Abstract
The nucleus is unique amongst cellular organelles in that it contains a myriad of discrete suborganelles. These nuclear bodies are morphologically and molecularly distinct entities, and they host specific nuclear processes. Although the mode of biogenesis appears to differ widely between individual nuclear bodies, several common design principles are emerging, particularly, the ability of nuclear bodies to form de novo, a role of RNA as a structural element and self-organization as a mode of formation. The controlled biogenesis of nuclear bodies is essential for faithful maintenance of nuclear architecture during the cell cycle and is an important part of cellular responses to intra- and extracellular events.
The different nuclear bodies in the cell nucleus have distinct components and are sites for different processes. But they share the ability to self-organize, form de novo in certain conditions, and use RNA as a structural element.
The mammalian cell nucleus contains a multitude of discrete suborganelles, referred to as nuclear bodies or nuclear compartments (reviewed in Dundr and Misteli 2001; Spector 2001; Lamond and Spector 2003; Handwerger and Gall 2006; Zhao et al. 2009). These bodies are an essential part of the nuclear landscape as they compartmentalize the nuclear space and create distinct environments within the nucleus (reviewed in Misteli 2007). Many nuclear bodies carry out specific nuclear functions, such as the synthesis and processing of pre-ribosomal RNA in the nucleolus, the storage and assembly of spliceosomal components in nuclear speckles, or the retention of RNA molecules in paraspeckles. The mechanisms by which nuclear bodies contribute to function are highly diverse. In some cases, a nuclear body may be host to a particular activity such as transcription; in other cases, a nuclear body seems to act indirectly by regulating the local concentration of its components in the nucleoplasm.
In many ways, nuclear bodies are similar to conventional cellular organelles in the cytoplasm. Like cytoplasmic organelles, they contain a specific set of resident proteins, which defines each structure molecularly. Although many nuclear bodies are spherical in shape, most can be characterized based on their unique morphology, particularly when analyzed by electron microscopy and by their nuclear distribution patterns. However, in stark contrast to conventional cytoplasmic organelles, nuclear bodies are not delineated by lipid membranes, and their structural integrity appears to be entirely mediated by protein–protein and possibly protein–RNA interactions. The absence of a demarcating lipid membrane points to unique mechanisms of biogenesis.
The mechanisms of nuclear body biogenesis are physiologically important for two reasons. First, proper biogenesis is essential for their faithful inheritance during cell division. Many nuclear bodies disassemble as cells enter mitosis and must then efficiently and rapidly reassemble at the end of mitosis to ensure full functionality of the daughter nuclei. Second, nuclear bodies may form in response to specific cellular activities. Coordinated disassembly and efficient biogenesis of nuclear bodies is thus critical for maintenance of cellular integrity, homeostasis, and responsiveness.
NUCLEAR BODIES ARE DYNAMIC STEADY-STATE STRUCTURES
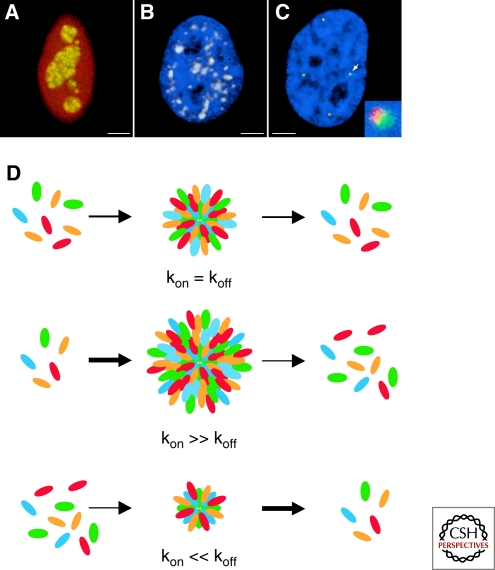
Nuclear bodies are a morphologically and molecularly highly diverse group of cellular structures (Table 1; Fig. 1A–C). At one end of the spectrum is the nucleolus, which is often found in less than 3 copies, and often as a single organelle, and may reach a size up to several micrometers. At the other end of the spectrum, Cajal bodies are rarely larger than 1.5 µm, and as many as 10 can be found in a cell. Size, shape, and number of a given nuclear body are often dependent on cell type, are variable between tissues, and may change in response to cellular conditions. Despite their diverse morphological appearances, nuclear bodies share several common features of organization (reviewed in Misteli 2001, 2007; Lamond and Spector 2003; Handwerger and Gall 2006; Bernardi and Pandolfi 2007; Pederson and Tsai 2009).
Table 1.
Biogenesis and inheritance of nuclear bodies.
| Putative functions | Defining components | Typical Size (μm) | Typical Number | Activity dependence | Nucleation template | De novo formation | Mode of Inheritance | |
|---|---|---|---|---|---|---|---|---|
| Nucleolus | Ribosome biogenesis | RNA pol I machinery rRNA processing/ ribosome assembly factors | 3–8 | 1–4 | + | DNA/RNA | + | disassembly, persist as mitotic nucleoli derived foci |
| Nuclear speckles | Storage and recycling of splicing factors | Pre-mRNA splicing factors | 2–3 | 20–50 | + | DNA/RNA | + | Disassemble/reassembly |
| Nuclear stress bodies | Regulation of transcription and splicing under stress | HSF1, HAP | 1–2 | 2–6 | + | RNA | Unknown | Unknown |
| Histone locus body | Histone gene synthesis | NPAT, FLASH,U7 snRNP | 0.2–1.2 | 2–4 | + | DNA/RNA | Unknown | Disassemble in metaphase, reform in G1/S-phase |
| Cajal body | Biogenesis, maturation and recycling of small RNAs | Coilin, SMN | 0.2–1.5 | 1–10 | − | DNA/RNA | + | Disassembly into mitotic CBs |
| PML nuclear body | Regulation of genome stability, DNA repair, control of transcription, viral defense | PML protein | 0.1–1 | 10–30 | − | DNA/RNA | + | Mitotic PML accumulations |
| Paraspeckles | mRNA regulation, RNA editing | NEAT1/MENε/βncRNAs PSP1, p54nrb/NONO | 0.2–1 | 2–20 | + | RNA | Unknown | Mitotic paraspeckles |
| Perinucleolar compartment | Post-transcriptional regulation of a subset of pol III RNAs | PTB | 0.2–1 | 1–2 | + | RNA | Unknown | Disassembly/reassembly |
Figure 1.
Structure and maintenance of nuclear bodies. The appearance and number of nuclear bodies is highly variable. Prominent examples include (A) the nucleolus (RNA polymerase I subunit RPA43 in green, rRNA processing enzyme fibrillarin in red); (B) nuclear speckles (splicing factor SC35 in red; DNA in blue); and (C) Cajal bodies (coilin in red) and histone locus bodies (histone genes transcription factor NPAT in green), which are closely associated with each other (arrow). Inset: higher magnification view. Scale bars: 2 µm. (D) Nuclear bodies are dynamic, steady-state structures exposed to a continuous flow of material in and out. The ratio of on/off rate determines the size, and possibly the shape, of the body. Minor changes in on/off-rate ratio can strongly influence nuclear body morphology.
A common property of nuclear bodies is their overall structural stability. When followed by time-lapse microscopy, individual nuclear bodies persist during the entire interphase between entry into G1 phase and exit from G2 phase. During this time, the bodies undergo dynamic motions within the nucleus, whose extent is generally inversely proportional to their size (Platani et al. 2000; Muratani et al. 2002; Dundr et al. 2007). The large bodies such as nucleoli and nuclear speckles, typically 2–3 µm in diameter, are largely immobile and only undergo limited local motion (Misteli et al. 1997a; Kruhlak et al. 2000; Olson and Dundr 2005). The smaller bodies such as Cajal bodies or PML nuclear bodies, with an average size of 500 nm–1 µm, undergo more extensive intranuclear motion and frequent fusion and fission events (Platani et al. 2002; Dellaire et al. 2006; Dundr et al. 2007; Chen et al. 2008).
Despite the overall structural stability, nuclear bodies are, however, intrinsically highly dynamic structures (Phair and Misteli 2000; Handwerger and Gall 2006). The vast majority of nuclear body components are proteins that are enriched in a nuclear body but they are also present, albeit at lower concentration, in the surrounding nucleoplasm (reviewed in Dundr and Misteli 2001). Key insights into the nature of nuclear bodies comes from photobleaching experiments that have demonstrated rapid dynamic exchange of a large number of the major nuclear body components with the surrounding nucleoplasm (Kruhlak et al. 2000; Phair and Misteli 2000; Snaar et al. 2000; Chen and Huang 2001; Weidtkamp-Peters et al. 2008). For example, the major nucleolar component fibrillarin or the marker Cajal body component coilin have residence times on the order of tens of seconds and several minutes, respectively, and it can be calculated that the entire pool of a given nuclear body protein is completely replaced within a few minutes (Phair and Misteli 2000; Dundr et al. 2004).
The absence of a delineating lipid boundary to distinguish nuclear bodies from their environment indicates that nuclear bodies are accumulations of proteins and RNAs and that the morphological appearance of a given nuclear body is largely determined by the sum of interactions of its components within the body. At present, no strictly architectural protein components of nuclear bodies are known. However, a role for dedicated protein–protein interactions in the structural maintenance of nuclear bodies is hinted at by the fact that several prominent nuclear body proteins have self-association properties (Hebert and Matera 2000). The major Cajal body protein coilin contains a self-interacting N-terminal domain, the PML protein forms homo-oligomers and the SMN protein, which is a major constituent of the gem nuclear bodies, oligomerizes readily with itself (Hebert and Matera 2000; Kentsis et al. 2002; Dundr et al. 2004; Morse et al. 2007). No covalent interactions between nuclear body components have, however, been identified to date as determinants of nuclear body structure, and it seems likely that most intrabody interactions are noncovalent and weak. Taken together, the overall structural stability and the rapid exchange of individual components make nuclear bodies dynamic steady-state protein/RNA structures, which form by dynamic interactions of their components in the nucleoplasm (Fig. 1D).
NUCLEAR BODY MAINTENANCE
The steady-state appearance of nuclear bodies is maintained by the continuous flow of molecules into and out of the body (Fig. 1D). The size, and possibly shape, of nuclear bodies is likely determined by the balance of on-rate relative to the off-rate of its components. An increase in on-rate or a drop in the off-rate will lead to an increase in size, whereas a decrease in on-rate or an increase in off-rate will lead to shrinkage (Fig. 1D). The molecular mechanisms that determine on/off rates of individual nuclear body components are poorly understood but likely involve post-translational modifications. This is best exemplified by SR protein splicing factors, which are major constituents of the nuclear speckles, which serve to store and assemble pre-mRNA splicing components. Several speckle components undergo a cycle of phosphorylation that coincides with their association with speckles (reviewed in Misteli 1997b). Many SR protein splicing factors exist in a hypo-phosphorylated state inside of speckles, where they are the target for speckle-resident SR-protein kinases of the Clk family. Hyper-phosphorylation of SR proteins then makes them competent to participate in the splicing reaction and at the same time leads to their dissociation from speckles. During the splicing reaction, the SR proteins are dephosphorylated, re-establishing their affinity for speckles, where they are recycled for a new round of splicing (reviewed in Misteli 1997b). A similar cyclical mechanism to control nuclear body association has been characterized for the nucleolar protein nucleostemin, which contains a GTP-binding domain that mediates nucleolar retention (Tsai and McKay 2005; Pederson and Tsai 2009). It is likely that similar, yet uncharacterized, mechanisms play key roles in controlling the association and dissociation, and thus shape and size, of many other nuclear bodies.
Control of nuclear body number is poorly understood. Even the number of nucleoli, which form around a fixed number of nucleolar organizing regions consisting of constant numbers of rDNA repeats on a defined number of chromosomes, varies greatly in different tissues and cell types. Some initial insight into number control comes from analysis of the Cajal body, where the marker protein coilin is capable of regulating the number of Cajal bodies per cell. This involves C-terminal phosphoserine residues that regulate its self-interaction activity (Shpargel et al. 2003; Hearst et al. 2009). Truncation or mutation of these phosphorylation sites leads to alteration in the number of Cajal bodies. In addition, expression of heterologous coilin proteins in different cellular backgrounds results in deregulation of Cajal body numbers. For example, mouse coilin forms numerous foci in human HeLa cells, but when expressed in mouse cells, the number of Cajal bodies does not increase. When frog coilin is expressed in either cell type, the number of Cajal bodies is highly up-regulated. In contrast, human coilin is unable to form foci in cells derived from coilin knockout mice (Shpargel et al. 2003).
Size and number of nuclear bodies are not fixed but are dependent on physiological conditions. The number of nucleoli correlates positively with cell proliferation. For example, in lymphocytes with high proteosynthetic activity, and thus a high requirement for ribosomal RNA (rRNA), nucleolar size increases (Hernandez-Verdun 2006). Cellular stress, such as inflammation and oncogenic stimulation, cause an increase in the number and size of PML nuclear bodies along with increased expression of PML protein (reviewed in Bernardi and Pandolfi 2007). Coilin in primary cells is hyperphosphorylated on C-terminal phosphoserine residues that reduce its self-interaction activity, and these cells lack Cajal bodies. In contrast, in transformed cells, which have a large number of Cajal bodies (CBs), coilin is hypo-phosphorylated and able to self-associate (Hearst et al. 2009). It seems that the phosphorylation status of coilin has an impact on Cajal body formation, and coilin is likely a target of a yet uncharacterized signal transduction pathway that might control Cajal body formation (Hearst et al. 2009).
ACTIVITY-DEPENDENT AND ACTIVITY-INDEPENDENT NUCLEAR BODIES
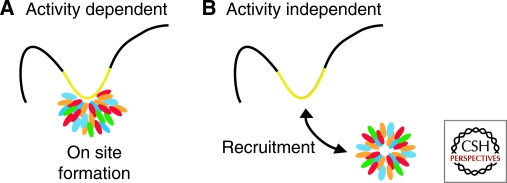
Nuclear bodies may be divided into two classes based on their mode of formation: activity-dependent nuclear bodies and activity-independent nuclear bodies. This classification is at this point somewhat tenuous, since unknown nuclear activities may be responsible for formation of a nuclear body assumed to form in an activity-independent fashion (Fig. 2). The former class includes bodies that form at sites of a particular nuclear function, such as transcription, and whose morphological appearance is strictly dependent on this activity (Table 1). The prototypical example for activity-dependent bodies is the nucleolus, which forms in response to transcriptional activity of clustered ribosomal DNA genes located on multiple chromosomes that cluster in the nucleolus (Shaw and Jordan 1995; Boisvert et al. 2007). The strict dependence of the morphological appearance of the nucleolus on ongoing transcription is evident by its rapid structural reorganization upon inhibition of rDNA transcription (Olson and Dundr 2005). Conversely, introduction of exogenous rDNA genes on plasmids into the nucleus is sufficient for the formation of new nucleoli (Oakes et al. 1998). Other activity-dependent nuclear bodies include the histone locus body, which forms around histone gene clusters in response to transcriptional activation of replication-dependent histone genes during S-phase (Bongiorno-Borbone et al. 2008), nuclear stress bodies that form at sites of satellite III repeat transcription in response to heat shock (Jolly et al. 2002), and nuclear speckles that undergo dramatic morphological changes by rounding up upon inhibition of transcription and they rapidly re-form at sites of heat shock-activated genes (reviewed in Misteli 2007; Hu et al. 2009).
Figure 2.
Classification of nuclear bodies based on activity. Nuclear bodies may either form (A) associated with a nuclear function, such as transcription (yellow), making their appearance dependent on the ongoing activity or (B) they may form independently of a function and subsequently associate with a particular nuclear site, such as an active gene locus (yellow).
On the other hand, there are nuclear bodies that seem to form without the requirement for a particular nuclear activity. Such apparently activity-independent bodies form in the nucleoplasm and may subsequently associate with particular nuclear locations (Table 1). Activity-independent bodies include PML nuclear bodies and Cajal bodies. Just like activity-dependent bodies, these structures are often associated with particular locations in the nucleus and sometimes with specific gene loci; however, these bodies probably do not form at these sites but rather are either recruited to them or activated genes are repositioned to associate with the preformed bodies (Carmo-Fonseca 2007). The clearest example of such recruitment comes from analysis of the interactions of the U2 snRNA gene cluster with the Cajal body (Dundr et al. 2007). Upon activation of the U2 snRNA genes, they undergo directed, actin-dependent motion to associate with a preformed Cajal body in the vicinity (Dundr et al. 2007).
NUCLEAR BODY TEMPLATES AND NUCLEATORS
Formation of a nuclear body requires an initial nucleation event. Nucleation serves to immobilize some of the freely diffusible key components of the body and to provide a template to recruit additional building blocks. In the case of activity-associated nuclear bodies, this function is carried out by the activities that drive the nuclear body formation. Examples for well-defined nucleation events are the nucleolus, where accumulation of nucleolar proteins on ribosomal DNA and pre-ribosomal RNA leads to the formation of the nucleolus or the recruitment of the histone pre-mRNA 3′-end processing factors in histone locus bodies. Immobilization of proteins in this way likely leads to recruitment of some of their interaction partners and to increased concentration of these proteins at the site of activity, ultimately resulting in the formation of a morphologically distinct suborganelle.
In the case of activity-independent bodies, other nucleators must exist. These architectural elements may either be structural scaffold proteins or RNAs. Such nucleators are likely fairly abundant components of a given nuclear body, and they should be necessary and sufficient for the formation of a nuclear body. No such nucleators of activity-independent nuclear bodies have been identified to date. A possibility is that putative nucleators of activity-independent bodies are part of an immobile nuclear matrix. However, this scenario is unlikely since most nuclear bodies are mobile and thus not permanently tethered to an underlying structure (Platani et al. 2002; Muratani et al. 2002; Görisch et al. 2004). The identification of nuclear body nucleators will be critically important for a full understanding of nuclear body biogenesis and function.
DE-NOVO FORMATION OF NBs
A critical test for whether a cellular structure is autonomous and self-sustaining is whether it can form de novo. Several nuclear bodies form de novo either under physiological or experimental conditions.
The nucleolus can be formed de novo by introducing extra rDNA minigenes on plasmids into a cell (Nierras et al. 1997; Oakes et al. 1998). Apparently functional mini-nucleoli with a tendency to coalesce into a single, larger nucleolus rapidly form around the ribosomal genes (Oakes et al. 1998). In addition, a large number of small nucleoli are formed in Xenopus oocytes as thousands of extrachromosomal rDNA gene copies are amplified during oogenesis (Mais et al. 2002). Similarly, nuclear speckles can form de novo upon re-activation of global transcription after inhibition. After reversal of a global transcription block, speckles expand from a condensed state and take on their typical irregular shape and new speckles form, associated with re-activated genes (Misteli et al. 1997; Shopland et al. 2003).
De novo formation is not limited to activity-dependent nuclear bodies. During viral infection, PML nuclear bodies form rapidly. Upon entry of a viral genome into the nucleus, several major components of the PML nuclear body, including PML protein, Sp100, and Daxx, rapidly associate with the foreign genome and form what appears to be fully functional PML nuclear bodies (Everett and Murray 2005). The formation of PML nuclear bodies at sites of viral transcription has been proposed to be a first-line antiviral defense, possibly via PML-mediated modulation of the interferon response (Everett and Chelbi-Alix 2007). A similarly physiological and possibly protective de novo formation event occurs in response to acute cellular stress. Heat shock induces the transcription of the normally silent satellite III repeats on human chromosome 9 (Jolly et al. 2002). Transcriptional activation of these sequences leads to accumulation of various RNA-binding proteins and several SR protein splicing factors to form de novo a morphologically distinct nuclear stress body (Denegri et al. 2001; Metz et al. 2004)
The strongest evidence for de novo formation of nuclear bodies comes from studies on the Cajal body. Transient overexpression of Cajal body components or the SMN protein, which is enriched in Cajal-body-associated structures called gems, leads to formation of Cajal bodies in cells that usually do not contain them (Sleeman et al. 2001). More direct evidence for de novo formation of Cajal bodies comes from tethering experiments in which components of the Cajal body were artificially immobilized on chromatin. Immobilization of individual Cajal body components invariably resulted in formation of a fully functional and morphologically intact Cajal body (Kaiser et al. 2008). Since the site of tethering was random and did not represent an active gene locus, these results demonstrate the capacity of Cajal bodies to form de novo and in a DNA sequence-independent manner in vivo.
A ROLE FOR RNA IN NUCLEAR BODY FORMATION
RNA is emerging as an important structural component of nuclear bodies. It may serve two roles in their biogenesis. For one, nascent RNAs may act as templates for the formation of nuclear bodies. Most activity-dependent nuclear bodies form around sites of transcription such as the nucleolus around rDNA, the histone locus bodies near histone gene clusters, or nuclear stress granules in response to activation of satellite III repeats upon heat stress. The nascent RNA transcripts generated at these loci act as a template to attract and retain RNA-interacting proteins, leading to the formation of a nuclear body.
An additional role of RNA in nuclear body biogenesis is its function as a dedicated architectural element. Many non-coding RNAs (ncRNAs) play important functions in nuclear RNA metabolic complexes such as tRNAs, snRNAs, or rRNAs. It had long been hypothesized that ncRNAs may also contribute to nuclear architecture (Huang and Spector 1996; Prasanth et al. 2005). This proposal was based on the finding that poly(A)-RNAs are retained for extended periods of time in nuclear speckles after inhibition of transcription, at a time when all exported RNAs should have left the nucleus (Huang and Spector 1996). The recent identification of the first architectural RNAs confirms the role of ncRNAs as structural elements of the nucleus. The NEAT1 (also known as MEN-ε/β), a long stable nuclear-retained polyadenylated ncRNA (Chen and Carmichael 2009; Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009; reviewed in Bond and Fox 2009), was found to be essential for the formation of paraspeckles. While other protein components of paraspeckles are also located diffusely throughout the nucleoplasm, NEAT1 is found almost exclusively in paraspeckles. Knockdown of these ncRNAs by antisense or RNAi induces the disruption of paraspeckles. Interestingly, knockdown of paraspeckle-specific proteins, p54nrb/NONO and PSF, which interact with NEAT1, induce similar paraspeckle disintegration, indicating that NEAT1–paraspeckle protein interactions are crucial for paraspeckle integrity (Chen and Carmichael 2009; Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009). Interestingly, paraspeckles are not present in human embryonic stem cells (hESCs) due to the lack of NEAT1 expression (Chen and Carmichael 2009). Expression of NEAT1 is activated when hESCs are induced to differentiate into trophoblasts, which is accompanied by the appearance of paraspeckles. Thus, it seems that paraspeckle formation is linked to the loss of pluripotency in hESCs. Moreover, NEAT1 is strongly up-regulated during muscle differentiation concomitant with an increase in the number of paraspeckles in differentiated myotubes, possibly pointing to a role in physiological control of paraspeckle number (Sunwoo et al. 2009). These findings make it likely that ncRNAs play a key role as structural components in nuclear bodies and open the door for identification of architectural ncRNAs in other nuclear bodies.
NUCLEAR BODY ASSEMBLY PATHWAYS
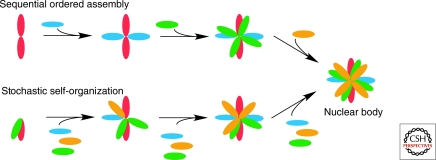
In theory, NBs can be assembled by two distinct mechanisms: (1) The biogenesis may involve a tightly controlled sequence of assembly steps or (2) it may occur via the largely stochastic and random interaction of individual components without a strict hierarchical order of assembly (Fig. 3). Recent experiments in which Cajal body components were irreversibly tethered to chromatin to assess their ability to nucleate a Cajal body have tested these possibilities and the results support a stochastic assembly model. Immobilization of individual CB components to an engineered random site in the genome was shown to be sufficient to initiate the formation of morphologically normal and functional Cajal bodies de novo (Kaiser et al. 2008). De novo Cajal bodies formed after tethering of structural components such as coilin or SMN, which are required for formation, since their depletion by RNAi abolishes Cajal body biogenesis. Importantly, Cajal body formation can also be initiated by other functional groups of Cajal body components, including spliceosomal snRNPs, the SMN protein complex, small nucleolar RNPs, and small Cajal-body specific RNPs, indicating the absence of a requirement for a strict sequence of assembly events. While these experiments used the activity-independent Cajal body as a model system, it remains to be seen whether formation of activity-dependent nuclear bodies have a stricter requirement for sequential assembly steps.
Figure 3.
Theoretical nuclear body assembly pathways. Assembly of nuclear bodies may either involve a hierarchical and ordered assembly pathway in which assembly occurs by association of nuclear body components in a strictly defined sequence of steps or, alternatively, in a stochastic manner in which components assemble in random order.
A ROLE FOR SELF-ORGANIZATION IN NUCLEAR BODY BIOGENESIS
Several features of nuclear bodies point to a role for self-organization in their biogenesis (reviewed in Misteli 2001, 2007). First, the intrinsically dynamic nature of all nuclear bodies in which a stable structure is formed by the dynamic exchange and interplay of multiple factors is a hallmark of self-organized structures. The dynamic nature of interactions within the body permits the rapid, combinatorial exploration of various binding partners to achieve the most stable network of interactions. The nature of this network ultimately determines the size and possibly the shape of the structure. The existence of nuclear bodies as dynamic steady-state structures combines two opposing needs of a nuclear body: the requirement for a stable, permanent structural entity and the requirement to respond rapidly to changing environmental conditions by changing its architecture. The dynamic nature of interactions facilitates rapid reorganization of the nuclear body structure, for example in response to environmental cues, by altering the interactions of a few nuclear body components. These relatively small changes have the potential to lead to large changes in structure.
A second indicator of the self-organizing nature of nuclear bodies is the finding that de novo assembly of Cajal bodies does not follow a strict and linear assembly pathway but rather that many, in fact the majority, of Cajal body components can trigger full assembly of the body (Kaiser et al. 2008). Although this observation alone does not demonstrate a role for self-organization, it is consistent with it. Despite the absence of experimental data to test the assembly pathways for other nuclear bodies, it is very likely that most activity-dependent nuclear bodies follow similar assembly principles once they are nucleated by an upstream component, often a nascent RNA such as the case for the nucleolus or the histone locus body.
A further hallmark of the self-organizing nature of nuclear bodies is their limited size. Self-organizing systems often are characterized by size criticality in which a structure does not grow beyond a certain size. This limiting size is set by intrinsic properties of the system, such as the combination of interactions within the structure or the flow of material through the structure, but it is generally independent of concentration. Concentration-independent size regulation is found in at least some nuclear bodies. Cajal bodies do not grow beyond a certain size when nucleated by overexpression of large amounts of components; neither do de novo-formed Cajal bodies nucleated by tethering grow beyond their typical size despite an excess of building components in the nucleus (Sleeman et al. 2001; Kaiser et al. 2008). Similarly, the size of most other nuclear bodies falls within a well-defined range and may only change in response to cellular cues, which likely affect the flow and the interactions of proteins within the structure, thus changing the intrinsic critical size of the body.
Self-organization is likely a driving force in nuclear body biogenesis. Similar properties such as a high rate of flow, size-criticality, and a high degree of plasticity are also hallmarks of other cellular compartments defined by membranes, such as the Golgi complex or the endosomal system, as well as of other cellular structures, such as the actin cytoskeleton. It is therefore likely that the self-organizing nature of nuclear bodies is not the exception but rather the rule in cellular biogenesis, and insights from nuclear body biogenesis will be applicable to understanding cellular biogenesis in general.
MITOTIC INHERITANCE OF NUCLEAR BODIES
The assembly and disassembly of nuclear bodies is of critical relevance for their inheritance during cell division. Some nuclear bodies that are present in high copy number persist through cell division in the form of mitotic bodies leading to the equal segregation of their components between daughter cells based on their stochastic distribution throughout the cell volume. Other nuclear bodies, including the activity-associated bodies, disassemble during cell division and must be rapidly re-assembled as cells enter G1 phase.
Nucleolus
At the onset of mitosis, when transcription is blocked by the mitosis-dependent phosphorylation of RNA polymerase I transcription factors and processing components, the nucleolus breaks down (Gautier et al. 1992; Leung et al. 2004; Boisvert et al. 2007). By the time cells reach prophase, processing of pre-rRNA is also inhibited, leading to accumulation of unprocessed and partially processed pre-rRNAs with many rRNA processing components in the periphery of condensed chromosomes and dispersed throughout the cytoplasm (Dundr and Olson 1998; Van Hooser et al. 2005). During anaphase, pre-rRNA processing intermediates with associated processing components appear in numerous highly mobile nucleolus-derived foci (NDFs) scattered throughout the cytoplasm (Dundr et al. 2000). These structures are the mitotic remnants of the interphase nucleolus. Once transcription resumes in early telophase, pre-rRNA intermediates with associated processing components are released from disintegrating NDFs and the surface of chromosomes. After the release they accumulate in the prenucleolar bodies (PNBs) in the nucleoplasm of newly formed daughter nuclei. As chromosomes decondense in late telophase, these components are sequentially released from PNBs to re-build nucleoli around transcriptionally active nucleolar organizing regions (NORs). Thus, pre-rRNA intermediates and processing components are recycled from one cell cycle to the next (Dundr et al. 2000; Savino et al. 2001; Boisvert et al. 2007; Prieto and McStay 2008). Moreover, the re-formation of nucleoli after mitosis is associated with resumption of RNA polymerase I transcription since microinjection of anti-RNA polymerase I antibodies into mitotic cells blocks nucleolus re-formation (Benavente et al. 1987). In addition, nucleolar re-formation depends on resumption of pre-rRNA processing. Inactivation of phosphorylation activity of cyclin B/Cdk1 in metaphase by the specific inhibitor roscovitine leads to reactivation of RNA polymerase I transcription but not pre-rRNA processing (Sirri et al. 2000). This strongly indicates that these two events are reactivated in a coordinated manner. Thus, NORs at the end of mitosis need to sequester components of both the transcription and processing machineries prior to the onset of transcription. This conclusion is supported by analysis of pseudo-NORs, engineered arrays containing multiple enhancer elements of ribosomal gene stably integrated in human cells (Prieto and McStay 2008). These pseudo-NORs are capable of triggering the formation of a nuclear structure without RNA polymerase I transcription but they only accumulate the RNA polymerase I transcription machinery plus factors required for early events in pre-rRNA maturation, but not later processing factors (Prieto and McStay 2007; 2008). The differential effects of early and late steps in rRNA synthesis on nucleolus morphology indicate that they make independent and additive contributions to nucleolar structure.
Nuclear Speckles
During early mitosis, nuclear speckles disassemble and their components scatter throughout the cytoplasm. Reassembly of nuclear speckles occurs in telophase when components aggregate into cytoplasmic speckles, referred to as mitotic interchromatin granule clusters (Ferreira et al. 1994; Prasanth et al. 2003). Once the nuclear envelope is formed in telophase, the RNA polymerase II transcription machinery enters the nucleus followed by pre-mRNA splicing components and other pre-mRNA processing factors (Prasanth et al. 2003; Bubulya et al. 2004). In late telophase, SR splicing factors accumulate briefly in nucleolus associated patches (NAPs) around active nucleolar organizing regions, whereas spliceosomal snRNPs are enriched in other regions of newly formed daughter nuclei. NAPs form very transiently and disintegrate when newly formed nuclear speckles appear in G1 phase. SR splicing proteins are hypo-phosphorylated in NAPs, which may be necessary for SR protein modification for their activation to a splicing-competent state, prior to their targeting to newly formed nuclear speckles (Bubulya et al. 2004). These findings indicate that the pre-mRNA splicing machinery is recruited to transcription sites upon its entry into daughter nuclei, and nuclear speckles form in an activity-dependent manner in early G1 phase in response to the establishment of a steady level of RNA polymerase II transcription.
Paraspeckles
Formation of paraspeckles is initiated after cell division. Once paraspeckles are formed, they remain stable throughout the cell cycle and persist in mitosis until anaphase, when they scatter throughout the cell (Fox et al. 2005). Disappearance of cytoplasmic paraspeckles in early telophase, when daughter nuclei are formed, is coordinated with the nuclear entry of PSP1, a marker protein of paraspeckles. Individual paraspeckles begin to form in early G1 phase until approximately 1 hour after RNA transcription is initiated (Fox et al. 2005). Importantly, blocking the initiation of RNA polymerase II transcription by the inhibitor DRB at the end of mitosis prevents formation of paraspeckles, suggesting that these structures are dependent on ongoing transcription (Fox et al. 2005; Clemson et al. 2009). The earliest paraspeckles are often positioned next to the NEAT1 gene locus and an increase in NEAT1 expression by ectopic plasmids leads to a corresponding increase in the number of paraspeckles, which is essentially unchanged upon PSP1 overexpression (Clemson et al. 2009). However, eventually additional paraspeckles form, and they appear to be nonrandomly distributed throughout the nucleoplasm, suggesting their association with specific, yet unidentified, gene loci.
Histone Locus Body
Histone locus bodies are nuclear bodies that form around the replication-dependent histone gene clusters, which are only active in S phase (Marzluff et al. 2008). Histone locus bodies persist until early prometaphase and disintegrate in metaphase until telophase. When diploid cells enter G1 phase, two histone locus bodies are formed and in S phase their number increases to four, reflecting the number of histone gene clusters (Zhao et al. 2000; Bongiorno-Borbone et al. 2008). It is neither known in mammalian cells whether histone locus bodies form synchronously with Cajal bodies, nor, whether histone locus bodies and Cajal bodies form as distinct nuclear bodies and then fuse or whether they assemble as a joint structure.
Cajal Body
The number of Cajal bodies differs during the cell cycle and is maximal in G1/S phase. Cajal bodies are frequently associated with snRNA genes and histone genes in a transcription-dependent manner (Frey and Matera 1995, 2001; Cioce and Lamond 2005; Matera and Shpargel 2006). At the onset of mitosis, Cajal bodies remain intact, and they scatter throughout the cytoplasm without physical contact with condensed chromosomes. The number and size of mitotic Cajal bodies remain approximately unchanged from metaphase to telophase. Importantly, mitotic Cajal bodies contain spliceosomal snRNPs through the course of mitosis even without transcription. Once the nuclear envelope is formed in daughter cells during early telophase, mitotic Cajal bodies disintegrate and coilin rapidly enters the nuclear interior, where it is initially diffusely distributed. Newly re-formed Cajal bodies appear in daughter cells later in G1 phase once transcription is fully established (Carmo-Fonseca et al. 1993; Ferreira et a. 1994).
PML Nuclear Body
As cells enter mitosis, the number of PML nuclear bodies decreases by aggregation into larger Mitotic Accumulations of the PML Protein (MAPPs) (Dellaire et al. 2006; Chen et al. 2008). The PML protein, a structural component of PML nuclear bodies, is de-SUMOylated during mitosis, which affects its ability to recruit PML nuclear body components and likely influences MAPP formation (Dellaire et al. 2006). MAPPs become more mobile by relaxing their tethering to chromatin regions, and they progressively lose key components such as Sp100, Daxx or SUMO-1, from prophase to telophase. A subset of MAPPs physically interacts with mitotic chromosomes, which likely provide a source of PML protein for PML nuclear body formation in newly forming daughter nuclei. Upon exit from mitosis, after the nuclear envelope is fully reformed, Sp100 and Daxx enter daughter nuclei first before their targeting into newly formed PML nuclear bodies in early G1 phase (Chen et al. 2008). The loss of MAPPs from the cytoplasm and the increase in PML nuclear body number indicates that components of MAPPs are being incorporated into PML bodies even though a large portion of PML aggregates persist in the cytoplasm during G1 phase, slowly decreasing in number over time (Dellaire et al. 2006; Chen et al. 2008).
PHYSIOLOGICAL BIOGENESIS OF NUCLEAR BODIES
Biogenesis of nuclear bodies is important during various physiological responses. The de novo biogenesis of heat-shock bodies is a dramatic example of formation of nuclear bodies as part of a specific cellular response. Cells respond to stressful conditions by widespread inhibition of transcription and activation of specific heat-shock gene expression programs. One of the cellular events during heat shock is the transcriptional activation of tandem arrays of repetitive noncoding satellite III (sat III) repeats located on chromosome 9q12 (Valgardsdottir et al. 2008). Upon induction, the sat III transcripts remain associated with the locus and act as scaffolds for recruitment of several transcription and splicing factors to form nuclear stress bodies (Denegri et al. 2001; Alastalo et al. 2003). Their formation is strictly dependent on the presence of sat III transcripts, and they rapidly disassemble upon return to physiological temperature (Metz et al. 2004). It has been proposed that formation of nuclear stress bodies provides a dynamic means to sequester and release specific protein factors in normal and/or stressed cells and that sat III transcripts may be involved in modulating epigenetic changes during the heat-shock response (Biamonti 2004).
PML nuclear body formation has been implicated in responses to viral infection (Everett and Chelbi-Alix 2007). DNA viruses commonly localize and replicate their genomes in proximity to PML nuclear bodies (reviewed in Maul 1998). In addition, PML bodies form de novo at the sites of viral genome deposition in the nucleus (Everett and Murray 2005). Association of sites of viral DNA replication with PML nuclear bodies indicates that these nuclear bodies may enhance viral DNA replication (Sourvinos and Everett 2002). The fact that many viral proteins accumulate in PML nuclear bodies and then mediate PML nuclear body disruption suggests antiviral activity associated with these domains. In fact, failure to disrupt PML nuclear bodies during infection with HSV-1 virus inhibits viral DNA replication (Burkham et al. 2001). In line with an antiviral role of PML nuclear body biogenesis, exogenous expression of PML isoform III or PML VI can impede infection by a number of RNA and DNA viruses (Everett and Chelbi-Alix 2007) and upon treatment of cells with interferon, which is produced in response to viral infection. PML nuclear bodies undergo a dramatic increase in size and number (Lavau et al. 1995), possibly as a consequence of interferon induced expression of several PML-nuclear body components, including PML itself (Chelbi-Alix et al. 1995; Fabunmi et al. 2001).
Although not well-characterized for most nuclear bodies, their appearance also changes during development. For example, in Drosophila, nurse nuclei of the youngest egg chambers contain a single HLB and a single CB, but as the egg chamber grows, the number of HLBs increases to 8–16 HLBs with one or two prominent CBs (Liu at al. 2009). In older egg chambers, CBs break up into multiple small bodies that eventually disappear, but the number and size of HLBs remain the same and they start to accumulate coilin. Thus, at the end of oogenesis, nurse cell nuclei contain prominent HLBs positive for coilin but lack typical CBs. It seems likely that most nuclear bodies undergo changes in shape and size as organisms develop.
CONCLUSIONS
Nuclear bodies are prominent features in the nuclear landscape. They carry out important nuclear functions and contribute to cellular responses. While their functions are gradually being uncovered, their biogenesis is poorly understood. Elucidation of the molecular mechanisms for the maintenance and formation is of great interest not only as part of ongoing efforts to understand the role of nuclear architecture in genome function, but also because nuclear bodies are unique amongst cellular organelles in that they are devoid of defining membrane, yet they are distinct and stable structures. Insights into their biogenesis will likely reveal how higher-order protein aggregates form in cells and might have implications for the assembly of large macromolecular complexes and other cellular organelles.
It is highly probable that the next few years will bring dramatic progress in our still rudimentary understanding of nuclear body biogenesis. The availability of new experimental tools promises to explore the molecular basis of nuclear-body architecture. Proteomic analysis of nuclear bodies will yield a thorough understanding of the composition of each body. Using these component lists, the role of each can be systematically probed by RNAi-based approaches, which will identify the components necessary for nuclear body establishment and maintenance. Conversely, the recently developed tethering approaches allow for the first time testing of what protein, and possibly RNA, components are sufficient to form nuclear bodies. The combination of these tools should yield a thorough picture of the mechanisms and principles involved in nuclear body formation. In parallel, an even greater challenge is the full understanding of the physiological role of nuclear bodies and their formation. This question is now also amenable to experimental investigation using similar tools in systems of differentiation and disease. Although the biogenesis of nuclear bodies is still mysterious, there is little doubt that this area is likely one with the most potential for future discoveries—and surprises.
Footnotes
Editors: Tom Misteli and David L. Spector
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Alastalo TP, Hellesuo M, Sandqvist A, Hietakangas V, Kallio M, Sistonen L 2003. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J Cell Sci 116: 3557–3570 [DOI] [PubMed] [Google Scholar]
- Benavente R, Rose KM, Reimer G, Hügle-Dörr B, Scheer U 1987. Inhibition of nucleolar reformation after microinjection of antibodies to RNA polymerase I into mitotic cells. J Cell Biol 105: 1483–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP 2007. Structure, dynamics and functions of promyelocytic leukemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 [DOI] [PubMed] [Google Scholar]
- Biamonti G 2004. Nuclear stress bodies: a heterochromatin affair? Nat Rev Mol Cell Biol 5: 493–498 [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De Laurenzi V 2008. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle 7: 2357–2367 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Bond CS, Fox AH 2009. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubulya PA, Prasanth KV, Deerinck TJ, Gerlich D, Beaudouin J, Ellisman MH, Ellenberg J, Spector DL 2004. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J Cell Biol 167: 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkham J, Coen DM, Hwang CB, Weller SK 2001. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J Virol 75: 2353–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Ferreira J, Lamond AI 1993. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis–evidence that the coiled body is a kinetic nuclear structure. J Cell Biol 120: 841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M 2007. How genes find their way inside the cell nucleus. J Cell Biol 179: 1093–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix MK, Pelicano L, Quignon F, Koken MH, Venturini L, Stadler M, Pavlovic J, Degos L, de Thé H 1995. Induction of the PML protein by interferons in normal and APL cells. Leukemia 9: 2027–2033 [PubMed] [Google Scholar]
- Chen D, Huang S 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol 153: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-L, Carmichael GG 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Kappel C, Beaudouin J, Eils R, Spector DL 2008. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. Mol Biol Cell 19: 3147–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M, Lamond AI 2005. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol 21: 105–131 [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP 2006. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J Cell Sci 119: 1034–1042 [DOI] [PubMed] [Google Scholar]
- Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G 2001. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell 12: 3502–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Olson MO 1998. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol Biol Cell 9: 2407–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO 2000. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol 150: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli M 2001. Functional architecture in the cell nucleus. Biochem J 356: 297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T 2004. In vivo kinetics of Cajal body components. J Cell Biol 164: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung M-H, John S, Upender M, Ried T, Hager G, Matera GA 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179: 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Murray J 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol 79: 5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89: 819–830 [DOI] [PubMed] [Google Scholar]
- Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN 2001. Interferon gamma regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies. J Cell Sci 114: 29–36 [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Carmo-Fonseca M, Lamond AI 1994. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol 126: 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI 2005. p54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16: 5304–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A 92: 5915–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG 2001. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 154: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T, Robert-Nicoud M, Guilly MN, Hernandez-Verdun D 1992. Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. J Cell Sci 102: 729–737 [DOI] [PubMed] [Google Scholar]
- Görisch SM, Wachsmuth M, Ittrich C, Bacher CP, Rippe K, Lichter P 2004. Nuclear body movement is determined by chromatin accessibility and dynamics. Proc Natl Acad Sci U S A 101: 13221–13226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Gall JG 2006. Subnuclear organelles: new insights into form and function. Trends Cell Biol 16: 19–26 [DOI] [PubMed] [Google Scholar]
- Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD 2009. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci 122: 1872–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG 2000. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell 11: 4159–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D 2006. The nucleolus: a model for the organization of nuclear functions. Histochem. Cell Biol 126: 135–148 [DOI] [PubMed] [Google Scholar]
- Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS 2009. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol 185: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL 1996. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol 133: 719–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Konecny L, Grady DL, Kutskova YA, Cotto JJ, Morimoto RI, Vourc’h C 2002. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J Cell Biol 156: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M 2008. De novo formation of a subnuclear body. Science 322: 1713–1717 [DOI] [PubMed] [Google Scholar]
- Kentsis A, Gordon RE, Borden KL 2002. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc Natl Acad Sci U S A 99: 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Lever MA, Fischle W, Verdin E, Bazett-Jones DP, Hendzel MJ 2000. Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J Cell Biol 150: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL 2003. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi PP, Pelicci PG, Dejean A 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11: 871–876 [PubMed] [Google Scholar]
- Leung AK, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, Daigle N, Zomerdijk J, Ellenberg J, Lamond AI 2004. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J Cell Biol 166: 787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-L, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG 2009. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Cell Biol 20: 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mais C, McStay B, Scheer U 2002. On the formation of amplified nucleoli during early Xenopus oogenesis. J Struct Biol 140: 214–226 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Shpargel KB 2006. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr Opin Cell Biol 18: 317–324 [DOI] [PubMed] [Google Scholar]
- Maul GG 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20: 660–667 [DOI] [PubMed] [Google Scholar]
- Metz A, Soret J, Vourc’h C, Tazi J, Jolly C 2004. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J Cell Sci 117: 4551–4558 [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL 1997aThe dynamics of a pre-mRNA splicing factor in living cells. Nature 387: 523–527 [DOI] [PubMed] [Google Scholar]
- Misteli TD, Spector L 1997b. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends in Cell Biol 7: 135–138 [DOI] [PubMed] [Google Scholar]
- Misteli T 2001. The concept of self-organization in cellular architecture. J Cell Biol 155: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T 2007. Beyond the sequence: cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Morse R, Shaw DJ, Todd AG, Young PJ 2007. Targeting of SMN to Cajal bodies is mediated by self-association. Hum Mol Genet 16: 2349–2358 [DOI] [PubMed] [Google Scholar]
- Muratani M, Gerlich D, Janicki SM, Gebhard M, Eils R, Spector DL 2002. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat Cell Biol 4: 106–110 [DOI] [PubMed] [Google Scholar]
- Nierras CR, Liebman SW, Warner JR 1997. Does Saccharomyces need an organized nucleolus? Chromosoma 105: 444–451 [PubMed] [Google Scholar]
- Oakes M, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol 143: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MO, Dundr M 2005. The moving parts of the nucleolus. Histochem. Cell Biol 123: 203–216 [DOI] [PubMed] [Google Scholar]
- Pederson T, Tsai RY 2009. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol 184: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609 [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI 2000. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol 151: 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Lamond AI, Swedlow JR 2002. Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Biol 4: 502–508 [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL 2003. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell 14: 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL 2005. Regulating gene expression through RNA nuclear retention. Cell 123: 249–263 [DOI] [PubMed] [Google Scholar]
- Prieto JL, McStay B 2007. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev 21: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B 2008. Pseudo-NORs: a novel model for studying nucleoli. Biochim Biophys Acta 1783: 2116–2123 [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T 2009. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 106: 2525–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino TM, Gébrane-Younès J, De Mey J, Sibarita JB, Hernandez-Verdun D 2001. Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol 153: 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG 1995. The nucleolus. Annu Rev Cell Dev Biol 11: 93–121 [DOI] [PubMed] [Google Scholar]
- Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB 2003. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol 162: 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Ospina JK, Tucker KE, Matera AG, Hebert MD 2003. Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci 116: 303–312 [DOI] [PubMed] [Google Scholar]
- Sirri V, Hernandez-Verdun D, Roussel P 2002. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol 156: 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman JE, Ajuh P, Lamond AI 2001. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 114: 4407–4419 [DOI] [PubMed] [Google Scholar]
- Snaar S, Wiesmeijer K, Jochemsen AG, Tanke HJ, Dirks RW 2000. Mutational analysis of fibrillarin and its mobility in living human cells. J Cell Biol 151: 653–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourvinos G, Everett RD 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J 21: 4989–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL 2001. Nuclear domains. J Cell Sci 114: 2891–2893 [DOI] [PubMed] [Google Scholar]
- Tsai RY, McKay RD 2005. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol 168: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, Riva S, Biamonti G 2008. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res 36: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Yuh P, Heald R 2005. The perichromosomal layer. Chromosoma 114: 377–388 [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P 2008. Dynamics of component exchange at PML nuclear bodies. J Cell Sci 121: 2731–2743 [DOI] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev 14: 2283–2297 [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL 2009. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev 19: 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]