Abstract
Summary: OmicsAnalyzer is a Cytoscape plug-in for visual omics-based network analysis that (i) integrates hetero-omics data for one or more species; (ii) performs statistical tests on the integrated datasets; and (iii) visualizes results in a network context.
Availability: Implemented in Java, OmicsAnalyzer runs with Cytoscape 2.6 and 2.7. Binaries, documentation and video walkthroughs are freely available at http://vrac.iastate.edu/~jlv/omicsanalyzer/
Contact: julied@iastate.edu; netscape@iastate.edu
1 INTRODUCTION
The widespread use of omics-related biotechnologies has led to many heterogeneous omics datasets. Software packages facilitate the interpretation of omics data by using graph models where nodes represent bio-elements and edges are bio-interactions between these elements (Junker et al., 2006). However, the complexity of biological systems and omics datasets require an integrative analytical environment, which allows network manipulation at different levels of detail, flexible mappings between multiple omics datasets coupled with data visualization and providing statistical analysis and system simulation and visualization of resulting analysis.
Cytoscape (Shannon et al., 2003), an open source platform software dedicated to network modeling in biology, provides a highly flexible environment for combining these elements. To meet the above requirements, OmicsAnalyzer (i) manipulates networks through interface connection with third-party plug-ins; (ii) maps omics datasets within or across species and manages the datasets; (iii) visualizes dynamic data using plots and animations; and (iv) analyzes data using built-in statistical functions. The integration of these key analysis functionalities is a key building block for omics data analysis in systems biology.
2 FUNCTIONALITY AND IMPLEMENTATION
The key features of OmicsAnalyzer are presented in three levels (Fig. 1A), which function both independently as standalone tools and communicate with each other to facilitate system-level analysis. The levels are accessible as tabs in the Cytoscape control panel. In the first level, Data Collection and Preprocessing, there are two modules: network generator and omics data integration. The network generator enables the user to select subnetworks of interest with respect to the network topology at different levels of detail such as cycle, dense module or network motif. OmicsAnalyzer manipulates the network by communicating with the Cytoscape platform or third-part plug-ins. Specific functionality includes: (i) selection of arbitrary subnetworks using a graphical editor provided by Cytoscape; and (ii) defining special interest networks based on network motifs, cycles, modules, shortest paths determined by existing Cytoscape plug-ins, such as SubgraphCreator. The Omics data integration module is based on our previously developed plug-in, OmicsViz (Xia and Dickerson, 2008). Data integration is performed in two ways. First, omics datasets can be linked using a common naming scheme between the data file and names defined in a user-selectable network attribute. Second, the user can provide a mapping file of homologous or orthologous terms to allow integration of omics datasets from different biotechnology platforms with networks and mapping of omics datasets across species (for detailed mapping rules, see Xia and Dickerson, 2008). The module can also map multiple omics datasets to multiple networks by selecting a dataset in under the data integration tab while viewing a network. The integration allows users to compare omics data across related networks to gain insights into biological dynamics and function.
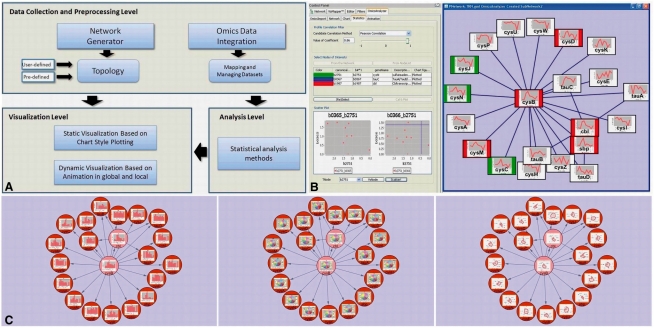
Fig. 1.
(A) Workflow of OmicsAnalyzer. It consists of three levels, the data collection and preprocessing level, visualization level and analysis level. Other four pictures show an example of OmicsAnalyzer functionalities. (B) In a subnetwork featured by a hub gene cysB of an E.Coli regulatory network, three nodes are selected as reference nodes (green, blue and red), their correlated node groups with respect to imported transcriptomics dataset are found by Pearson's correlation calculation and highlighted in red and green, respectively. Scatterplots for two pairs of genes are also shown. (C) The subnetwork and the associated microarray dataset are visualized in three different chart-based new network views.
At the statistical level, OmicsAnalyzer offers a variety of statistical analysis methods for imported omics datasets that are mapped onto biochemical networks. Users can visually mine one or more datasets integrated on a single network for relationships or other patterns using our node scatter plots. It also provides a user-defined correlation coefficient (Pearson's, Kendall and Spearman's rank) selection criterion, which highlights and clusters nodes meeting the criterion with respect to a reference node. The resulting clusters are visually differentiated using colors automatically assigned to corresponding reference nodes. For interactive analysis, when one reference node is selected, its group (correlated nodes) is highlighted.
The visualization level provides different methods to display data and network. OmicsAnalyzer uses node view components to describe data. For integrated visualization of multiple omics datasets associated with one node, it produces several types of chart pictures using the JFreeChart library (Gilbert and Morgner, 2007). Users can switch between line, bar, spider web and pie chart styles to examine the omics datasets simultaneously. To visualize the dynamics of time series datasets, OmicsAnalyzer provides local and global animation functions. In local mode, users can focus on specific set of nodes where a line chart animation is shown in a panel. In global mode, it uses node color to map measurement values at one time or experimental condition. Users can control the play of animation and investigate the entire network dynamics in omics datasets. Multiple datasets can be read in at one time and the user can select which ones they would like see in a scrollable list.
3 USAGE EXAMPLE
A brief usage example is shown Figure 1, which uses an Escherichia coli K-12 regulatory pathway as an input network (Covert et al., 2004; Fig. 1 in Samal and Jain, 2008) and omics data (provided by Ishii et al., 2007) as imported data. In the network, a gene, cysB, serves as a hub gene with high connectivity. Here, OmicsAnalyzer created a subnetwork module which contains nodes with direct links to cysB. In the subnetwork, three reference nodes are picked out to find the corresponding correlated group. After setting Pearson's correlation method and 0.86 as coefficient threshold, the resulting two groups are visualized in the network panel. Each group is highlighted colorfully according to background color of their reference node (Fig. 1B). For specific nodes of interest, OmicsAnalyzer offers scatterplots to examine hidden relationships in the datasets. At the lower left corner of Fig. 1B, two genes tauA (b0365) and tauB (b0366), which might be related to gene cysN (b2751) in the dataset are selected to apply the scatter plotting. User can easily see the expression profile of gene tauB is more related to gene cysN than tauA. More details about the functionalities and a tutorial of OmicsAnalyzer are available online. Figure 1C shows visualizations of the associated transcriptomics data in three chart styles in network context.
ACKNOWLEDGEMENTS
We would like to thank the Cytoscape development team and Jie Li, Pan Du, Wengang Zhou and Narasimha Rao for their technical support and help, and also Jennifer L. Reed and Areejit Samal, for providing the E.coli metabolic network.
Funding: National Science Foundation (grants IOS-0500461, DBI-0543441, EEC-0813570 and IIS-0612240).
Conflict of Interest: none declared.
REFERENCES
- Covert MW, et al. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- Gilbert D, Morgner T. JFreeChart, a free java class library for generating charts. 2007 Available at: http://www.jfree.org/ (last accessed date 2009) [Google Scholar]
- Ishii N, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- Junker BH, et al. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal A, Jain S. The regulatory network of E. coli metabolism as a Boolean dynamical system exhibits both homeostasis and flexibility of response. BMC Syst. Biol. 2008;2:21. doi: 10.1186/1752-0509-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Dickerson JA. OmicsViz: Cytoscape plug-in for visualizing omics data across species. Bioinformatics. 2008;24:2557–2558. doi: 10.1093/bioinformatics/btn473. [DOI] [PMC free article] [PubMed] [Google Scholar]