulation technology, whole genome amplification and high-throughput sequencing have now made it possible to sequence the genome of an individual cell. The bioinformatic analysis of these genomes, however, is far more complicated than the analysis of those generated using traditional, culture-based methods. In order to simplify this analysis, we have developed SmashCell (Simple Metagenomics Analysis SHell-for sequences from single Cells). It is designed to automate the main steps in microbial genome analysis—assembly, gene prediction, functional annotation—in a way that allows parameter and algorithm exploration at each step in the process. It also manages the data created by these analyses and provides visualization methods for rapid analysis of the results.
Availability: The SmashCell source code and a comprehensive manual are available at http://asiago.stanford.edu/SmashCell
Contact: eoghanh@stanford.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The rapid evolution of DNA sequencing platforms has had a dramatic, beneficial impact on the study of microbial ecology and population genetics. So far, these benefits have mostly come from shotgun community metagenomics that provides a high-level overview of the taxonomic and functional composition of microbial communities [see Arumugam et al. (2010) for details]. However, this approach is limited in its ability to yield complete genome sequences as well as the fine-scale genetic variation that defines population substructures within these communities. One possible solution uses a combination of single-cell manipulation technologies, multiple-displacement amplification (MDA) and high-throughput sequencing to generate single-cell amplified genomes (SAGs). This approach has already been used to characterize the genomes of uncultivated microbes (Marcy et al., 2007; Woyke et al., 2009) and as the throughput of the associated technologies increase it should become possible to obtain high-resolution profiles of populations or communities.
However, it is more difficult to produce a high-quality assembly and functional annotation from a SAG than from the output of traditional methods due to limitations inherent in sample preparation and sequencing (detailed below). To overcome this requires an iterative, exploratory approach that transforms the traditional linear process of genome assembly, gene prediction and functional annotation into a tree-like structure, each branch defined by a different choice of algorithm or parameters, one of which will be chosen as the final version (Fig. 1A). This approach is not easily achieved using existing tools, which take an assembled genome as their input and do not allow parameterization of subsequent steps (for a comparison with existing tools see Supplementary Table). In order to automate the process and deal with the resulting combinatorial increase in data we have created SmashCell. While developed for use on SAGs many of its analyses are equally applicable to traditional microbial genome sequences and low-complexity metagenomes.
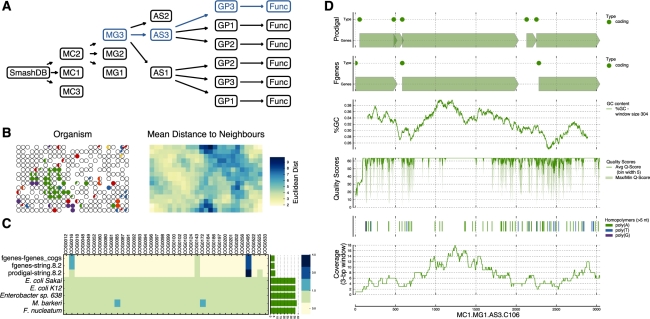
Fig. 1.
(A) The data model used in SmashCell is designed to reduce redundancy and facilitate the comparison of results using different parameters and/or algorithms [MC: metagenome collection, MG: metagenome (equivalent to a SAG), AS: assembly, GP: gene prediction, FUNC: functional annotation]. (B) K-mer frequency statistics supplement sequence similarity information to identify potential contaminants. This shows a self-organizing map (SOM) trained on the tetramer frequencies of an assembly. The left panel shows a series of pie charts highlighting the taxonomic identity (determined by best hit in GenBank, those with no hits are uncoloured) of the contigs assigned to each neuron. The right panel shows the U-matrix of the SOM. (C) The abundance of single-copy COGs can be used to assess genome completeness, the presence of contamination and the quality of the assembly. (D) SmashCell uses different graphs to aid in parameter and algorithm selection. Here the results from two different gene prediction algorithms are presented, along with GC-content, quality scores and read depth.
2 FEATURES
SmashCell automates the steps common to most genome analyses—assembly, gene prediction and functional annotation—and addresses some of the challenges posed by SAGs. For instance, it is difficult to isolate a single cell for sequencing without including some environmental DNA, in effect creating a metagenome. As a result, SmashCell includes both sequence similarity and k-mer based tools to identify potential contaminants, the latter being especially useful when the target genome and/or contaminants are not closely related to existing genome sequences (see Fig. 1B for details). Another challenge with SAGs is the orders of magnitude variance in MDA product abundance along the genome, which creates several obstacles to obtaining high-quality annotation. First, it hampers genome assembly, as most algorithms are designed for lower and more evenly distributed read depth. Secondly, it vastly increases the amount of sequencing required to obtain a complete genome sequence. To address the first challenge, SmashCell includes scripts to downsample overrepresented regions of the SAG and to address the second, SmashCell uses the STRING database (Jensen et al., 2009) to obtain counts of single-copy orthologous groups (Fig. 1C), which can then be used to estimate genome completeness. In addition to these SAG-specific features, SmashCell contains genome visualization and other tools (Fig. 1D) that are generally applicable to genomic and metagenomic data. SmashCell uses the same basic data model as SmashCommunity [designed for shotgun community sequencing; Arumugam et al. (2010)]. As a result, several of the analyses available in SmashCell can be run on data generated by SmashCommunity and vice versa. Documentation for these and many more features are available on the SmashCell website.
3 DESIGN AND IMPLEMENTATION
SmashCell is a framework written in Python that provides a variety of analysis tools that can be used either from the command line or from within other Python scripts. The main function of SmashCell is to automate the common steps in genome analysis in a way that facilitates parameter and algorithm exploration. Using the data model shown in Figure 1A, SmashCell manages the files and data associated with each of these steps, reducing redundancy and providing a layer of abstraction that simplifies access to these data. SmashCell also uses generic databases to provide a common format for assembly and gene prediction information, allowing it to work with a variety of third-party assemblers and gene prediction algorithms. In order to facilitate the exploration of genomic data, SmashCell automatically generates many different types of graphs (e.g. see Fig. 1B–D) and provides wrappers for exploratory statistical techniques.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank S. Pamp and P. Blainey for assistance with testing.
Funding: Human Frontiers Science Program (LTF to E.D.H); National Institutes of Health (1R01HG004863 and Director's Pioneer Award to D.A.R); Thomas C. and Joan M. Merigan Endowment at Stanford University (to D.A.R); European Community FP7 [MetaHIT].
Conflict of Interest: none declared.
REFERENCES
- Arumugam M, et al. SmashCommunity: a metagenomic annotation and analysis tool. 2010 doi: 10.1093/bioinformatics/btq536. submitted. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl Acad. Sci. USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, et al. Assembling the marine metagenome, one cell at a time. PloS ONE. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.