Abstract
Motivation: G → A hypermutation is an innate antiviral defense mechanism, mediated by host enzymes, which leads to the mutational impairment of viruses. Sensitive and specific identification of host-mediated G → A hypermutation is a novel sequence analysis challenge, particularly for viral deep sequencing studies. For example, two of the most common hepatitis B virus (HBV) reverse transcriptase (RT) drug-resistance mutations, A181T and M204I, arise from G → A changes and are routinely detected as low-abundance variants in nearly all HBV deep sequencing samples.
Results: We developed a classification model using measures of G → A excess and predicted indicators of lethal mutation and applied this model to 325 920 unique deep sequencing reads from plasma virus samples from 45 drug treatment-naïve HBV-infected individuals. The 2.9% of sequence reads that were classified as hypermutated by our model included most of the reads with A181T and/or M204I, indicating the usefulness of this model for distinguishing viral adaptive changes from host-mediated viral editing.
Availability: Source code and sequence data are available at http://hivdb.stanford.edu/pages/resources.html.
Contact: ereuman@stanfordalumni.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Hepatitis B virus (HBV) is a double-stranded DNA virus that infects more than 500 million people worldwide and is a leading cause of mortality as a result of cirrhosis and hepatocellular carcinoma. In the past 12 years, five nucleoside analogs have been licensed for HBV treatment. These drugs are capable of fully suppressing HBV replication but rarely eradicate infection because HBV is converted intra-cellularly into a stable covalently closed circular DNA form.
HBV replicates via an RNA intermediate, and its polymerase enzyme has a high mutation rate similar to other enzymes with reverse transcriptase activity. It thus generates a quasispecies of innumerable related virus variants from which drug resistant viruses can arise (Preikschat et al., 1999). Because HBV drug resistance is one of the obstacles to successful anti-HBV therapy, current treatment guidelines recommend HBV genotypic resistance testing for patients who experience primary or secondary virological failure while receiving nucleoside therapy (Lok et al., 2007).
Deep sequencing is increasingly performed on plasma samples from clinical trials to determine the clinical significance of low-abundance human immunodeficiency virus (HIV) and hepatitis C virus drug-resistance mutations prior to starting antiviral treatment (Vrancken et al., 2010). We and others have recently described the use of ultra-deep pyrosequencing (UDPS) with the 454 Life Sciences/Roche Genome Sequencer FLX platform (Margeridon-Thermet et al., 2009; Solmone et al., 2009) to detect low-abundance HBV variants. We previously showed that nucleoside analog drug-resistance mutations not detected by direct PCR sequencing could be detected by UDPS in as few as 1.0% of sequence reads. We also observed that many samples contained sequence reads with unusually high numbers of guanine (G) to adenine (A) changes relative to the direct PCR sequence, consistent with the recently described phenomenon of G → A hypermutation.
G → A hypermutation results from an innate antiviral defense mechanism mediated by the activity of host enzymes belonging to the apolipoprotein B RNA-editing catalytic polypeptide-like 3 (APOBEC3) family of cytidine deaminases (Cullen, 2006). These enzymes are capable of causing extensive deamination of cytidine bases to uridine in negative-stranded DNA, resulting in G → A hypermutation in positive-stranded DNA. Although APOBEC-mediated G → A hypermutation was first reported to act upon HIV (Sheehy et al., 2002), it has also been reported to act upon HBV (Noguchi et al., 2005; Suspène et al., 2005), other retroviruses, and retrotransposons (Cullen, 2006).
Because extensive G → A editing leads to mutational impairment of viruses, distinguishing hypermutated sequence reads from non-hypermutated reads is necessary for accurate analysis of viral quasispecies. This is particularly important in deep sequencing studies designed to detect low levels of nucleoside analog drug-resistant viruses because two of these, A181T and M204I, primarily result from G → A substitutions. Indeed, the creation of sensitive and specific methods for identifying APOBEC-mediated G → A hypermutation is a novel sequence analysis challenge.
We previously described an ad hoc method for identifying G → A hypermutation in HBV sequences (Margeridon-Thermet et al., 2009). Now, we develop a data-derived method using a probabilistic model based on the expectation–maximization (EM) algorithm, using a large number of sequences from untreated individuals.
2 METHODS
2.1 Patients and sequences
Baseline plasma samples were obtained from 45 HBV-infected nucleoside analog-naïve individuals with informed consent. Viral DNA was extracted from the plasma and sequenced both by direct PCR (Sanger) sequencing and by UDPS using the 454 Roche Genome Sequencer FLX platform. Raw UDPS reads were processed for quality prior to analysis. Direct PCR sequences were submitted to GenBank. The GenBank Accession IDs, HBV genotypes and further details of the sequencing and quality analysis protocols are given in Supplementary Data I and II.
UDPS reads with identical nucleic acid sequences were grouped into a single unique read. Each unique read was aligned to a reference HBV RT sequence corresponding to the HBV genotype of the sample as determined by the direct PCR sequence (Rozanov et al., 2004). Sequence alignment was performed using Pyromap, a Smith–Waterman-based local alignment program that uses the pyrosequencing quality scores for optimal placement of nucleotide insertions and deletions (Wang et al., 2007). The technical mismatch error rate was estimated at 0.1% per base by sequencing multiple plasmid HBV RT DNA clones. In previous studies, we have shown that this technical error rate is a combination of PCR and pyrosequencing error (Varghese et al., 2010).
2.2 Sequence analysis
Aligned UDPS sequence reads were examined for potential indications of G → A hypermutation: (i) an apparent excess of A's in positions at which the direct PCR sequence contained a G; and (ii) stop codons and atypical amino acid mutations. The first category included the number of G → A substitutions divided by the number of G nucleotides in the direct PCR sequence (‘G → A burden’) and the number of G → A substitutions divided by the number of all substitutions (‘G → A preference’) (Rose and Korber, 2000).
The second category included stop codons in the RT gene, both those occurring within the reading frame encoding the RT enzyme and those in the overlapping 1+ reading frame encoding the HBV surface S protein. The second category also included the number of atypical amino acid mutations resulting from G → A substitutions; our criteria for atypical mutations are described in Supplementary Data III. A stop codon within an open reading frame mutationally inactivates a protein, and therefore may be more likely to result from a host defense mechanism such as APOBEC-mediated G → A hypermutation than a viral escape mutation. Likewise, a mutation that has not previously been reported in a direct PCR sequence indicates that the mutation may reduce viral fitness and be more likely to represent a host-mediated change.
Two additional variables, the total number of G → A substitutions and the difference between the number of G → A substitutions and the number of A → G substitutions, were also considered, but an analysis of variance indicated a high (>0.85) correlation between these variables and at least one other listed above, making them redundant as classifiers in the following statistical analysis.
2.3 Statistical analysis
Five variables described above—G → A burden, G → A preference, RT stop codons, S stop codons, and atypical G → A mutations—were used to define naïve Bayes classifiers in a classification model based on the EM algorithm (Dempster et al., 1977). The model used Bayes' Theorem to assign probabilities of hypermutation to each UDPS sequence read.
We defined a training set by assigning all UDPS sequences an initial classification using an ad hoc filter of strong indicators of G → A hypermutation. Sequences that contained at least two G → A substitutions were preliminarily classified as hypermutants if they contained at least one stop codon in the RT or S genes, or if they had a G → A burden of at least 10% combined with a G → A preference of at least 75%. The training set hypermutants therefore consisted of the sequences most likely to represent viruses with G → A substitution rates much higher than expected, compared with a published HBV nucleotide substitution model (Fares and Holmes, 2002), and the sequences most likely to reflect mutationally inactivated viruses.
The number of hypermutants divided by the total number of sequences in the training set was used to define P(H), the probability that a sequence chosen at random is hypermutated. Based on the conditional probability distributions of the five classifiers, we iteratively calculated a revised probability of hypermutation for each UDPS sequence read, P(H)Read. At each iteration, sequences were classified as hypermutated if P(H)Read was ≥0.9. The overall probability of hypermutation and the conditional probability distribution of each classifier were then updated to reflect the new sequence classifications. Classification was revised in this manner until the model converged. The final P(H)Read≥0.9 was used as a filter to distinguish hypermutated from non-hypermutated sequences. Probability density comparisons were graphed using the R library sm and the function sm.density.compare.
3 RESULTS
UDPS yielded 325 920 unique sequence reads from 45 nucleoside analog-naïve HBV-infected individuals. The mean read length was 194 nucleotides (range: 100–290); all reads shorter than 200 nucleotides resulted from excluding nucleotides 5′ to the start or 3′ to the end of HBV RT. The mean sequence coverage per nucleotide was 3094 (range: 1982–5206). Reads had a mean 1.11 nucleotide differences from the corresponding direct PCR sequences (range: 0–35), including a mean 0.22 G → A substitutions.
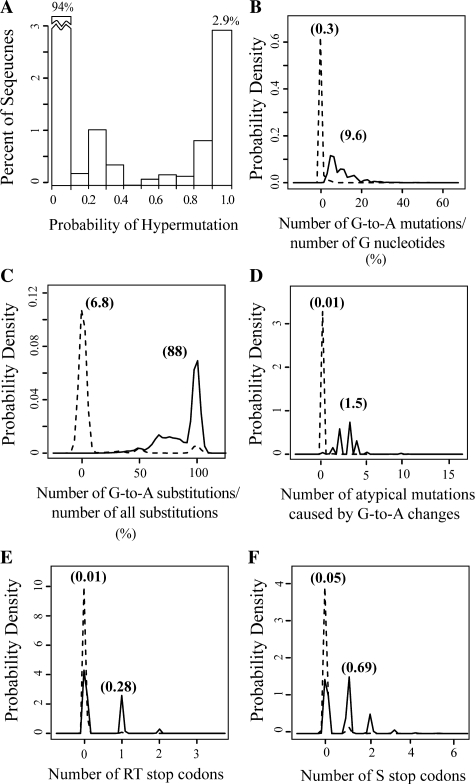
The EM-derived hypermutation filter classified 2.9% of unique UDPS sequence reads (9359) as hypermutated based on a final P(H)Read≥0.9. An additional 2.9% of sequences (9434) had a P(H)Read between 0.1 and 0.9. The remaining 94.2% of sequences (307 127) had a P(H)Read≤0.1. Figure 1A shows the final distribution of hypermutation probabilities, and Figures 1B–F show the final distributions of the five classifiers among the two classes of sequences, as well as their mean values.
Fig. 1.
Final distributions after application of our EM-based model. (A) The distribution of hypermutation probabilities P(H)Read among the full set of sequence reads. In all, 94% of sequences had hypermutation probabilities of ≤10% and 2.9% of sequences had probabilities ≥90%. (B–F) Probability densities of each of the five classifiers, for sequences classified as non-hypermutated (dashed lines) or hypermutated (solid lines).
Hypermutated sequences were characterized by G → A burdens ≥5%, G → A preferences ≥75% and two or more indications of potential lethal editing, including stop codons and atypical G → A mutations. Non-hypermutated sequences with P(H)Read≤0.1 were characterized by low G → A burdens, low G → A preferences and no stop codons or atypical G → A mutations. Sequences with intermediate probabilities were characterized by few G → A changes and one indication of potential lethal editing.
Samples from all 45 individuals contained one or more hypermutated reads, constituting 0.2–9.3% of unique UDPS reads. The Friedman rank sum test (Rice, 2007) showed that the sample of origin (d.f. = 44) had a more significant contribution to the level of hypermutation in a read than the primer pair used for PCR amplification (d.f. = 3), suggesting that hypermutation was a characteristic of individual plasma samples rather than a PCR artifact.
Following classification, we examined the upstream and downstream dinucleotide context of G → A substitutions within each class to assess whether hypermutation resulted from the preferential action of one enzyme in the APOBEC3 (A3A-H) family. The results of this analysis are described in Supplementary Data IV.
No direct PCR sequence contained an RT drug-resistance mutation. Among hypermutated UDPS sequence reads, the G → A mutations M204I (7.6%) and A181T (2.9%) were the most common drug-resistance mutations, followed by M204V (0.03%), S202G (0.03%), M250V (0.02%), V173L (0.01%) and T184I (0.01%), which are not caused by G → A substitutions. No other drug-resistance mutations occurred in hypermutated sequences. M204I and A181T occurred in 0.09 and 0.04% of non-hypermutated sequences, respectively. The full list of drug-resistance mutations and atypical mutations found in each class of sequences is given in Supplementary Data III.
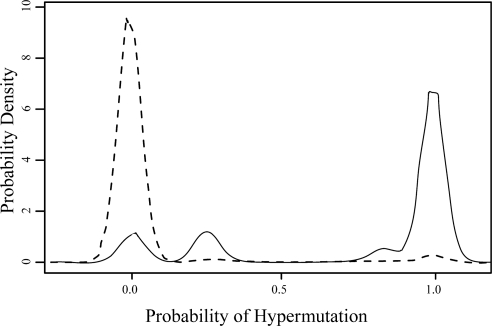
Figure 2 compares the hypermutation probability density of sequences containing A181T and/or M204I to that of sequences containing neither mutation. For the purposes of this analysis, we examined all A181T mutations (GCN → ACN) and the subset of M204I mutations caused by G → A substitutions (ATG → ATA).
Fig. 2.
Probabilities of hypermutation (P(H)Read) of sequence reads containing A181T and/or M204I (solid line) versus sequences containing neither mutation (dashed line). Most UDPS reads containing A181T and/or M204I had high hypermutation probabilities, while the reverse was true of reads containing neither mutation.
Although these mutations were not hypermutation classifiers, sequences with one or both mutations were significantly more likely to be classified as hypermutated, and had significantly higher values of each classifier, than sequences with neither mutation (P < 1E-5; Student's t-test). Prior to filtering out hypermutants, one or both of these mutations were present in ≥1% of all UDPS reads from nine individuals. After application of the filter, neither mutation occurred at a frequency of >0.5% in any individual.
4 DISCUSSION
UDPS provides insight into the evolutionary dynamics of the emergence of viral drug resistance and may eventually prove useful in clinical diagnostic testing. However, the biological and clinical significance of HBV UDPS results cannot be optimally interpreted without being able to distinguish hypermutated from non-hypermutated sequence reads. APOBEC-mediated G → A hypermutation results from a host defense mechanism against viral genomes and retroelements, and therefore mutations caused by this mechanism have different biological and clinical significance from mutations resulting from viral adaptation.
Three groups have developed methods for identifying hypermutated HIV-1 sequences (Rose and Korber, 2000; Kijak et al., 2007; Gifford et al., 2008). To our knowledge, no such method has been developed for HBV or for deep sequencing reads. Identifying G → A hypermutation in HBV UDPS reads is particularly challenging because such reads are shorter and therefore contain fewer informative sites than direct PCR sequences. Moreover, in contrast to HIV, HBV hypermutation could potentially be mediated by six of the seven enzymes in the APOBEC3 family (A3A-C and F-H) and does not occur in a consistent dinucleotide context (Köck and Blum, 2008; Henry et al., 2009).
In this study, we have demonstrated the use of a novel EM-based model in assigning probabilistic labels to UDPS reads in order to classify them as hypermutated or non-hypermutated, revealing a striking distinction between the characteristics of the two classes.
The discriminatory ability of our model benefited from the overlapping reading frames of HBV, with RT stop codons resulting in non-functional viruses and S stop codons resulting in viruses incapable of cell-to-cell spread (Locarnini and Warner, 2007). Our model also benefited from the use of an HBV variant database we recently constructed to identify previously unreported RT mutations resulting from G → A mutations, which may also be indicators of host-mediated viral editing.
The high concordance among the G → A excess classifiers and the numbers of stop codons and atypical RT mutations support the hypothesis that both categories of classifiers reflect a host defense mechanism rather than a mechanism of viral adaptation. Indeed, our results underestimate the deleterious effect of hypermutation because each sequence read of 200 nucleotides represents just 6% of the 3100 nucleotide HBV genome; extension of our analysis to the entire HBV genome could reveal additional mutations that, in combination, would indicate inviable viruses. Our filter is likely to have even greater discriminatory power on data from UDPS platforms with longer reads, including the Titanium series of reagents for 454 UDPS (Roche Applied Sciences).
The G → A hypermutation filter enabled us to address one of the motivating factors of this study: determining which UDPS reads containing the drug-resistance mutations A181T and/or M204I were likely to be hypermutated and presumably from non-functional viruses. Because the 45 samples we analyzed were from NRTI-naïve individuals, we were not surprised that the majority of unique UDPS reads with these mutations were hypermutated. Indeed, following application of our filter, the proportions of reads with A181T and/or M204I dropped to <0.1%, approaching the proportions of reads with other common drug-resistance mutations.
It should be noted, however, that G → A hypermutation rarely occurs at high enough levels to be detected by direct PCR sequencing. The presence of A181T and M204I in such sequences nearly always indicates clinically significant drug resistance. However, in deep sequencing reads from nucleoside-treated individuals or individuals with unknown treatment histories, our filter will be essential for accurately distinguishing between drug-resistant and inviable viruses. Although multiply infected hepatocytes can theoretically yield mosaic viruses in which only parts of the HBV genome is hypermutated or in which hypermutated RT genes are packaged by non-hypermutated envelope proteins (Suspène et al., 2005), the likelihood that either type of recombinant virus would successfully infect new hepatocytes is low.
The G → A hypermutation filter described here will provide researchers with a vital tool for distinguishing hypermutated from non-hypermutated HBV deep sequencing reads. The flexibility of the algorithm used in our model allows users to alter several parameters (such as the initial ad hoc classification system and the hypermutation probability cut-offs). Although we have focused on the utility of this filter for identifying nucleoside analog drug-resistance mutations (particularly A181T and M204I) in viruses from nucleoside-naive individuals, the filter will also provide a means of identifying biologically and clinically relevant drug-resistance mutations in nucleoside-treated individuals, as well as filtering out inviable viruses in studies unrelated to drug resistance.
Funding: National Institutes of Health (AI068581 and AI46148 to E.C.R., S.M.-T., T.L. and R.W.S.; R01GM086884 to S.H.).
Conflict of Interest: K.B.-E. and E.S.S. are employees of and hold stock in Gilead Sciences, Inc.
Supplementary Material
REFERENCES
- Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 2006;80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster AP, et al. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Series B Methodol. 1977;39:1–38. [Google Scholar]
- Fares MA, Holmes EC. A revised evolutionary history of hepatitis B virus (HBV) J. Mol. Evol. 2002;54:807–814. doi: 10.1007/s00239-001-0084-z. [DOI] [PubMed] [Google Scholar]
- Gifford RJ, et al. Sequence editing by Apolipoprotein B RNA-editing catalytic component and epidemiological surveillance of transmitted HIV-1 drug resistance. AIDS. 2008;22:717–725. doi: 10.1097/QAD.0b013e3282f5e07a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M, et al. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PLoS ONE. 2009;4:e4277. doi: 10.1371/journal.pone.0004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijak GH, et al. HyperPack: a software package for the study of levels, contexts, and patterns of APOBEC-mediated hypermutation in HIV. AIDS Res. Hum. Retroviruses. 2007;23:554–557. doi: 10.1089/aid.2006.0279. [DOI] [PubMed] [Google Scholar]
- Kock J, Blum HE. Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J. Gen. Virol. 2008;89:1184–1191. doi: 10.1099/vir.0.83507-0. [DOI] [PubMed] [Google Scholar]
- Locarnini S, Warner N. Major causes of antiviral drug resistance and implications for treatment of hepatitis B virus monoinfection and coinfection with HIV. Antivir. Ther. 2007;12(Suppl. 3):H15–H23. [PubMed] [Google Scholar]
- Lok AS, et al. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- Margeridon-Thermet S, et al. Ultra-deep pyrosequencing of hepatitis B virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naive patients. J. Infect. Dis. 2009;199:1275–1285. doi: 10.1086/597808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi C, et al. G to A hypermutation of hepatitis B virus. Hepatology. 2005;41:626–633. doi: 10.1002/hep.20580. [DOI] [PubMed] [Google Scholar]
- Preikschat P, et al. Hepatitis B virus genomes from long-term immuno-suppressed virus carriers are modified by specific mutations in several regions. J. Gen. Virol. 1999;80(Pt 10):2685–2691. doi: 10.1099/0022-1317-80-10-2685. [DOI] [PubMed] [Google Scholar]
- Rice JA. Mathematical Statistics and Data Analysis. 3rd. Thomson Brooks/Cole; 2007. p. 469. [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G –> A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Rozanov M, et al. A web-based genotyping resource for viral sequences. Nucleic Acids Res. 2004;32:W654–W659. doi: 10.1093/nar/gkh419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Solmone M, et al. Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen. J. Virol. 2009;83:1718–1726. doi: 10.1128/JVI.02011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène R, et al. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese V, et al. Nucleic acid template and the risk of a PCR-Induced HIV-1 drug resistance mutation. PLoS ONE. 2010;5:e10992. doi: 10.1371/journal.pone.0010992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrancken B, et al. Covering all bases in HIV research: unveiling a hidden world of viral evolution. AIDS Rev. 2010;12:89–102. [PubMed] [Google Scholar]
- Wang C, et al. Characterization of mutation spectra with ultra-deep pyro-sequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.