Abstract
Imaging has become an indispensable tool in the study of cancer biology and in clinical prognosis and treatment. The rapid advances in high resolution fluorescent imaging at single cell level and MR/PET/CT image registration, combined with new molecular probes of cell types and metabolic states, will allow the physical scales imaged by each to be bridged. This holds the promise of translation of basic science insights at the single cell level to clinical application. In this article, we describe the recent advances in imaging at the macro- and micro-scale and how these advances are synergistic with new imaging agents, reporters, and labeling schemes. Examples of new insights derived from the different scales of imaging and relevant probes are discussed in the context of cancer progression and metastasis.
Multi-photon microscopy and other techniques can be used to monitor the behavior and fate of tumor cells in vivo. This can be correlated with gene expression profiles to define “invasion signatures” and new prognostic markers.
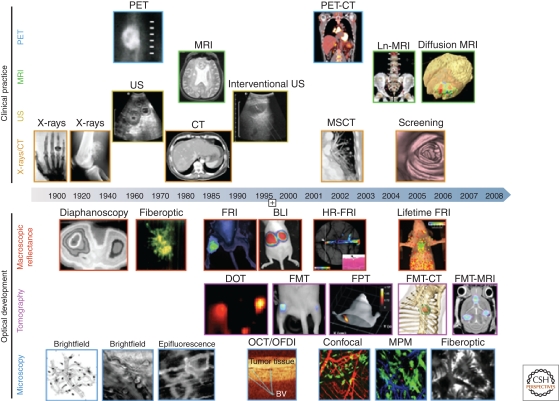
Imaging has become an indispensable tool in cancer research, clinical trials and medical practice. The last three decades have seen an explosive growth in the number and applications of different imaging technologies (Fig. 1). Imaging systems can be grouped by the energy used to derive visual information (X-rays, positrons, photons, sound waves), the spatial resolution attained (macro-, meso-, microscopic), or the type of information obtained (anatomic, physiological, molecular/cellular). Macroscopic imaging systems providing anatomic and physiological information are now in widespread clinical and preclinical use (computed tomography, CT; magnetic resonance imaging, MRI; ultrasound, US), while molecular imaging systems are either in clinical (positron emission tomography, PET; single-photon emission computed tomography, SPECT) or experimental use (fluorescence reflectance imaging, FRI; fluorescence-mediated tomography, FMT; bioluminescence imaging, BLI; laser-scanning confocal microscopy, LSCM; multiphoton microscopy, MPM). Ultimately, it is hoped that some of the molecular imaging systems will allow clinicians to not only see where a tumor is located in the body, but also to visualize the expression and activity of specific molecules (e.g., receptors, protein kinases, proteases), cells (e.g., T cells, macrophages, stem cells), and biological processes (e.g., apoptosis, angiogenesis, metastasis) that influence tumor behavior and/or responsiveness to therapeutic drugs.
Figure 1.
Imaging technologies used in oncology. Several macroscopic imaging technologies (above date line) are in routine clinical use and have advanced tremendously in their capabilities to obtain anatomic and functional information. Microscopic and other intravital optical techniques (below date line) have evolved over the last decade and now allow experimental studies of genetic, molecular, and cellular events in vivo (reproduced with permission from Nature).
Perhaps the biggest growth area is fluorescence imaging, with different microscopic and macroscopic technologies being adapted to in vivo use. Indeed, we are on the verge of being able to address some big questions in molecular oncology: How does the molecular machinery of signaling pathways interact in real time; what are the kinetics and flux rates of such networks; what are the differences between networks in malignant cells and normal tissues; can we exploit differences to make less toxic and more efficacious drugs; what are the “hubs” that will translate into most efficient read-outs of cancer development and therapeutic efficacy; and what is the spatial and temporal extent of tumor microenvironments that cause metastasis? In this article, we highlight applications of imaging technologies for breast cancer. Some recent review articles provide more in-depth information on clinical imaging technologies (Neves and Brindle 2006; Torigian et al. 2007) and cellular nanoimaging (Deisseroth et al. 2006; Soon et al. 2007).
MACROSCOPIC IMAGING TECHNOLOGIES
Table 1 summarizes the type, spatial resolution, depth penetration, imaging time, and cost of currently available common systems. MRI, PET, SPECT, and CT are useful for routine clinical practice and for testing therapeutic drug efficacy in therapeutic trials. Adaptations of these systems at much higher spatial resolutions have become available for use in experimental mouse models and have enabled the development of newer translational imaging probes. Because each individual technology has its unique strengths and limitations, hybrid imaging platforms such as PET-CT, FMT-CT, FMT-MRI, and PET-MRI are being developed to improve data reconstruction and visualization.
Table 1.
Overview of imaging systems
| Technique | Resolution | Depth | Time | Quantitation | Multi channel imaging | Imaging agents | Target | Cost | Primary small animal use | Clinical use |
|---|---|---|---|---|---|---|---|---|---|---|
| MR imaging | 10–100 µm | No limit | Min–hours | Absolute | No1 | Paramagnetic chelates, magnetic particles | A, P, M | $$$ | Versatile imaging modality with high soft tissue contrast | Yes |
| CT imaging | 50 µm | No limit | Min | Absolute | No | Iodine | A, P, M2 | $$ | Primarily for vascular, lung, and bone imaging | Yes |
| Ultrasound imaging | 50 µm | Cm | Sec–min | Absolute | No | Microbubbles | A, P, M2 | $$ | Vascular and interventional imaging | Yes |
| PET imaging | 1–2 mm | No limit | Min–hours | Absolute | No | F-18, Cu-64, C-11 and Ga-68 labeled compounds | P, M | $$$ | Versatile imaging modality with many different tracers | Yes |
| SPECT imaging | 1–2 mm | No limit | Min–hours | Absolute | Two | Tc-99m, In-111, I-131-labeled compounds, Ga-67, Tl-201 | P, M | $$ | Commonly used to image labeled antibodies, peptides, or perfusion, etc. | Yes |
| Fluorescence reflectance imaging (FRI) | 1 mm | <1 cm | Sec–min | Relative | Multiple | Photoproteins, fluorochromes | P, M | $ | Rapid screening of molecular events in surface-based disease | Yes |
| Fluorescence mediated tomography (FMT) | 1 mm | 2–3 cm | Mins | Absolute | Multiple | Near infrared, fluorochromes | P, M | $$ | Quantitative imaging of targeted or “smart” fluorochrome reporters | In development |
| Bioluminescence imaging | Several mm–cm | cm | Sec–min | Relative | Multiple | Luciferins, coelenterazines, luminol | M | $$ | Gene expression, cell and bacterial tracking, protein processing, and MPO activity | Potentially in development |
| Intravital microscopy (e.g., confocal, multiphoton) | 1 µm | <400–800 µm | Sec–hours | Relative | Multiple | Photoproteins, fluorochromes | A, P, M | $$$ | All of the above at higher resolutions but at limited depths and coverage | In development (endoscopy, skin) |
The Resolution and Cost columns refer to high-resolution, small animal imaging systems and are different for clinical imaging systems. Quantitation: “absolute” and “relative” refer to techniques that generate signals that are depth independent and dependent, respectively. “Relative” quantitation techniques typically require extensive controls; however, some of them (e.g., multiphoton microscopy, MPM) can be used to derive truly quantitative parameters (e.g., cell velocity, interaction time). Target: area(s) that a given imaging modality interrogates; (A) anatomic, (P) physiologic, (M) molecular. Cost of system: ($) <100 K, ($$)100–300 K, ($$$) >300 K.
1Dual channel imaging has been described. 2A limited number of molecularly targeted agents have been described. (Adapted from Rudin and Weissleder, 2003, Nat Rev Drug Discov, 2, 123–31.)
Magnetic resonance imaging (MRI) is a noninvasive, tomographic imaging modality that offers exquisite soft tissue contrast. MRI is based on the manipulation of the inherent nuclear magnetic moment of endogenous nuclei (most commonly 1H in H2O). Images are obtained by exposing nuclei to a static magnetic field, and within that static field, perturbing a steady-state equilibrium with time and space varying magnetic fields. After perturbation, all nuclei relax by two unique and co-dependent relaxation mechanisms: T1 (spin-lattice relaxation) and T2 (spin–spin relaxation). Recent advances in MRI equipment (higher field strengths, optimized pulse sequences, and better coil design, especially for the breast) have made this modality a procedure of choice for imaging many cancers. Coupled with the use of small molecule paramagnetic agents and magnetic nanoparticles, different tumor processes can now be probed. Imaging of angiogenesis, apoptosis, and specific targeting are all within the realm of experimental imaging. Dynamic contrast enhanced MRI (DCE-MRI) is being increasingly used clinically to detect breast cancers in dense or postoperative breast tissue, for invasive breast cancers, for screening in patients with a >20% greater lifetime risk of developing breast cancer (Saslow et al. 2007), for relapse monitoring, to detect metastatic disease, and for therapy evaluation (Costelloe et al. 2009; Lehman et al. 2009).
Positron emission tomography (PET) imaging detects positrons originating from the decay of systemically administered radiotracers containing 18F, 11C, or 13N. A considerable number of cancer-relevant metabolic substrates, drugs, and antibodies have thus been labeled (Kumar et al. 2008; Miller et al. 2008; Dunphy and Lewis 2009). Clinically, one of the most successful tracers is fluorodeoxyglucose (18FDG), and 18FDG PET-CT is now rapidly becoming the key clinical tool for the staging and assessment of cancer recurrence (Ben-Haim and Ell 2009). PET-CT has also gained widespread acceptance as an important tool for demonstrating early responses to intervention and therapy. In contrast to normal differentiated cells, which rely primarily on mitochondrial oxidative phosphorylation to generate the energy needed for cellular processes, many cancer cells switch to aerobic glycolysis, have increased metabolic demands, and internalize 18FDG. Metabolic pathways active in proliferating cells are directly controlled by signaling pathways involving known oncogenes and tumor suppressor genes. Our current understanding of how glycolysis, oxidative phosphorylation, the pentose phosphate pathway, and amino acid metabolism are interconnected in proliferating cells is slowly emerging (Vander Heiden et al. 2009). Small molecules that disrupt PI3K signaling lead to decreased glucose uptake by tumors as measured by 18FDG-PET, and the ability to inhibit tumor 18FDG uptake correlates with tumor regression (Engelman et al. 2008). The primary use of 18FDG PET-CT imaging in breast cancer today is thus for cancer staging (Ben-Haim and Ell 2009; Foster et al. 2009) and to monitor treatment response in clinical trials (Ellis et al. 2009).
Fluorescence imaging systems rely on photographic principles to collect low-light level images. Tomographic fluorescence systems (fluorescence molecular tomography, FMT) are quantitative and use transillumination to reconstruct 3D maps of fluorochromes based on sophisticated reconstruction algorithms (Ntziachristos et al. 2005). FMT is often combined with CT or MRI for improved photon reconstruction and image visualization (Grimm et al. 2005; McCann et al. 2009; Nahrendorf et al. 2009). A considerable number of near infrared-emitting fluorochromes and imaging agents have been developed for FMT imaging (Table 2) (Jaffer et al. 2009). Tomographic fluorescence systems have also been adapted to reconstruct fluorescent proteins (fluorescence protein tomography, FPT) (Zacharakis et al. 2005). Newer FPT systems can image model organisms at mesoscopic resolution (Vinegoni et al. 2008). In breast cancer research, the primary use of FMT systems is for therapy evaluation, tumor/metastasis detection, imaging of microcalcifications or proteases, and hybrid molecular/anatomic imaging (Bremer et al. 2005; Montet et al. 2005; Bhushan et al. 2008). Fluorescence reflectance imaging (FRI) systems consist of an excitation source, filters, and a charge coupled device (CCD) camera to obtain planar images. They are useful for imaging events in surface tumors (xenografts), surgically exposed organs, or for intra-operative imaging, but have limited depth resolution beyond 3–5 mm from the surface and, unlike FMT, are not quantitative.
Table 2.
Overview of agents
| Class | Subtype | Examples | Substrate | Primary use | Ex/Em | Turn-on/shift | Clinical | Modality | Limitations |
|---|---|---|---|---|---|---|---|---|---|
| Genetic tags | GFP | eGFP | NA | Fusion, reporter | 484/507 | Yes | No | IVM | Limited to proteins; no human use |
| RFP | mCherry | NA | Fusion, reporter | 587/610 | Yes | No | IVM | lower QY compared to GFP | |
| mRaspberry | NA | Fusion, reporter | 598/625 | Yes | No | IVM, FMT | lower QY compared to GFP | ||
| Katushka | NA | Fusion, reporter | 588/635 | Yes | No | IVM, FMT | lower QY compared to GFP | ||
| IFP | Biliverdin | Fusion, reporter | 684/708 | No | IVM, FMT | Emerging | |||
| Photo- conversion | Dendra | NA | Cytoplasmic, | 490/507 green | No | IVM | No human use | ||
| Cell position | 553/573 red | ||||||||
| Luciferases | Firefly | Luciferin | Reporter | broad em | Yes | No | BLI | Limited to proteins; no human use | |
| Renilla | Coelenterazine | Reporter | broad em | Yes | No | BLI | Limited to proteins; no human use | ||
| Gausia | Coelenterazine | Reporter | broad em | Yes | No | BLI | Limited to proteins; no human use | ||
| Antibodies | IgG | EGFR | NA | Cell surface marker | TD | Possible | Yes | PET, IVM, FMT | Delivery barriers; limited efficacy in vivo |
| Her2/neu | NA | Cell surface marker | TD | Possible | Yes | PET, IVM, FMT | Delivery barriers; limited efficacy in vivo | ||
| Fragments | Fab | NA | Cell surface marker | TD | Possible | Yes | PET, IVM, FMT | Delivery barriers; kinetics | |
| Diabody | NA | Cell surface marker | TD | Possible | Yes | PET, IVM, FMT | Delivery barriers; kinetics | ||
| Minibody | NA | Cell surface marker | TD | Possible | Yes | PET, IVM, FMT | Delivery barriers; kinetics | ||
| Small molecules | Metabolites | FDG, FLT, acetate | NA | Metabolism | TD | No | Yes | PET | Limited number agents |
| Steroids | NA | Reporter | TD | No | Yes | PET | Limited number agents | ||
| Hyperpolarized Molecules | NA | Metabolism | — | — | Yes | MRI | Time limitation | ||
| Drugs | Taxol-NBD | NA | Intracellular targets | TD | Possible | No | PET, IVM, FMT | Limited number of high affinity agents | |
| Prodrugs | Cathepsins, luminol | NA | Enzymes | TD | Yes | Yes | FMT, BLI | Limited number of high affinity agents | |
| Peptides | RGD, collagen | NA | Receptors | TD | No | Yes | PET, IVM, FMT | Limited number of high affinity agents | |
| Tags | CFSE, VT680 | NA | Cell tags | TD | Yes | No | IVM, FMT | Ex vivo labeling | |
| Cy dyes | NA | Generic labeling | TD | Yes | Planned | IVM, FMT | |||
| Environment | pH | NA | Hypoxia, endosomes | TD | Yes | No | IVM, FMT | Limited number of high affinity agents | |
| Nanotechnology | Q-dot | CdSe | NA | Cell surface marker | TD | Yes | No | IVM, FMT | Possible delivery barriers |
| Magnetic | MION | NA | Macrophages | TD | DMR | Yes | MRI | Possible delivery barriers | |
| CLIO | NA | Cell surface marker | TD | DMR | No | MRI | Experimental use only | ||
| Ferumoxytol | NA | Macrophages | TD | DMR | Yes | MRI | Possible delivery barriers | ||
| Polymers | Prosense | NA | Proteasses | TD | Yes | Planned | FMT | Possible delivery barriers | |
| Angiosense | NA | Vascualrity | TD | No | No | FMT | Experimental use only | ||
| Bionano | Phage | NA | Cell surface marker | TD | No | No | FMT, MRI | Experimental use only | |
| Bioorthogonal | Staudinger | triarylphosphine | Azide | Proteins, carbohydrate | TD | No | No | IVM | Limited in vivo experience |
| 3,2 click | alkyne | Azide | Proteins | TD | No | No | FMT, IVM, PET | Limited in vivo experience; reaction slow | |
| 4,6 Click | Norbornene | Tetrazine | Many targets | TD | No | No | FMT, IVM, PET | Emerging | |
| TCO | Tetrazine | All targets | TD | Yes | Planned | FMT, IVM, PET | Emerging |
TD = depends on attached fluorochrome (VT, Alexa, Cy, etc.) or other tracer (18F, 11C).
Bioluminescence imaging (BLI) has emerged as a useful experimental imaging technique for small animals. The imaging signal depends on expression levels of a luciferase, the presence of an exogenously administered substrate (a luciferin), ATP, O2, and depth. Numerous luciferase/luciferin pairs have been harnessed for in vivo imaging: firefly/luciferin pairings, in particular, have been used on account of their high wavelength and quantum yields; Renilla reniformis/coelenterazine and Gaussia luciferase/coelenterazine have also been used because of their flash kinetics and ability to generate photons outside cells (Table 2). In these experiments, luciferin (>100 mg/kg i.p) is injected immediately prior to data acquisition. Unlike fluorescence techniques, there is no inherent background with bioluminescence, which makes this technique highly sensitive. However, the method currently does not allow absolute quantification of target signal. Rather, BLI's primary uses are in binary mode (yes/no luciferase expression) or as an imaging tool to follow the same animal under identical conditions (including positioning) to provide semiquantitative biological data (Gross and Piwnica-Worms 2005a). BLI is primarily used in experimental breast cancer research for imaging gene expression, therapy evaluation (Kalra et al. 2009; Shah et al. 2008; Szafran et al. 2009; Viola et al. 2008; Wang et al. 2009), or for imaging peroxidase activity in vivo (Chen et al. 2004; Gross et al. 2009). Bioluminescence microscopy is a recently emerging technology and is now gaining increasing interest among cell biologists for its ability to image cells under low light, eliminating the need for fluorescent molecules (Kammerloher 2008).
MICROSCOPIC IMAGING TECHNIQUES
Types
Several microscopic imaging approaches, previously established for cell imaging, have recently been adapted to in vivo imaging. Confocal microscopy uses a pinhole in front of a detector to collect photons coming from in-focus points within the sample. Confocal microscopy setups have become more widespread as they are user-friendly and less expensive. However, penetration depths are typically limited to <100 µm. Multiphoton microscopy (MPM) imaging systems typically achieve depth resolutions of 500–800 µm, yield tomographic z-stacks in multiple channels (typically 3–5), and are increasingly being used for intravital imaging (Wang et al. 2002; Halin et al. 2005; Wyckoff et al. 2009). With the advancements in multiphoton microscopy and the development of both fluorescent xenograft and transgenic tumor models in mice, our understanding of the behavior of tumor cells has been greatly enhanced (Condeelis and Segall 2003; Sahai 2007). Due to the ability of the multiphoton microscope to penetrate deep into solid tissue (Condeelis and Segall 2003; Andresen et al. 2009) and provide single cell resolution, tumor progression during metastasis can now be followed at the single cell level (Wang et al. 2002, 2007a; Sahai et al. 2005). The ability to image in multiple colors allows the simultaneous imaging of stromal and tumor cells, which has led to a better understanding of the role of the microenvironment in metastasis (Sahai et al. 2005; Wyckoff et al. 2007).
Techniques and Mammary Models
For imaging of mammary glands and tumors, the most common technique for preparing an optical path is called the “skin-flap” method. This technique involves exposing the tissue directly to the microscope objective by making a small incision, and folding the skin back to remove the stratum germanitivum portion of the skin from the optical light path to minimize light scattering (Jain et al. 2002; Condeelis and Segall 2003; Hoffman 2005). This method normally allows for up to 6 hours of imaging in one animal, requiring the compilation of data from several animals to complete the analysis. Through hydration, physiologic monitoring, and careful control of temperature, imaging times of up to 24 hours can be achieved (Egeblad et al. 2008). To extend the duration of imaging in a single animal over multiple days or weeks, a reversible skin flap (Yang et al. 2002), which is repeatedly opened and closed, has been used. However, this practice runs the risk of inducing an inflammatory response in the tissue being imaged. Another approach to extend duration is to use a dorsal skinfold chamber (Lehr et al. 1993) in which the tumor is grown in the space between the skin and glass coverslip on the back of the mouse. This preparation is convenient for imaging over extended times (Jain et al. 2002). However, a significant disadvantage is that most tumors must be grown ectopically and tumor size and shape is constrained by the size of the chamber, which may produce artifacts in the tumor microenvironment.
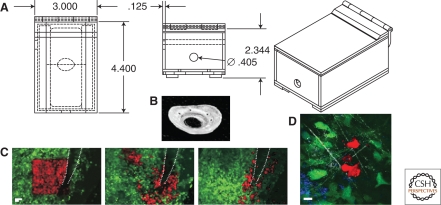
Recently, several new techniques have been described that extend the duration of intravital imaging of the same mammary tissue from hours to days without the above disadvantages (Gligorijevic et al. 2009). These new approaches offer information on cell–cell and cell–extracellular matrix interactions, as well as fate mapping of individual tumor cells and cell populations. An extension of this is the recently developed combined approach using a mammary imaging window and photo-conversion in vivo to visualize and quantify invasion and intravasation in orthotopic mammary tumors (Fig. 2) (Kedrin et al. 2008). The Mammary Imaging Window (MIW) allows imaging of the same tumor over multiple imaging sessions spanning multiple days. Positioning of the animal and maintaining the orientation over several sessions is achieved via a stereotactic imaging box (Fig. 2A). The MIW (Fig. 2B) is sutured into the skin over the tumor tissue, preventing the tissue from drying or becoming infected and allowing imaging of the tumor at the orthotopic site without constraining tumor size and shape due to its soft material.
Figure 2.
Combination of MIW and photoconversion allows monitoring of the behavior of tumor cells in specific microenvironments. (A) An imaging box allows for reproducible animal positioning and environmental control (bottom, side, and full views). A mouse is placed in the box and the MIW is secured in place between two sliding doors on the bottom of the box. Anesthesia flow is established. (B) Photograph of the form fitting MIW. The MIW is cast in silicon molds from polyester resin. Scale bar, 4 mm. (C) Tumor cell behavior is controlled by the microenvironment. Images are shown of an area photoconverted next to a major blood vessel (white dotted line) taken at 0 h (left panel), 6 h (middle), and 24 h (right panel) after photoconversion. Photoconverted tumor cells disappear or move closer to the blood vessel at 24 h and appear in the lung (not shown). Scale bar, 50 µm (see Kedrin et al. 2008). (D) Four channel multiphoton imaging allows the visualization of interactions between macrophages and tumor cells in specific microenvironments. Images were taken after photoconversion of Dendra2 tumor cells. Channels collected: collagen (white, SHG), tumor cells (green and red, Dendra2), and macrophages (blue, AlexaFluor647 dextran 10 K). Scale bar, 10 µm. Figures courtesy of Bojana Gligorijevic of the Gruss-Lipper Biophotonics Center.
New Multiphoton Microscope Designs
In order to visualize many of the new proteins and injectable probes, it will be necessary to develop new “broad band” multiphoton platforms. In general, the far red probes cannot be imaged using currently available commercial systems. Several of these far red probes are discussed to illustrate the technical design issues in dealing with their excitation and emission.
For example, the excitation peaks of mKate2 and mCherry are at 588 nm, which suggests a maximal multiphoton cross section at ∼1176 nm, while tdTomato peaks at 554 nm (suggests a maximal multiphoton cross section ∼1108 nm). The 2-fold greater transmission depth of excitation light in these multi-photon wavelength ranges makes these probes of value for deep tissue imaging. To cover the tuning range from 1100–1600 nm, various investigators (Kao 2004; Andresen et al. 2009; Vadakkan et al. 2009; Wyckoff et al. 2009) use a Ti:Sapphire pumped Optical Parametric Oscillator (OPO) as an extension of the typical Ti-Sapphire laser light source used in multiphoton microscopes (Pelouch et al. 1992). The gap between the Ti-Sapphire and OPO light sources, which spans 1050–1100 nm, can be eliminated through use of the second harmonic of the OPO idler beam (Makarov et al. 2008). Considering the high cost of femtosecond lasers and the large amount of power they generate, a sensible design is to split the light path from the Ti:Sapphire laser to use 90% of the laser power for pumping the OPO and the other 10% for the excitation of lower wavelengths.
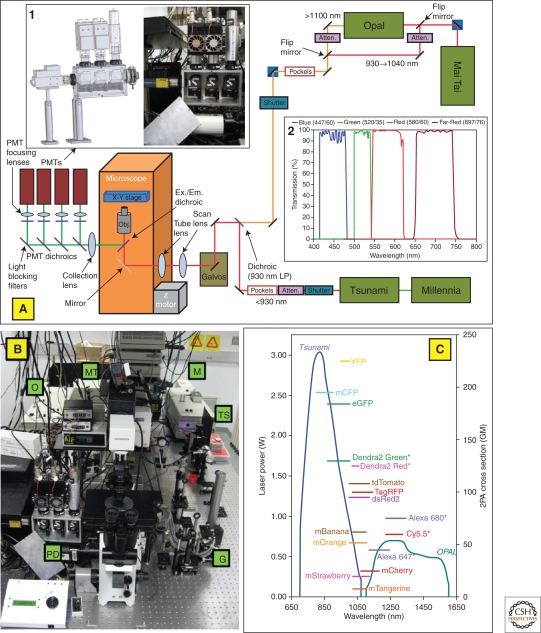
However, if the goal is to image rapid events such as cell motility in multiple colors, two separate laser systems are required to simultaneously excite fluorophores in the range of 650–1600 nm. This is the approach taken by the Condeelis laboratory as illustrated in Figure 3 (Wyckoff et al. 2009). Because this system utilizes two separate laser light sources, it does not suffer from the same restrictions of tunability and intensity control found in systems that split light from one femtosecond source both for pumping an OPO and illuminating a sample.
Figure 3.
Custom-built two-laser multiphoton microscope (TLMPM) provides excitation at 650–1040 nm and 1100–1600 nm and collection in four distinct channels. Optical Layout of TLMPM. Wavelengths <950 nm are excited by the Tsunami laser and 960–1040 nm by the Mai-Tai laser. For excitation at 1100–1600 nm, a flip mirror in the light path switches the Mai-Tai from being an illumination source to a pump beam for the OPO. Fluorescence is collected in one of four PMT detectors. Figures courtesy of David Entenberg of the Gruss-Lipper Biophotonics Center.
The use of the OPO-based extension of wavelengths makes room for the use of future far-red fluorescent proteins and other probes, excited from 1100–1600 nm (∼700–800 nm in the single photon microscopes). As previously demonstrated, red-shifted probes are particularly useful in overcoming green autofluorescence and light scattering in tissues (Muller-Taubenberger and Anderson 2007).
Support Software for Multiphoton 4D Imaging
Until recently, most image processing tools available were not optimized for intravital or 4D imaging. They were commonly designed for two-dimensional images or time-lapse sequences without measurement of axial movements or ability to follow movement of the same cell throughout a Z-stack. Image processing software design is complicated by difficulty in the display of 4D (3D information over time) images as well as difficulty in analysis of 3D data due to decreased resolution in the z versus the x and y dimensions. Also, results of intravital imaging often include slight drifts in one or more of the x, y, z dimensions as well as irregular animal breathing artifacts. As the number of groups imaging in vivo or in vitro 3D cultures is growing, solving these issues is becoming increasingly important and initial steps have been made. For example, to follow the trafficking of T-cells, Miller et al. (2003) have developed a color-coded scheme based on different axial positions, and analyzed cell movement in lateral versus axial directions. More recent examples of immune cell motility analyses (Mempel et al. 2004; Peters et al. 2008) use the commercially available Imaris or Volocity packages combined with custom Matlab scripts to analyze motility in 4D. However, all reported techniques are based on the ability of software to automatically segment and threshold images based on fluorescence intensity. Unfortunately, this is a task that cannot be fully automated for cells in solid tissues such as tumors where the cells are packed close to each other in three dimensions. These reasons make automated segmentation and thresholding of single cells in tumors an extremely challenging task.
To this end, there has been an effort to develop software that facilitates the tracking of tumor cells in three dimensions and over time (Wyckoff et al. 2009). To fully leverage existing capabilities and minimize development time, this software was developed as a plug-in for the open-source image processing package, ImageJ (Abramoff et al. 2004). The plug-in is designed to allow users to easily outline and track the location of cellular features as the features progress throughout time and change in depth within the 4 dimension data. The marking of these features can be done with a variety of types of selection tools, including point, polygon, or freehand selections. Hence, a particular cellular feature is followed and marked throughout time and depth of the image set, to form a track. Multiple features may be followed in a single set of images by creating a multitude of tracks. In addition, if reference points can be identified within the 4D data, their locations may serve as the basis of a reference track that may be used to compensate overall x,y,z drift within the image set (Wyckoff et al. 2009).
IMAGING AGENTS, REPORTERS, AND LABELING
Normal and cancer cells cannot be easily distinguished by in vivo imaging with few exceptions. Consequently, molecules and cells of interest need to be labeled to become visible by imaging. There are three principle strategies: (1) encoding with genetic reporters (e.g., photoproteins or PET and MR detectable reporter genes), (2) radiotracer, fluorochrome, or magnetically tagged affinity molecules (e.g., labeled antibodies or small molecules), and (3) biorthogonal reporter strategies. Table 2 summarizes the different labeling techniques, types of affinity molecules, primary use, and some of the limitations of each.
Genetic reporter strategies using fluorescent (green and red-shifted fluorescent proteins), photoconvertible fluorescent proteins, bioluminescent (Firefly, Click beetle, Renilla, and Gaussia, and other luciferases), or other fusion proteins (Herpes simplex virus-1 thymidine kinase, transferin receptor, somatostatin receptor type 2, dopamine receptor type 2, human Na/I symporter) are largely limited to mouse models of cancer but have enjoyed widespread use in basic sciences (Gross and Piwnica-Worms 2005b; Giepmans et al. 2006). Optical reporter genes are cloned into promoter/enhancer regions or engineered as fusion proteins and thus allow longitudinal studies of biological processes. Such reporters are particularly valuable in genetically engineered mouse models with temporal and tissue-specific control of activation of oncogenes and/or tumor-suppressors, because they permit the study of oncogenic transformation and to assess drug action, resistance, and toxicity (Sharpless and Depinho 2006; Ventura et al. 2007). In recent years, there has been an increasing number of new and improved fluorescent proteins with higher brightness and red-shifted wavelengths, making them desirable probes for MPM and FMT imaging (Drobizhev et al. 2009). Fluorescent proteins in the orange and red part of the visible spectra (Shaner et al. 2004) have been improved (Shaner et al. 2008) and were followed by development of far-red emitting tdKatushka, mKate, and mKate2 (Shcherbo et al. 2007, 2009). Most of these proteins can be photoconverted using single- or multiphoton illumination (Kremers et al. 2009), which allows for additional applications for fate mapping of single cells and populations in vivo, and for long duration imaging experiments by optical microscopy (Kedrin et al. 2008).
Injectable imaging agents with specificity for molecular targets have the advantage of being useable translationally from mouse to man but currently exist for <5% of cancer targets of interest. This is largely because in vivo imaging agents face much more stringent design criteria compared to in vitro reagents. The basic underlying difficulty is to design agents with high target-to-background ratios in vivo. Ideal agents should contribute minimally to background signal (e.g., nonspecific tissue extravasation, internalization into macrophages, renal or hepatic elimination obscuring adjacent organs, in vivo delivery barriers), but should yield high local concentrations at intended molecular target sites. For these reasons, current in vivo agent designs often harness amplification strategies using chemical (multivalency, quenching, covalent target binding, magnetic relaxation switching, uncaging, relaxivity changes, FRET) or biological means (cellular trapping, enzymatic conversion, pre-targeting). All of the injectable imaging agents rely on reporter molecules such as fluorochromes (indocyanines, quantum dots), radiotracers (18F, 11C, 111In, 99mTc), or magnetic labels (Gd-chleates, magnetic nanoparticles, hyperpolarized molecules) to be detected. Hyperpolarization is a new technique that is being increasingly employed to improve the signal-to-noise ratio in MR imaging and is showing promise for advancing the identification of biomarkers and metabolic alterations for a variety of diseases in real time (Golman et al. 2003).
Bioorthogonal ligation strategies for coupling materials in the presence of complex biological milieu are of great interest to imaging because of their selectivity, easier delivery, and timing insensitivities. In general, these strategies can result in highly selective labeling of proteins and can be combined with pre-targeting approaches. To date, only a few bioorthogonal reactions have been reported, the most popular being the Staudinger ligation and the {3+2} cycloaddition “click” reaction between azides and alkynes (Prescher et al. 2004). Several ring strained dienophile derivatives that do not require a copper catalyst and are much faster, have recently been reported for imaging and offer exciting new technical possibilities (Devaraj et al. 2008, 2009a,b).
APPLICATIONS IN CANCER BIOLOGY AND CLINICAL ONCOLOGY
The following sections review the role ascribed to cellular and molecular players in tumor progression based on in vivo studies, and highlight some of the main applications and recent advances in tumor imaging at single cell resolution. Several other review articles have covered the use of established clinical imaging technologies (Quon and Gambhir 2005; Neves and Brindle 2006; Torigian et al. 2007).
Studying Invasion and Metastasis by MPM
In general, to take full advantage of the capabilities of the multiphoton microscope it is necessary to have animal models with multiple fluorescently encoded cell types that are relevant to the biological problem of interest. For example, using MPM imaging, a paracrine loop between tumor cells and macrophages has been observed in mammary tumors in vivo (Wyckoff et al. 2004, 2007). This was possible because of the development of a MMTV-PyMT × MMTV-iCre/CAG-CAC-EGFP transgenic mouse in which only the tumor cells are fluorescently labeled (Ahmed et al. 2002). When imaged by MPM, following an intravenous injection of Texas red-labeled dextran, which the macrophages endocytose, it was found that tumor cell invasion and intravasation require the interaction between a tumor cell and a macrophage and that intravasation of tumor cells occurs at the site where a peri-vascular macrophage is located on a blood vessel (Wang et al. 2004; Wyckoff et al. 2004). By correlating MPM with expression profiling of live cells captured with the in vivo invasion assay described below (Wang et al. 2004; Wyckoff et al. 2004), the interaction between tumor cells and macrophages was shown to be mediated by a paracrine signaling loop.
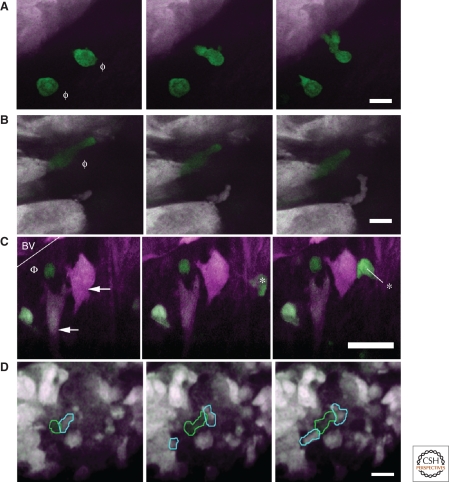
These results have been confirmed by the more recent development of two-color fluorescent mice with mammary tumors in which the genotype, MMTV-PyMT × MMTV-iCre/CAG-CAC-ECFP × c-fms-GFP, allows for imaging the interactions between CFP-labeled mammary tumor cells and GFP-labeled macrophages simultaneously and at single cell resolution (Fig. 4) (Wyckoff et al. 2009). This approach allows one to circumvent the possibility that injection of markers, such as Texas red, only marks a sub-population of cells (Wyckoff et al. 2007; Egeblad et al. 2008). In these 2-color tumors, macrophages are observed to move at speeds ranging from 1–8 µm/min and interact with tumor cells to initiate their migration (Fig. 4A,B). In addition, tumor cells and macrophages are observed to move toward each other creating cell groups (Fig. 4C) that migrate collectively (Fig. 4D). Such collective behavior is consistent with signaling between tumor cells and macrophages involving a paracrine loop (Wyckoff et al. 2004).
Figure 4.
Multiphoton imaging of mammary tumors in two color mice. The genotype of this mouse is MMTV-PyMT × MMTV-iCre/CAG-CAC-ECFP × c-fms-GFP where mice expressing CFP in tumor cells (white) and GFP in macrophages (green) allows the visualization of interactions between tumor cells and macrophages in living tumors. (A, B) Tumor cells interacting with macrophages in the invasion microenvironment. In the two sequences of still frames (left to right in time over 8 min) from two time lapse movies (A, B), interactions between tumor cells and macrophages (Ф) are observed during the macrophage-mediated initiation of tumor cell migration. Scale bars, A and B = 10 µm. (C) Tumor cells interact with peri-vascular macrophages in a relay pattern. In the first three panels (0, 4, 8 min) taken from a movie, a peri-vascular macrophage (Ф) adjacent to a blood vessel (line marks the wall of blood vessel [BV]) is attracting two tumor cells (arrows), which in turn attract another macrophage (* in second and third frame) all converging on the peri-vascular macrophage (pseudo coloring of tumor cells is purple; macrophages are green). (D) Tumor cells and their accompanying macrophages collectively migrate in single file in mammary tumors. In this sequence of still frames taken at 0, 7, and 14 min, a tumor cell (yellow outline) is followed closely by a macrophage (green outline), which in turn is followed by a tumor cell. This type of migration is believed to result from relay chemotaxis involving the EGF/CSF1 paracrine loop. Scale bars, C and D = 25 µm. Figure courtesy of Jeffrey Wyckoff, Gruss-Lipper Biophotonics Center.
A role for macrophages in the invasion of the mammary fat pad by the terminal end bud epithelial cells in normal developing mammary glands has been observed using MPM (Ingman et al. 2006). This interesting result suggests that tumor cell invasion and metastasis may share strategies for macrophage-mediated epithelial cell migration during normal morphogenesis. This insight begs for more extensive parallel analysis of normal morphogenesis of the mammary gland in comparison to mammary tumor cell behavior, to detect further similarities between morphogenesis and tumor invasion and metastasis.
These results illustrate how subpopulations of macrophages can be differentiated and their spatial relationships with tumor cells defined. In the future, activity probes and biosensors for both macrophage and tumor cell invasive activity (Nahrendorf et al. 2007; Blum et al. 2009; McIntyre and Matrisian 2009), such as markers for podosomes and invadopods, respectively, can be used to image invasive activity at the sites of invasion and intravasation (Oser et al. 2009; Sameni et al. 2009). Furthermore, genetically encoded probes for multiphoton imaging of additional cell types are in development by many laboratories, including all-cell fluorescent mice (Egeblad et al. 2008; Tran Cao et al. 2009; Yang et al. 2009) and mice with fluorescent dendritic cells, activated T-cells, and endothelial cells surrounding blood vessels (Motoike et al. 2000; Amoh et al. 2005; Egeblad et al. 2008). These new animal models will make it possible to explore the consequences of the interactions between tumor cells and stromal cells during tumor progression and between stromal cells and normal epithelial cells during normal morphogenesis in vivo.
Correlation of Tumor Cell Behavior In Vivo with Gene Expression and Clinical Markers of Metastatic Risk
A new technology is to correlate MPM with expression profiling of tumor cells within the microenvironments imaged to identify pathways and genes that code for the behaviors of tumor cells in vivo. For example, it was found that paracrine chemotaxis of tumor cells toward macrophages is essential for invasion in primary mammary tumors of mice while paracrine chemotaxis of tumor cells toward peri-vascular macrophages is essential for intravasation (Wyckoff et al. 2004, 2007; Goswami et al. 2005). The point at which tumor cells migrate through the endothelium of blood vessels was identified to be the site of docking on the blood vessel of at least one peri-vascular macrophage (Wyckoff et al. 2007).
The in vivo invasion assay (reviewed in Wyckoff et al. 2009) was developed to collect the tumor cells that were observed in the multiphoton microscope to migrate toward and intravasate into blood vessels (Wyckoff et al. 2004, 2007). The in vivo invasion assay has been coupled to expression profiling of small numbers of invasive cells to reveal the identities of the genes correlated with the survival, adjuvant-resistance, and chemotaxis to macrophage-supplied epidermal growth factor (EGF), for all phenotypes of these migratory tumor cells (Goswami et al. 2004; Wang et al. 2004, 2007a). These genes fall into well-defined pathways and are coordinately regulated in metastatic tumor cells (Condeelis et al. 2005; Wang et al. 2005, 2007a). These pathways are collectively called the Invasion Signature.
MPM has been used to test the importance, in invasion and metastasis, of the pathways identified in the Invasion Signature, including the ZBP1 (Z-DNA-binding protein 1 [Wang et al. 2004; Lapidus et al. 2007]), ROCK (Wyckoff et al. 2006), Mena (Pilippar et al. 2008), cofilin (Wang et al. 2006, 2007b), and EGF receptor (Xue et al. 2006; Kedrin et al. 2009) pathways. The results of these studies confirm that the motility pathways are synergistic to create tumor cells that have passed through epithelial-mesenchymal transformation and are capable of chemotaxis to EGF and penetration of basement membrane barriers using invadopodia (reviewed in Oser et al. 2009). They also led to a new insight that the motility pathways of invasive mammary tumor cells converge through the Mena pathway, making the protein Mena a potentially important marker for prognosis and therapy (Philippar et al. 2008; Goswami et al. 2009).
As described earlier, MPM had demonstrated that invasive carcinoma cells in mouse and rat mammary tumors intravasate when associated with peri-vascular macrophages. This thus serves to identify a tumor microenvironment of intravasation as an anatomical landmark in tumors (Wyckoff et al. 2007; Kedrin et al. 2008). Using this MPM-defined anatomical landmark composed of an invading carcinoma cell marked by Mena overexpression, and a peri-vascular macrophage as a guide, it was possible to define a triple stain marker for use in anatomic pathology with antibodies against Mena, macrophages, and endothelial cells to find the same landmark in human breast tumors (Robinson et al. 2009). In humans, this landmark for intravasation is called TMEM (Tumor Microenvironment of Metastasis) and its density is correlated with metastatic risk in breast cancer patients (Robinson et al. 2009). This work illustrates the power of combining multi-photon imaging at single cell resolution with mouse models of breast cancer to develop new insights into the mechanisms of human breast cancer metastasis and new prognostic markers for clinical use.
Fate Mapping of Tumor Cells at High Resolution In Vivo
Due to changes in tissue shape during growth and in the case of tumors, cell migration, a way of marking tumor cells, is essential for identifying the same cells in multiple imaging sessions, which may be separated by days. To address this, tumor cells have been used that express the photoconvertible protein Dendra2 throughout the cytoplasm, allowing them to be photoconverted from green to red using blue light (405 nm single photon or 810 nm 2-photon). This approach was used to quantify and compare metastatic behaviors of cells in different microenvironments within the same tumor (Kedrin et al. 2008). In Figure 2C, tumor cell behavior in the microenvironment surrounding a major blood vessel is shown at 0, 6, and 24 hours after photoconversion. The time window for visualizing a group of photoconverted cells in areas surrounding major blood vessels is limited here to 24 hours by the high cell motility and intravasation rates of the tumor cells when near a large blood vessel. However, in areas where only microvessels are present, cell invasion of the surrounding environment can be monitored for up to seven days due to the relative absence of intravasation (Gligorijevic et al. 2009). In addition, the composition of the microenvironment responsible for such dramatic events can be investigated by four channel MPM, using the microscope design described earlier (Fig. 3), of dendra-photoconverted tumor cells, extracellular matrix, and macrophages, as shown in Figure 2D.
In a different approach (Perentes et al. 2009), serial multiphoton imaging sessions were combined with the dorsal chamber model to study the mechanism of collagen fiber remodeling by tumor-associated fibroblasts. Collagen was utilized as an internal landmark to recognize and image the same tumor region in imaging sessions over a 9-day period. As mentioned, collagen fibrils are visible by second harmonic generation (SHG) imaging without any additional labeling. Based on the SGH signal from fibers, images were corrected for different animal orientations and tissue changes over serial imaging sessions and aligned based on similar bulk distribution of collagen fibers. Individual fibers were then analyzed for a decrease in length and an increase in overlap with fibroblasts.
In order to analyze cell motility of several subpopulations of myeloid cells in mammary tumors, Egeblad et al. (2008) developed an optimized version of the “skin-flap” technique, which allows for a single time-lapse imaging session extending for up to 27 hours by careful temperature, anesthesia, and hydration control. Use of a spinning disc confocal microscope allowed very fast image acquisition. Software was used to control the fully motorized stage, which collected images of several adjacent fields, resulting in the mosaic view of five combined fields in four-channels and three Z-sections at 45 frames/hour. However, this approach is limited to imaging of approximately 2 cell diameters in depth into the tissue. It is also not suitable for dimmer probes as only 1% of the excitation light reaches the sample (Nakano 2002).
Mitosis
A number of morphological cell markers such as chromatin-fluorescent fusion proteins (e.g., H2B-RFP) and fluorescent spindle proteins (e.g., tubulin-GFP, centrin-GFP) have been described, primarily for cellular imaging in vitro (Kapoor et al. 2000; Yarrow et al. 2003; Orth et al. 2008). More recently, however, some of these constructs have been used for in vivo imaging of mitosis and mitotic arrest following systemic administration of antimitotics such as taxol or kinesin-5 inhibitors. The major challenge for imaging these cancer-cell-associated traits has been physiological movement and the need to image over prolonged periods of time. Newer immobilization techniques, respiratory compensation algorithm, and motion suppression software as described earlier are facilitating these explorations.
Apoptosis and Therapeutic Response
Real-time imaging of apoptosis (cell death) is a coveted application to assess cytotoxic drug efficacy and to potentially monitor toxicity of these drugs on normal tissues. Successful new generation drugs will likely exploit differences (rather than similarities) in activating the apoptotic machinery between malignant and normal cells. The ability to image such effects in vivo could thus have far reaching implications for therapeutic efficacy and toxicity assessment. Strategies to image cell death include the use of recombinant photoprotein reporters activatable by caspase-3-cleavage (Bullok and Piwnica-Worms 2005) or as reporters of the mitochondrial outer membrane permeabilization (MOMP) (Spencer et al. 2009). Alternative approaches to image apoptosis at the single cell level include labeling of cells with fluorochromes in the cytoplasm and nucleus and monitoring of cytoplasm/nucleus fluorescence (Cf/Nf) ratio over time. Dead cells typically show decreased Cf/Nf ratio due to loss of cell membrane integrity and DNA fragmentation (Mempel et al. 2006). An alternative includes visualization of cell morphology as cell body condensation and membrane blebbing are characteristic of cells entering apoptosis. These approaches have been used successfully to elucidate in vivo dynamics of T lymphocyte-mediated killing and the mechanisms employed by suppressor T lymphocytes to blunt cytotoxic functions (Mempel et al. 2006; Mrass et al. 2006).
Macroscopic imaging technologies often rely on fluorescent (Ntziachristos et al. 2004) or radiolabeled (Blankenberg et al. 2006) ligands such as Annexin V with high affinity for apoptotic cells since the morphology of individual cells cannot be resolved. In the clinic, analysis of PET glucose metabolism (18FDG) is also progressively exploited as a surrogate for treatment efficacy (Engelman et al. 2008; Avril et al. 2005; Ben-Haim and Ell 2009; Vander Heiden et al. 2009).
Intraoperative Imaging
Complete surgical resection of neoplasia remains one of the most efficient tumor therapies. However, malignant cell clusters are often left behind during surgery due to the inability to visualize and differentiate them against host tissue. Given the significant advances in microscopy and probe development detailed earlier, strategies are now being developed to harness some of these for clinical “intraoperative imaging” at cellular resolution levels. Emerging applications include better definition of tumor margins and identifying small metastases to improve accuracy of resections, sentinel lymph node mapping, or simply defining anatomy during surgical intervention. Most systems are based on reflectance imaging at video rate combined with the use of near infrared fluorochromes. Tumor margins have been visualized using different fluorescent strategies that include targeting of cancer cells (Koyama et al. 2007), or the proteolytic or phagocytic activities of peripheral host cells (Kirsch et al. 2007). Sentinel nodal mapping has been improved through the use of near infrared fluorochrome-labeled albumin (Tanaka et al. 2006), red-shifted quantum dots (Kim et al. 2004; Frangioni et al. 2007), and fluorescent nanoparticles also detectable by other imaging modalities (Pittet et al. 2006). In a recent study, multicolor fluorescence intraoperative live microscopy (“color-FILM”) has been introduced as a method to bring the power of modern experimental microscopy to bear on clinical surgical oncology (Thurber et al. 2009).
ACKNOWLEDGMENTS
The authors wish to thank Bojana Gligorijevic, David Entenberg, Jeffrey Wyckoff, and Mikael Pittet for contributions of text and figures to this review. The work described herein was supported by CA100324, CA113395, and CA126511 (J.C.), and CA92782, EB004626, CA119349, CA86355, and CA126515 (R.W.). Ralph Weissleder has an equity stake and is a consultant to VisEn, a manufacturer of fluorescent imaging probes and systems. The authors report no other conflicts of interest relevant to this publication.
Footnotes
Editors: Mina J. Bissell, Kornelia Polyak, and Jeffrey Rosen
Additional Perspectives on Mammary Gland Biology available at www.cshperspectives.org
REFERENCES
- Abramoff MD, Magelhaes PJ, Ram SJ 2004. Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Ahmed F, Wyckoff J, Lin EY, Wang W, Wang Y, Henninghausen L, Miyazaki J, Jones J, Pollard JW, Condeelis JS, et al. 2002. GFP expression in the mammary gland for imaging of mammary tumor cells in transgenic mice. Cancer Res 62: 7166–7169 [PubMed] [Google Scholar]
- Amoh Y, Yang M, Li L, Reynoso J, Bouvet M, Moossa AR, Katsuoka K, Hoffman RM 2005. Nestin-linked green fluorescent protein transgenic nude mouse for imaging human tumor angiogenesis. Cancer Res 65: 5352–5357 [DOI] [PubMed] [Google Scholar]
- Andresen V, Alexander S, Heupel WM, Hirschberg M, Hoffman RM, Friedl P 2009. Infrared multiphoton microscopy: Subcellular-resolved deep tissue imaging. Curr Opin Biotechnol 20: 54–62 [DOI] [PubMed] [Google Scholar]
- Avril N, Sassen S, Schmalfeldt B, Naehrig J, Rutke S, Weber WA, Werner M, Graeff H, Schwaiger M, Kuhn W 2005. Prediction of response to neoadjuvant chemotherapy by sequential F-18-fluorodeoxyglucose positron emission tomography in patients with advanced-stage ovarian cancer. J Clin Oncol 23: 7445–7453 [DOI] [PubMed] [Google Scholar]
- Ben-Haim S, Ell P 2009. 18F-FDG PET and PET/CT in the evaluation of cancer treatment response. J Nucl Med 50: 88–99 [DOI] [PubMed] [Google Scholar]
- Bhushan KR, Misra P, Liu F, Mathur S, Lenkinski RE, Frangioni JV 2008. Detection of breast cancer microcalcifications using a dual-modality SPECT/NIR fluorescent probe. J Am Chem Soc 130: 17648–17649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg FG, Vanderheyden JL, Strauss HW, Tait JF 2006. Radiolabeling of HYNIC-annexin V with technetium-99m for in vivo imaging of apoptosis. Nat Protoc 1: 108–110 [DOI] [PubMed] [Google Scholar]
- Blum G, Weimer RM, Edgington LE, Adams W, Bogyo M 2009. Comparative assessment of substrates and activity based probes as tools for non-invasive optical imaging of cysteine protease activity. PLoS One 4: e6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer C, Ntziachristos V, Weitkamp B, Theilmeier G, Heindel W, Weissleder R 2005. Optical imaging of spontaneous breast tumors using protease sensing ‘smart’ optical probes. Invest Radiol 40: 321–327 [DOI] [PubMed] [Google Scholar]
- Bullok K, Piwnica-Worms D 2005. Synthesis and characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. J Med Chem 48: 5404–5407 [DOI] [PubMed] [Google Scholar]
- Chen W, Tung C, Weissleder R 2004. Imaging reactive oxygen species in arthritis. Mol Imaging 3: 159–162 [DOI] [PubMed] [Google Scholar]
- Condeelis J, Segall J 2003. Intravital imaging of cell movement in tumours. Nat Rev Cancer 3: 921–930 [DOI] [PubMed] [Google Scholar]
- Condeelis J, Singer RH, Segall JE 2005. The great escape: When cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol 21: 695–718 [DOI] [PubMed] [Google Scholar]
- Costelloe CM, Rohren EM, Madewell JE, Hamaoka T, Theriault RL, Yu TK, Lewis VO, Ma J, Stafford RJ, Tari AM, et al. 2009. Imaging bone metastases in breast cancer: Techniques and recommendations for diagnosis. Lancet Oncol 10: 606–614 [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ 2006. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci 26: 10380–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj N, Weissleder R, Hilderbrand S 2008. Tetrazine-based cycloadditions: Application to pretargeted live cell imaging. Bioconjug Chem 19: 2297–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj NK, Keliher EJ, Thurber GM, Nahrendorf M, Weissleder R 2009a. 18F labeled nanoparticles for in vivo PET-CT imaging. Bioconjug Chem 20: 397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj NK, Upadhyay R, Haun JB, Hilderbrand SA, Weissleder R 2009b. Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/trans-cyclooctene cycloaddition. Angew Chem Int Ed Engl 48: 7013–7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobizhev M, Tillo S, Makarov NS, Hughes TE, Rebane A 2009. Absolute two-photon absorption spectra and two-photon brightness of orange and red fluorescent proteins. J Phys Chem B 113: 855–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy M, Lewis J 2009. Radiopharmaceuticals in preclinical and clinical development for monitoring of therapy with PET. J Nucl Med 50Suppl. 1:106S–121S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M, Ewald AJ, Askautrud HA, Truitt ML, Welm BE, Bainbridge E, Peeters G, Krummel MF, Werb Z 2008. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech 1: 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Gao F, Dehdashti F, Jeffe DB, Marcom PK, Carey LA, Dickler MN, Silverman P, Fleming GF, Kommareddy A, et al. 2009. Lower-dose vs high-dose oral estradiol therapy of hormone receptor-positive, aromatase inhibitor-resistant advanced breast cancer: A phase 2 randomized study. JAMA 302: 774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. 2008. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14: 1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JA, Abdolrasulnia M, Doroodchi H, McClure J, Casebeer L 2009. Practice patterns and guideline adherence of medical oncologists in managing patients with early breast cancer. J Natl Compr Canc Netw 7: 697–706 [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Kim SW, Ohnishi S, Kim S, Bawendi MG 2007. Sentinel lymph node mapping with type-II quantum dots. Methods Mol Biol 374: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY 2006. The fluorescent toolbox for assessing protein location and function. Science 312: 217–224 [DOI] [PubMed] [Google Scholar]
- Gligorijevic B, Kendrin D, Segall JE, Condeelis J, van Rheenen J 2009. Dendra2 photoswitching through the mammary imaging window. J Vis Exp pii: 1278.doi: 103791/1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golman K, Olsson LE, Axelsson O, Månsson S, Karlsson M, Petersson JS 2003. Molecular imaging using hyperpolarized 13-C. Br J Radiol 76: S118–S127 [DOI] [PubMed] [Google Scholar]
- Goswami S, Wang W, Wyckoff JB, Condeelis JS 2004. Breast cancer cells isolated by chemotaxis from primary tumors show increased survival and resistance to chemotherapy. Cancer Res 64: 7664–7667 [DOI] [PubMed] [Google Scholar]
- Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS 2005. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res 65: 5278–5283 [DOI] [PubMed] [Google Scholar]
- Goswami S, Philippar U, Sun D, Patsialou A, Avraham J, Wang W, Di Modugno F, Nistico P, Gertler F, and Condeelis J 2009. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin Exp Metastasis 26: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm J, Kirsch DG, Windsor SD, Kim CF, Santiago PM, Ntziachristos V, Jacks T, Weissleder R 2005. Use of gene expression profiling to direct in vivo molecular imaging of lung cancer. Proc Natl Acad Sci U S A 102: 14404–14409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Piwnica-Worms D 2005a. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods 2: 607–614 [DOI] [PubMed] [Google Scholar]
- Gross S, Piwnica-Worms D 2005b. Spying on cancer: Molecular imaging in vivo with genetically encoded reporters. Cancer Cell 7: 5–15 [DOI] [PubMed] [Google Scholar]
- Gross S, Gammon ST, Moss BL, Rauch D, Harding J, Heinecke JW, Ratner L, Piwnica-Worms D 2009. Bioluminescence imaging of myeloperoxidase activity in vivo. Nat Med 15: 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halin C, Rodrigo Mora J, Sumen C, von Andrian UH 2005. In vivo imaging of lymphocyte trafficking. Annu Rev Cell Dev Biol 21: 581–603 [DOI] [PubMed] [Google Scholar]
- Hoffman RM 2005. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer 5: 796–806 [DOI] [PubMed] [Google Scholar]
- Ingman WV, Wyckoff J, Gouon-Evans V, Condeelis J, Pollard JW 2006. Macrophages promote collagen fibrillogenesis around terminal end buds of developing mammary gland. Dev Dyn 235: 3222–3229 [DOI] [PubMed] [Google Scholar]
- Jaffer F, Libby P, Weissleder R 2009. Optical and multimodality molecular imaging: Insights into atherosclerosis. Arterioscler Thromb Vasc Biol 29: 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Munn L, Fukumura D 2002. Dissecting tumour pathophysiology using intravital microscopy. Nat Rev Cancer 2: 266–276 [DOI] [PubMed] [Google Scholar]
- Kalra J, Warburton C, Fang K, Edwards L, Daynard T, Waterhouse D, Dragowska W, Sutherland BW, Dedhar S, Gelmon K, et al. 2009. QLT0267, a small molecule inhibitor targeting integrin-linked kinase (ILK), and docetaxel can combine to produce synergistic interactions linked to enhanced cytotoxicity, reductions in P-AKT levels, altered F-actin architecture and improved treatment outcomes in an orthotopic breast cancer model. Breast Cancer Res 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerloher W 2008. Bioluminescence microscopy for cellular level circadian analysis in the suprachiasmatic nucleus. Nat Methods 5 url: http://www.nature.com/nmeth/journal/v5/n11/full/nmeth.f.226.html [Google Scholar]
- Kao F 2004. The use of optical parametric oscillator for harmonic generation and two-photon UV fluorescence microscopy. Microsc Res Tech 63: 175–181 [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 150: 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J 2008. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods 5: 1019–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, Wyckoff J, Boimel PJ, Coniglio SH, Hynes NE, Arteaga CL, Segall JE 2009. ERBB1 and ERBB2 have distinct functions in tumor cell invasion and intravasation. Clin Cancer Res 15: 3733–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, et al. 2004. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol 22: 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, Nielsen GP, Quade BJ, Chaber CJ, Schultz CP, et al. 2007. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med 13: 992–997 [DOI] [PubMed] [Google Scholar]
- Koyama Y, Hama Y, Urano Y, Nguyen DM, Choyke PL, Kobayashi H 2007. Spectral fluorescence molecular imaging of lung metastases targeting HER2/neu. Clin Cancer Res 13: 2936–2945 [DOI] [PubMed] [Google Scholar]
- Kremers GJ, Hazelwood KL, Murphy CS, Davidson MW, Piston DW 2009. Photoconversion in orange and red fluorescent proteins. Nat Methods 6: 355–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Dhanpathi H, Basu S, Rubello D, Fanti S, Alavi A 2008. Oncologic PET tracers beyond [(18)F]FDG and the novel quantitative approaches in PET imaging. Q J Nucl Med Mol Imaging 52: 50–65 [PubMed] [Google Scholar]
- Lapidus K, Wyckoff J, Mouneimne G, Lorenz M, Soon L, Condeelis JS, Singer RS 2007. ZBP1 enhances cell polarity and reduces chemotaxis. J Cell Sci 120: 3173–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman CD, DeMartini W, Anderson BO, Edge SB 2009. Indications for breast MRI in the patient with newly diagnosed breast cancer. J Natl Compr Canc Netw 7: 193–201 [DOI] [PubMed] [Google Scholar]
- Lehr HA, Leunig M, Menger MD, Nolte D, Messmer K 1993. Dorsal skinfold chamber technique for intravital microscopy in nude mice. Am J Pathol 143: 1055–1062 [PMC free article] [PubMed] [Google Scholar]
- Makarov NS, Drobizhev M, Rebane A 2008. Two-photon absorption standards in the 550–1600 nm excitation wavelength range. Opt Express 16: 4029–4047 [DOI] [PubMed] [Google Scholar]
- McCann CM, Waterman P, Figueiredo JL, Aikawa E, Weissleder R, Chen JW 2009. Combined magnetic resonance and fluorescence imaging of the living mouse brain reveals glioma response to chemotherapy. Neuroimage 45: 360–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JO, Matrisian LM 2009. Optical proteolytic beacons for in vivo detection of matrix metalloproteinase activity. Methods Mol Biol 539: 155–174 [DOI] [PubMed] [Google Scholar]
- Mempel TR, Henrickson SE, von Andrian UH 2004. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427: 154–159 [DOI] [PubMed] [Google Scholar]
- Mempel TR, Pittet MJ, Khazaie K, Weninger W, Weissleder R, von Boehmer H, von Andrian UH 2006. Regulatory T cells reversibly suppress cytotoxic T cell function independent of effector differentiation. Immunity 25: 129–141 [DOI] [PubMed] [Google Scholar]
- Miller MJ, Wei SH, Cahalan MD, Parker I 2003. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc Natl Acad Sci U S A 100: 2604–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PW, Long NJ, Vilar R, Gee AD 2008. Synthesis of 11C, 18F, 15O, and 13N radiolabels for positron emission tomography. Angew Chem Int Ed Engl 47: 8998–9033 [DOI] [PubMed] [Google Scholar]
- Motoike T, Loughna S, Perens E, Roman BL, Liao W, Chau TC, Richardson CD, Kawate T, Kuno J, Weinstein BM, et al. 2000. Universal GFP reporter for the study of vascular development. Genesis 28: 75–81 [DOI] [PubMed] [Google Scholar]
- Montet X, Ntziachristos V, Grimm J, Weissleder R. 2005. Tomographic fluorescence mapping of tumor targets. Cancer Res 65: 6330–6336 [DOI] [PubMed] [Google Scholar]
- Mrass P, Takano H, Ng LG, Daxini S, Lasaro MO, Iparraguirre A, Cavanagh LL, von Andrian UH, Urtl HC, Haydon PG, et al. 2006. Random migration precedes stable target cell interactions of tumor-infiltrating T cells. J Exp Med 203: 2749–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Taubenberger A, Anderson KI 2007. Recent advances using green and red fluorescent protein variants. Appl Microbiol Biotechnol 77: 1–12 [DOI] [PubMed] [Google Scholar]
- Nahrendorf M, Sosnovik DE, Waterman P, Swirski FK, Pande AN, Aikawa E, Figueiredo JL, Pittet MJ, Weissleder R 2007. Dual channel optical tomographic imaging of leukocyte recruitment and protease activity in the healing myocardial infarct. Circ Res 100: 1218–1225 [DOI] [PubMed] [Google Scholar]
- Nahrendorf M, Waterman P, Thurber G, Groves K, Rajopadhye M, Panizzi P, Marinelli B, Aikawa E, Pittet MJ, Swirski FK, et al. 2009. Hybrid in vivo FMT-CT imaging of protease activity in atherosclerosis with customized nanosensors. Arterioscler Thromb Vasc Biol 29: 1444–14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A 2002. Spinning-disk confocal microscopy—a cutting-edge tool for imaging of membrane traffic. Cell Struct Funct 27: 349–355 [DOI] [PubMed] [Google Scholar]
- Neves A, Brindle K 2006. Assessing responses to cancer therapy using molecular imaging. Biochim Biophys Acta 1766: 242–261 [DOI] [PubMed] [Google Scholar]
- Ntziachristos V, Schellenberger EA, Ripoll J, Yessayan D, Graves E, Bogdanov A Jr, Josephson L, Weissleder R 2004. Visualization of antitumor treatment by means of fluorescence molecular tomography with an annexin V-Cy5.5 conjugate. Proc Natl Acad Sci U S A 101: 12294–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos V, Ripoll J, Wang LV, Weissleder R 2005. Looking and listening to light: The evolution of whole-body photonic imaging. Nat Biotechnol 23: 313–320 [DOI] [PubMed] [Google Scholar]
- Orth JD, Tang Y, Shi J, Loy CT, Amendt C, Wilm C, Zenke FT, Mitchison TJ 2008. Quantitative live imaging of cancer and normal cells treated with Kinesin-5 inhibitors indicates significant differences in phenotypic responses and cell fate. Mol Cancer Ther 7: 3480–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, Desmarais V, van Rheenen J, Koleske AJ, Condeelis J 2009. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol 186: 571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelouch WS, Powers PE, Tang CL 1992. Ti:sapphire-pumped, high-repetition-rate femtosecond optical parametric oscillator. Opt Lett 17: 1070–1072 [DOI] [PubMed] [Google Scholar]
- Perentes JY, McKee TD, Ley CD, Mathiew H, Dawson M, Padera TP, Munn LL, Jain RK, Boucher Y 2009. In vivo imaging of extracellular matrix remodeling by tumor-associated fibroblasts. Nat Methods 6: 143–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NC, Egen JG, Secundino N, Debrabant A, Kimblin N, Kamhawi S, Lawyer P, Fay MP, Germain RN, Sacks D 2008. In vivo imaging reveals an essential role for neutrophils in leishmaniasis transmitted by sand flies. Science 321: 970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim HD, Gaimpieri S, Wang Y, Goswami S, Wyckoff JB, Lauffenburger DA, et al. 2008. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell 15: 813–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet MJ, Swirski FK, Reynolds F, Josephson L, Weissleder R 2006. Labeling of immune cells for in vivo imaging using magnetofluorescent nanoparticles. Nat Protoc 1: 73–79 [DOI] [PubMed] [Google Scholar]
- Prescher JA, Dube DH, Bertozzi CR 2004. Chemical remodelling of cell surfaces in living animals. Nature 430: 873–877 [DOI] [PubMed] [Google Scholar]
- Quon A, Gambhir S 2005. FDG-PET and beyond: Molecular breast cancer imaging. J Clin Oncol 23: 1664–1673 [DOI] [PubMed] [Google Scholar]
- Robinson BD, Sica GL, Liu Y-F, Rohan TE, Gertler FB, Condeelis JS, Jones JG 2009. Tumor microenvironment of metastasis (TMEM) in human breast carcinoma: A potential prognostic marker linked to hematogenous dissemination. Clin Cancer Res 15: 2433–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E 2007. Illuminating the metastatic process. Nat Rev Cancer 7: 737–749 [DOI] [PubMed] [Google Scholar]
- Sahai E, Wyckoff J, Philippar U, Segall JE, Gertler F, Condeelis J 2005. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameni M, Cavallo-Medved D, Dosescu J, Jedeszko C, Moin K, Mullins SR, Olive MB, Rudy D, Sloane BF 2009. Imaging and quantifying the dynamics of tumor-associated proteolysis. Clin Exp Metastasis 26: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslow D, Boetes C, Burke W, Harms S, Leach MO, Lehman CD, Morris E, Pisano E, Schnall M, Sener S, et al. 2007. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin 57: 75–89 [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, et al. 2007. Bright far-red fluorescent protein for whole-body imaging. Nat Methods 4: 741–746 [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, et al. 2009. Far-red fluorescent tags for protein imaging in living tissues. Biochem J 418: 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Hingtgen S, Kasmieh R, Figueiredo JL, Garcia-Garcia E, Martinez-Serrano A, Breakefield X, Weissleder R 2008. Bimodal viral vectors and in vivo imaging reveal the fate of human neural stem cells in experimental glioma model. J Neurosci 28: 4406–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5: 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless N, Depinho R 2006. The mighty mouse: Genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov 5: 741–754 [DOI] [PubMed] [Google Scholar]
- Soon L, Braet F, Condeelis J 2007. Moving in the right direction-nanoimaging in cancer cell motility and metastasis. Microsc Res Tech 70: 252–257 [DOI] [PubMed] [Google Scholar]
- Spencer S, Gaudet S, Albeck JG, Burke JM, Sorger PK 2009. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran AA, Folks K, Warram J, Chanda D, Wang D, Zinn KR 2009. Death receptor 5 agonist TRA8 in combination with the bisphosphonate zoledronic acid attenuated the growth of breast cancer metastasis. Cancer Biol Ther 8: 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E, Choi HS, Fujii H, Bawendi MG, Frangioni JV 2006. Image-guided oncologic surgery using invisible light: Completed pre-clinical development for sentinel lymph node mapping. Ann Surg Oncol 13: 1671–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber G, Figueiredo J, Weisslede R 2009. Multicolor fluorescent intraoperative live microscopy (FILM) for surgical tumor resection. PLOS One 4: e8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torigian DA, Huang SS, Houseni M, Alavi A 2007. Functional imaging of cancer with emphasis on molecular techniques. CA Cancer J Clin 57: 206–224 [DOI] [PubMed] [Google Scholar]
- Tran Cao HS, Reynoso J, Yang M, Kimura H, Synder CS, Hoffman RM, Bouvet M 2009. Development of the transgenic cyan fluorescent protein (CFP)-expressing nude mouse for ‘Technicolor’ cancer imaging. J Cell Biochem 107: 328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadakkan TJ, Culver JC, Gao L, Anhut T, Dickinson ME 2009. Peak multiphoton excitation of mCherry using an optical parametric oscillator (OPO). J Fluoresc 19: 1103–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M, Cantley L, Thompson C 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661–665 [DOI] [PubMed] [Google Scholar]
- Vinegoni C, Pitsouli C, Razansky D, Perrimon N, Ntziachristos V 2008. In vivo imaging of Drosophila melanogaster pupae with mesoscopic fluorescence tomography. Nat Methods 5: 45–47 [DOI] [PubMed] [Google Scholar]
- Viola RJ, Provenzale JM, Li F, Li CY, Yuan H, Tashjian J, Dewhirst MW 2008. In vivo bioluminescence imaging monitoring of hypoxia-inducible factor 1α, a promoter that protects cells, in response to chemotherapy. AJR Am J Roentgenol 191: 1779–1784 [DOI] [PubMed] [Google Scholar]
- Wang H, Cao F, De A, Cao Y, Contag C, Gambhir SS, Wu JC, Chen X 2009. Trafficking mesenchymal stem cell engraftment and differentiation in tumor-bearing mice by bioluminescence imaging. Stem Cells 27: 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wyckoff JB, Frohlich VC, Oleynikov Y, Hüttelmaier S, Zavadi J, Cermak L, Bottinger EP, Singer RH, White JG, et al. 2002. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res 62: 6278–6288 [PubMed] [Google Scholar]
- Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS 2004. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res 64: 8585–8594 [DOI] [PubMed] [Google Scholar]
- Wang W, Goswami S, Sahai E, Wyckoff JB, Segall JE, Condeelis JS 2005. Tumor cells caught in the act of invading: Their strategy for enhanced cell motility. Trends Cell Biol 15: 138–145 [DOI] [PubMed] [Google Scholar]
- Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS 2006. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol 173: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wyckoff JB, Goswami S, Wang Y, Sidani M, Segall JE, Condeelis JS 2007a. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res 67: 3505–3511 [DOI] [PubMed] [Google Scholar]
- Wang W, Eddy R, Condeelis J 2007b. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer 7: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J 2004. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res 64: 7022–7029 [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E 2006. Rock- and myosin-dependent matrix deformation enables protease-independent tumour-cell invasion in vivo. Curr Biol 16: 1515–1523 [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Wang Y, Lin EY, Li J-F, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J 2007. Direct visualization of macrophage mediated intravasation of carcinoma cells in mouse mammary tumors. Cancer Res 67: 2649–2656 [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Gligorijevic B, Entenberg D, Segall J, Condeelis J 2009. High-resolution multiphoton imaging of tumors in vivo. In Live Cell Imaging (ed. Spector DL, Goldman RD), pp. 441–462 Cold Spring Harbor Laboratory Press, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Wyckoff J, Liang F, Sidani M, Violini S, Tsai KL, Zhang ZY, Sahai E, Condeelis J, Segall JE 2006. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res 66: 192–197 [DOI] [PubMed] [Google Scholar]
- Yang M, Baranov E, Wang JW, Jiang P, Wang X, Sun FX, Bouvet M, Moossa AR, Penman S, Hoffman RM 2002. Direct external imaging of nascent cancer, tumor progression, angiogenesis, and metastasis on internal organs in the fluorescent orthotopic model. Proc Natl Acad Sci U S A 99: 3824–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Reynoso J, Bouvet M, Hoffman RM 2009. A transgenic red fluorescent protein-expressing nude mouse for color-coded imaging of the tumor microenvironment. J Cell Biochem 106: 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrow JC, Feng Y, Perlman ZE, Kirchhausen T, Mitchison TJ 2003. Phenotypic screening of small molecule libraries by high throughput cell imaging. Comb Chem High Throughput Screen 6: 279–286 [DOI] [PubMed] [Google Scholar]
- Zacharakis G, Kambara H, Shih H, Ripoll J, Grimm J, Saeki Y, Weissleder R, Ntziachristos V 2005. Volumetric tomography of fluorescent proteins through small animals in vivo. Proc Natl Acad Sci U S A 102: 18252–18257 [DOI] [PMC free article] [PubMed] [Google Scholar]