Abstract
Proteins generally must fold into precise three-dimensional conformations to fulfill their biological functions. In the cell, this fundamental process is aided by molecular chaperones, which act in preventing protein misfolding and aggregation. How this machinery assists newly synthesized polypeptide chains in navigating the complex folding energy landscape is now being understood in considerable detail. The mechanisms that ensure the maintenance of a functional proteome under normal and stress conditions are also of great medical relevance, as the aggregation of proteins that escape the cellular quality control underlies a range of debilitating diseases, including many age-of-onset neurodegenerative disorders.
Chaperones such as Hsp70 prevent protein misfolding and aggregation. They are up-regulated when cells are stressed but decline during aging, which may lead to increased cellular levels of toxic aggregates.
Proteins are the conformationally most complex and versatile biological macromolecules. They are responsible for almost all cellular functions. It is thus not surprising that cells have evolved an elaborate machinery to ensure the proper quality of their proteome. Pioneering experiments by Anfinsen over four decades ago showed that proteins can fold spontaneously in vitro, providing the fundamental insight that the linear sequence of the polypeptide chain contains all necessary information to specify a protein’s three-dimensional structure (Anfinsen 1973). These findings suggested that protein folding inside cells may also be a spontaneous process. However, since the late 1980s it has become clear that in the cell, a large fraction of newly synthesized proteins require assistance from molecular chaperones to reach their folded states efficiently and at a biologically relevant time-scale (Hartl 1996). Many of these components are so-called heat shock or stress proteins. Their major role is to prevent protein misfolding and aggregation, both under normal conditions and under the influence of stresses that increase the concentrations of improperly folded proteins. Together with the ubiquitin proteasome system and other machinery for the clearance of misfolded species, the chaperone system constitutes the central element of cellular protein quality control or proteostasis. In this article we are reviewing the principles of chaperone action in the cytoplasm and the mechanisms that allow cells to manage conditions of cytoplasmic conformational stress. Understanding the precisely balanced nature of the proteostasis network and the reasons for its gradual failure during aging is of great medical interest in light of the growing number of age-of-onset diseases that are caused by protein aggregation.
FOLDING AND AGGREGATION
Although the physical principles of protein folding have been studied intensely for almost 50 years, how the final fold (and the folding process) is determined by the amino acid sequence remains one of the most important problems in biology (Fersht 2008). Several ideas to explain the mechanisms of folding have been proposed from experiment and simulation. The famous Levinthal paradox indicated that folding cannot occur by a stochastic exploration of all possible conformations, for even a small protein would need a biologically unrealistic time frame to do so (Levinthal 1968). Indeed, the attractive and repulsive forces between neighboring amino acid residues favor certain conformations of individual amino acids in the polypeptide chain, thereby dramatically reducing the conformational space and the number of possible folding pathways available. Folding typically does not occur as an all-at-once process but involves intermediates. Their formation is considered the rule for larger proteins >100 amino acids (∼90% of all proteins in a cell), which have a significant tendency to rapidly collapse in aqueous solution into compact non-native conformations (Brockwell and Radford 2007; Bartlett and Radford 2009). Such intermediates either represent on-pathway “stepping stones” toward the native state or kinetically stable, misfolded conformations that may require substantial reorganization before the native state can be reached. The formation of metastable, non-native interactions during folding is interpreted as a consequence of the ruggedness of the funnel-shaped folding energy landscape (Fig. 1) (Onuchic and Wolynes 2004; Jahn and Radford 2005; Lindberg and Oliveberg 2007). The propensity to populate such misfolded states increases with topologically complex fold types that are stabilized by long-range interactions (e.g., α/β domain architectures) or when proteins contain multiple domains that are separate in the native state but may interact during folding (Netzer and Hartl 1997; Wright et al. 2005).
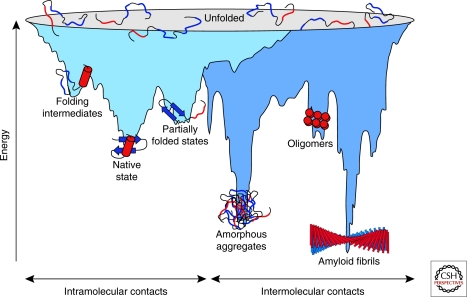
Figure 1.
Energy landscape of protein folding and aggregation. The light blue surface shows the multitude of conformations “funneling” to the native state via intramolecular contacts and the dark blue area the conformations moving toward amorphous aggregates or amyloid fibrils via intermolecular contacts. Both parts of the energy surface overlap. Aggregate formation can occur from intermediates populated during de novo folding or by destabilization of the native state and is normally prevented by molecular chaperones. Cell-toxic oligomers may occur as off-pathway intermediates of amyloid fibril formation. (Reprinted, with permission of Macmillan Publishers Ltd., from Hartl and Hayer-Hartl 2009.)
Partially folded or misfolded states often tend to aggregate because these forms typically expose hydrophobic amino acid residues and regions of unstructured polypeptide backbone, features that are largely buried in the native state. Like intramolecular folding, aggregation, i.e., the association of two or more non-native protein molecules, is driven by hydrophobic forces and predominantly results in the formation of amorphous structures that lack long-range order (Fig. 1). Alternatively, aggregation can lead to the formation of ordered, fibrillar assemblies called amyloid, in which β-strands run perpendicular to the long fibril axis (cross-β structure) (Fig. 1). Although apparently restricted to a subset of proteins under physiological conditions, these thermodynamically very stable structures are accessible to many proteins under denaturing conditions, largely independent of sequence, suggesting that their formation is an inherent property of the polypeptide chain (Dobson 2003). Importantly, this type of protein aggregation often involves the formation of less ordered, oligomeric intermediates, which are thought to be highly toxic for eukaryotic cells (Haass and Selkoe 2007), giving rise to neurodegenerative diseases, systemic amyloidoses, and other disorders.
FOLDING IN THE CELL AND THE CONCEPT OF MOLECULAR CHAPERONES
Aggregation is a concentration dependent process and is greatly enhanced in the highly crowded environment of the cell (Zimmerman and Trach 1991; Ellis and Minton 2006), which results in excluded volume effects that substantially enhance the affinities between folding intermediates. The translation process further increases the risk of misfolding and aggregation, because incomplete polypeptide chains are unable to fold into stable native conformations. Additionally, the exit channel of the large ribosomal subunit, which is approximately 100 Å long but at most 20 Å wide (Ban et al. 2000), largely precludes folding beyond the formation of α-helical elements (Woolhead et al. 2004; Lu and Deutsch 2005) and thus prevents the carboxy-terminal 40–60 amino acid (aa) residues of the chain to engage in tertiary interactions. As a consequence, productive folding can only occur after a complete protein or at least a domain (∼50 to 300 aa in length) has emerged from the ribosome, consistent with recent simulations of nascent chain folding (Elcock 2006). Because translation is relatively slow (∼25–120 s for a 500 aa protein), nascent chains are exposed in partially folded, aggregation-sensitive states for prolonged periods of time, especially in eukaryotic cells where polypeptide chains are on average longer and translation is slower than in prokaryotes. Moreover, non-native intra- or interchain contacts formed during translation could block folding on completion of synthesis and favor aggregation. Although the specific relative arrangement of ribosomes in poly-ribosome complexes helps to reduce interchain contacts (Brandt et al. 2009), molecular chaperones interact cotranslationally with many if not most nascent polypeptides and inhibit their premature (mis)folding and aggregation.
We define a molecular chaperone as any protein that interacts, stabilizes, or helps a non-native protein to acquire its native conformation, but is not present in the final functional structure (Hartl 1996; Hartl and Hayer-Hartl 2009). Chaperones are involved in a multitude of cellular functions, including de novo folding, refolding of stress-denatured proteins, oligomeric assembly, intracellular protein transport, and assistance in proteolytic degradation. The chaperones that participate broadly in protein biogenesis, such as the Hsp70s and chaperonins (Hsp60s), promiscuously recognize hydrophobic amino acid side-chains exposed by non-native proteins and promote folding through ATP-regulated binding and release cycles. Chaperone binding blocks aggregation, whereas transient release of bound hydrophobic regions is necessary for folding to proceed. It is important to realize that chaperones act not by contributing steric information to the folding process, but rather by optimizing the efficiency of folding. Notably, a number of essential proteins, such as actins and tubulins, encounter high energetic barriers in folding and are unable to reach their native states in the absence of chaperones. Their folding is mediated by barrel-shaped chaperonins, a specialized chaperone class. Because mutations often disrupt a protein’s ability to fold, it follows that the chaperone system is also important in buffering such deleterious mutations (Maisnier-Patin et al. 2005; Tang et al. 2006). This buffering function is thought to be critical in the evolution of new protein functions and phenotypic traits (Rutherford and Lindquist 1998; Kerner et al. 2005; Maisnier-Patin et al. 2005; Tokuriki and Tawfik 2009).
Different classes of structurally unrelated chaperones have been described (Chang et al. 2007; Tang et al. 2007). Their members are often known as stress proteins or heat shock proteins (Hsp), as they are up-regulated by cells under conditions of conformational stress in which the concentrations of aggregation-prone folding intermediates increase. Chaperones are usually classified according to their molecular weight (Hsp40, Hsp60, Hsp70, Hsp90, Hsp100 and the so-called small Hsps). With some exceptions, members of all these classes are present throughout the three kingdoms of life.
THE CYTOSOLIC CHAPERONE MACHINERY
The chaperone pathways acting in protein folding in the cytosol follow general organizational principles (Frydman 2001; Hartl and Hayer-Hartl 2002). In all three domains of life, a first tier of ribosome-associated chaperones function in stabilizing nascent polypeptides on ribosomes and initiating their folding, whereas a second tier of components act downstream in completing the folding process (Fig. 2) (Langer et al. 1992; Albanese et al. 2006). Both systems cooperate in coherent pathways.
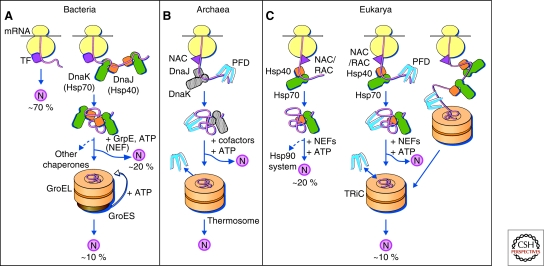
Figure 2.
Protein folding in the cytosol. (A) Models for the chaperone-assisted folding of newly synthesized polypeptides in the cytosol. Bacteria (left panel). TF, trigger factor; N, native protein. Nascent chains probably interact generally with TF, and most small proteins (∼70% of total) may fold rapidly on synthesis without further assistance. Longer chains interact subsequently with DnaK and DnaJ (Hsp70 system) and fold on one or several cycles of ATP-dependent binding and release (∼20% of total). About 10% of chains transit the chaperonin system (GroEL and GroES) for folding. (B) Archaea (middle panel). PFD, prefoldin; NAC, nascent chain-associated complex. Note that only some archaeal species contain DnaK/DnaJ. (C) Eukarya (right panel). Like TF, NAC (nascent chain associated complex) probably interacts generally with nascent chains, but the role of NAC in folding is not yet clear. About 20% of chains reach their native states in a reaction assisted by RAC (ribosome associated complex), Hsp70, and Hsp40. A fraction of these must be transferred to Hsp90 for folding. About 10% of chains are co- or posttranslationally passed on to the chaperonin TRiC in a reaction mediated by Hsp70 and PFD, both of which interact directly with TRiC. PFD recognizes the nascent chains of certain TRiC substrates, including actin and tubulins. (Modified, with permission of The American Association for the Advancement of Science, from Hartl and Hayer-Hartl 2002.)
RIBOSOME-ASSOCIATED CHAPERONES
This group of chaperones includes bacterial trigger factor (TF) (Fig. 2A) and eukaryotic ribosome-associated complex (RAC) (Fig. 2B,C), both of which dock on to the large ribosomal subunit close to the polypeptide exit site (Kramer et al. 2009). RAC consist of the Hsp70 homologs Ssb1/2, Ssz1, and the Hsp40 homolog zuotin in Saccharomyces cerevisiae or of Hsp70L1/Mpp11, homologs of Ssz1/zuotin, in higher eukaryotes (Hundley et al. 2005; Otto et al. 2005). Additionally, archaea and eukarya contain the nascent chain-associated complex, NAC (Chang et al. 2007; Tang et al. 2007; Kramer et al. 2009) (Fig. 2B,C). Among these, the function of TF is best understood. TF is an abundant ∼50 kDa protein, consisting of an amino-terminal ribosome-binding domain, a peptidyl-prolyl isomerase (PPIase) domain, and a carboxy-terminal domain, the latter being positioned in the structure between the amino and PPIase domains (Ferbitz et al. 2004) (Fig. 3A, left panel). All three domains participate in binding to hydrophobic elements of nascent chains. The amino-domain mediates the interaction with the ribosome (Kaiser et al. 2006; Lakshmipathy et al. 2007; Merz et al. 2008). The carboxy-domain, containing two armlike protrusions, is the major binding region for hydrophobic nascent chain segments (Merz et al. 2006; Lakshmipathy et al. 2007). Ribosome binding causes a conformational aperture of TF, presumably activating TF for nascent chain interaction (Fig. 3A, right panel) (Baram et al. 2005; Schlunzen et al. 2005; Kaiser et al. 2006). Chain release from TF is ATP-independent and is probably facilitated by the tendency of the bound polypeptide to bury hydrophobic regions. This may occur to some extent within the protected environment of TF (Ferbitz et al. 2004; Merz et al. 2008). However, when a nascent chain exposes strongly hydrophobic segments, TF may leave the ribosome while remaining associated with the elongating chain (Fig. 3A, right panel) (Agashe et al. 2004). The eventual dissociation of TF facilitates folding or polypeptide transfer to downstream chaperones, such as the Hsp70 protein DnaK. Evidence has also been presented for an alternative role of TF in the assembly of certain ribosomal proteins (Martinez-Hackert and Hendrickson 2009). In this case, TF is thought to transiently stabilize the folded structure of a nonassembled subunit predominantly through electrostatic interactions.
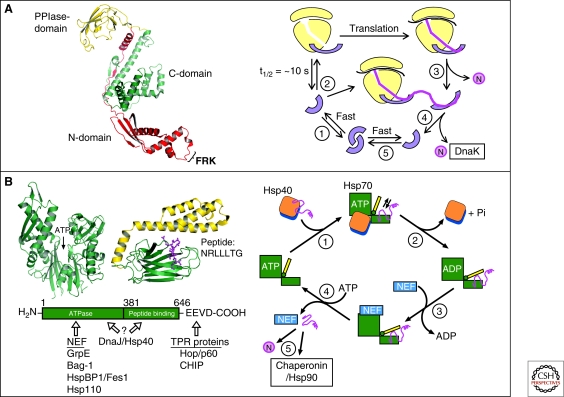
Figure 3.
Chaperone paradigms: Trigger factor and Hsp70 system. (A) Trigger factor (TF): The left panel shows the domain structure of TF, including the FRK ribosome-binding loop in the amino-domain (PDB 1W26) (Ferbitz et al. 2004), and the right panel shows a model of the TF reaction cycle (modified from Kaiser et al. [2006]). (1) Free TF is in rapid equilibrium between monomeric and dimeric states. (2) TF monomer binds to nontranslating ribosomes with a KD of ∼1 µM and a mean residence time of approximately 10 seconds. Ribosome binding causes a conformational expansion of TF and may activate TF for interaction with nascent chains. (3) Nascent chains that interact weakly with TF may begin to compact cotranslationally in the vicinity of TF. Release of TF from nascent chain coincides with TF dissociation from the ribosome and allows completion of folding to native state (N). (4) Structurally more complex proteins may strongly interact with TF. TF remains bound to nascent chain after dissociating from the ribosome and a new TF molecule can enter at the ribosome. Eventual chain dissociation from TF facilitates transfer to DnaK or folding. (5) Released TF enters the monomer-dimer pool. (B) Hsp70 system. Left panel: Structures of the ATPase domain (PDB 1DKG) and the peptide-binding domain (PDB 1DKZ) (Zhu et al. 1996) of Hsp70 shown representatively for Escherichia coli DnaK. The α-helical lid of the peptide binding domain is shown in yellow and the extended peptide substrate as a ball-and-stick model in pink. ATP indicates the position of the nucleotide binding site. The amino acid sequence of the peptide is indicated in single-letter code. The interaction of prokaryotic and eukaryotic cofactors with Hsp70 is shown schematically. Residue numbers refer to human Hsp70. Only the Hsp70 proteins of the eukaryotic cytosol have the COOH-terminal sequence EEVD that is involved in binding of tetratricopeptide repeat (TPR) cofactors (Scheufler et al. 2000). Right panel: Hsp70 reaction cycle. NEF, nucleotide exchange factor (GrpE in case of E. coli DnaK; Bag, HspBP1, and Hsp110 in case of eukaryotic cytosolic Hsp70). N, native protein. (A,B: Modified with permission from Hartl and Hayer-Hartl 2009.)
THE HSP70 CHAPERONE FAMILY
All cells, except certain archaea, contain constitutive and stress-inducible cytosolic Hsp70s that do not bind directly to the ribosome, such as bacterial DnaK, yeast Ssa1-4, and the Hsc70/Hsp70 proteins of higher eukaryotes (Chang et al. 2007). Besides co- or post-translationally assisting in the folding of approximately 20% of newly synthesized polypeptides (Teter et al. 1999; Thulasiraman et al. 1999), these components have a variety of additional functions, including protein trafficking and assistance in the proteolytic degradation of terminally misfolded proteins. Hsp70s generally function in cooperation with Hsp40 or J-proteins and with various nucleotide exchange factors (NEFs). As a common structural feature, all Hsp70 family members consist of an amino-terminal ATPase and a carboxy-terminal substrate binding domain, the latter being composed of a β-sandwich subdomain and an α-helical lid segment (Zhu et al. 1996) (Fig. 3B, left panel). The β-sandwich recognizes extended, approximately seven-residue segments enriched with hydrophobic amino acids (Rüdiger et al. 1997). Such segments occur on average every 50–100 residues in proteins but are typically only exposed in nascent or non-native chains. The α-helical lid and a conformational change in the β-sandwich domain regulate the affinity state for peptide in an ATP-dependent manner (Mayer et al. 2000; Pellecchia et al. 2000). In the ATP-bound state, the lid adopts an open conformation, allowing peptide binding. Hydrolysis of ATP to ADP is strongly accelerated by Hsp40, leading to lid closure and stable peptide binding (Fig. 3B, right panel). Interaction of substrate with Hsp70 is mediated by the so-called J-domain that is present in all Hsp40s (Mayer et al. 2000). Hsp40s also interact directly with unfolded polypeptides and can recruit Hsp70 to protein substrates (Fig. 3B, right panel) (Young et al. 2003). Following ATP-hydrolysis, various NEFs (GrpE in bacteria; Bag, HspBP1, or Hsp110 in eukaryotes) bind to the Hsp70 ATPase domain and catalyze ADP-ATP exchange, which results in lid-opening and substrate release, thereby completing the reaction cycle (Fig. 3B, right panel).
Hsp70-mediated folding and prevention of aggregation may be explained by a process of kinetic partitioning: Binding of Hsp70 to non-native substrate hinders aggregation by transiently shielding exposed hydrophobic segments. Release allows fast-folding molecules (or domains) to bury hydrophobic residues, whereas molecules that need longer than a few seconds to fold will rebind to Hsp70 and avoid aggregation. A subset of proteins that are unable to partition to fast-folding trajectories on Hsp70 cycling remain stabilized in a nonaggregated state and must be transferred into the specialized environment of the chaperonin cage for folding (Langer et al. 1992; Kerner et al. 2005).
THE CHAPERONINS
Chaperonins are large double-ring complexes of approximately 800 kDa. Two groups of chaperonin exist (Horwich et al. 2007; Tang et al. 2007): Group I chaperonins (also called Hsp60s) occur in bacteria (GroEL), mitochondria, and chloroplasts. They have seven-membered rings and functionally cooperate with Hsp10 proteins (GroES in bacteria), which form the lid of the folding cage. The group II chaperonins in archaea (thermosome) and in the eukaryotic cytosol (TRiC/CCT) have eight- or nine-membered rings. They are independent of Hsp10 factors, as their lid function is built into the chaperonin ring in the form of specialized α-helical extensions. Unlike the Hsp70s, the chaperonins promote folding through ATP-regulated cycles of global protein encapsulation.
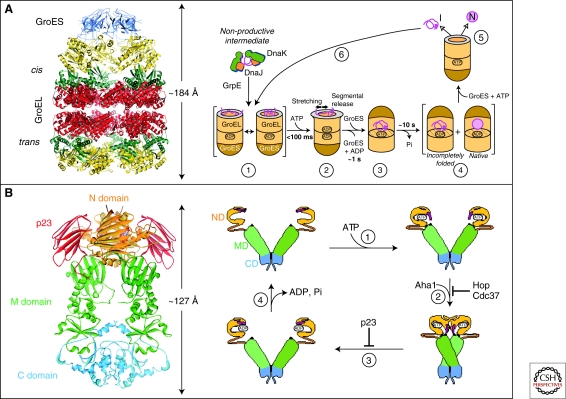
The GroEL/GroES chaperonin system of E. coli has been studied most extensively (Hartl 1996; Hartl and Hayer-Hartl 2002; Horwich et al. 2007). GroEL interacts with at least approximately 250 different cytosolic proteins. Most of these are between 20 and 50 kDa in size and have complex α/β or α+β domain topologies (Kerner et al. 2005). Such proteins are stabilized by many long-range interactions and tend to populate kinetically trapped folding intermediates (Gromiha and Selvaraj 2004). The GroEL double-ring is composed of approximately 57 kDa subunits that consist of an equatorial ATPase domain, an intermediate hinge-domain, and an apical substrate-binding domain. The apical domains expose hydrophobic amino acid residues for substrate binding toward the ring center. GroES is a heptameric ring of approximately 10 kDa subunits that covers the ends of the GroEL cylinder (Fig. 4A, left panel) (Xu et al. 1997; Horwich et al. 2007). Recent biophysical measurements have shown that GroEL-bound substrates populate non-native states that lack stable tertiary interactions (Fig. 4A, right panel) (Horst et al. 2005; Hillger et al. 2008; Sharma et al. 2008). Folding depends on the global encapsulation of the substrate in the chaperonin ring by GroES (Fig. 4A, right panel) (Brinker et al. 2001; Tang et al. 2006; Horwich et al. 2007). GroES binding is ATP-regulated and causes a dramatic conformational change of the system that leads to the formation of a cage with a highly hydrophilic, net-negatively charged inner wall. Encapsulated protein up to approximately 60 kDa is free to fold in this environment for 10–15 seconds, the time needed for ATP hydrolysis in the GroES-bound ring (cis-ring). Protein substrate leaves the cage on GroES dissociation, which is induced by ATP binding in the opposite ring (trans-ring). Not-yet folded substrate rapidly rebinds to GroEL for further folding attempts (Fig. 4A, right panel). Proteins that exceed the size limit of the chaperonin cage (∼60 kDa) either use the Hsp70 system for folding (Agashe et al. 2004; Kerner et al. 2005) or may reach native state through binding and release from GroEL without encapsulation (Chaudhuri et al. 2001).
Figure 4.
Chaperone paradigms: Chaperonins and Hsp90 system. (A) The GroEL-GroES chaperonin. Left panel: Crystal structure of the asymmetric GroEL-GroES complex (PDB 1AON) (Xu et al. 1997). Cis, the GroES-bound chamber of GroEL. Trans, the opposite GroEL ring. Right panel: Working model summarizing the conformational changes in a substrate protein on transfer from DnaK/DnaJ (Hsp70 system) to GroEL and during GroEL/GroES-mediated folding. Note that binding of a second substrate molecule to the open ring of GroEL in steps 4 and 5 as well as the transient formation of a symmetrical GroEL-GroES2 complex is omitted for simplicity. (1) Substrate protein may be delivered to GroEL by DnaK/DnaJ in a nonaggregated, but kinetically trapped state. On binding to GroEL it undergoes local unfolding to an ensemble of expanded and more compact conformations. (2) ATP-dependent domain movement of the apical GroEL domains result in stretching of tightly bound regions of substrate and in release and partial compaction of less stably bound regions. (3) Compaction is completed on substrate encapsulation by GroES. (4) Folding in the chaperonin cage. (5) Substrate release on GroES dissociation. (6) Rebinding of incompletely folded states. N, native state; I, folding intermediate. (Reprinted, with permission of Macmillan Publishers Ltd., from Hartl and Hayer-Hartl 2009.) (B) The Hsp90 system. Left panel: Crystal structure of the Hsp90 dimer from S. cerevisiae with the regulator p23 bound to the amino-domain (PDB 2CG9) (Ali et al. 2006). Right panel: ATPase cycle of Hsp90. On ATP binding (1) the amino-terminal ATPase domain (ND) of Hsp90 undergoes a conformational change leading to the closure of the ATP lid. After lid closure, the first 24 amino acid residues of each Hsp90 monomer dimerize and the first β-strand and α-helix swap to associate with the ND of the other monomer (2). Furthermore, in each monomer, the ND contacts the corresponding M-domain (MD). This metastable conformation is committed for ATP hydrolysis (3). This results in a compaction of the Hsp90 dimer, in which the individual monomers twist around each other. After hydrolysis (4), the NDs dissociate and both monomers separate amino-terminally. Various cofactors regulate this cycle: Cdc37, which delivers certain kinase substrates to Hsp90, inhibits the ATPase activity, and Hop, a TPR-protein that joins Hsp70 to Hsp90, inhibits ND dimerization. Aha1 stimulates ATP hydrolysis, whereas p23 stabilizes the dimerized form of Hsp90 before ATP hydrolysis. These factors are thought to adjust the kinetic properties of the cycle to achieve certain conformational transitions in Hsp90-bound client proteins, as well as their release from Hsp90. (Modified, with permission, from Wandinger et al. 2008.)
Enclosing unfolded protein, one molecule at a time, avoids aggregation during folding. In addition, an effect of steric confinement likely modulates the folding energy landscape. Notably, larger substrates fully occupy the limited volume of the GroEL/GroES nano-cage (Clare et al. 2009). Although it has been suggested that the chaperonin merely functions as a passive aggregation-prevention device (Apetri and Horwich 2008), other studies provided evidence that encapsulation can substantially accelerate folding over the rate of spontaneous folding (measured in the absence of aggregation) (Brinker et al. 2001; Tang et al. 2006; Chakraborty et al. 2010). This rate acceleration may be attributed to steric confinement entropically destabilizing misfolded states and promoting their conversion to more compact, native-like conformations, consistent with confinement theory (Baumketner et al. 2003; Hayer-Hartl and Minton 2006). Mutational analysis showed that the polar residues of the cavity wall are critical in this process (Wang et al. 2002; Tang et al. 2006; Tang et al. 2008). These polar residues are expected to promote folding by accumulating ordered water molecules in their vicinity, thereby generating a local environment in which a substrate protein is forced to bury exposed hydrophobic residues more effectively (England et al. 2008). Other elements of the chaperonin mechanism may also contribute to accelerating folding. For example, it has been suggested that repeated unfolding events in successive binding and release cycles may help to reverse kinetically trapped states (“iterative annealing”) (Thirumalai and Lorimer 2001; Lin et al. 2008).
The group II chaperonin in the eukaryotic cytosol is called TRiC (for TCP-1 ring complex) or CCT (for chaperonin containing TCP-1). It is hetero-oligomeric and consists of eight paralogous subunits per ring (Frydman 2001). The general domain structure of the group II chaperonins is similar to that of GroEL (Ditzel et al. 1998). However, all group II chaperonins deviate from GroEL in that their apical domains contain finger-like protrusion, which act as a built-in lid and replace the function of GroES. These segments open and close in an ATP-dependent reaction cycle (Meyer et al. 2003). Class II chaperonins interact functionally with the co-chaperone prefoldin (Geissler et al. 1998; Vainberg et al. 1998; Leroux et al. 1999; Siegers et al. 1999; Siegert et al. 2000), which functions in substrate transfer to chaperonin (Martin-Benito et al. 2002). Substrate transfer to TRiC is also mediated by Hsp70, consistent with the general sequence of chaperone interactions in the cytosolic folding pathway (Langer et al. 1992; Frydman et al. 1994) (Fig. 2). Hsp70 appears to fulfill this role by interacting directly with TRiC (Cuellar et al. 2008).
TRiC/CCT interacts with approximately 10 % of newly synthesized cytosolic proteins (Thulasiraman et al. 1999; Gavin et al. 2002; Spiess et al. 2004), including actin and tubulins as the most abundant substrates (Gao et al. 1992; Yaffe et al. 1992). TRiC substrates do not share any apparent similarity in sequence or structure, except for a certain preponderance of proteins with WD40 β-propeller domains (Ho et al. 2002; Valpuesta et al. 2002). Several substrates are 100–120 kDa in size, suggesting that TRiC may be able to assist the co-translational folding of individual domains of larger proteins, consistent with its capacity to interact with nascent polypeptide chains (Frydman et al. 1994; Etchells et al. 2005). As in the case of GroEL, protein folding by TRiC involves ATP-dependent substrate encapsulation in the chaperonin cavity (Meyer et al. 2003; Shimon et al. 2008). The TRiC reaction cycle is much slower than that of GroEL, potentially providing a substantially longer period of protein encapsulation and folding in the chaperonin cage.
THE Hsp90 SYSTEM
Hsp90 is another important chaperone system that functions downstream of Hsp70 in the final structural maturation and conformational regulation of a number of signaling molecules and transcription factors (Pearl and Prodromou 2006; Zhao and Houry 2007). Hsp90 cooperates in this process with a multitude of regulators and cochaperones, many of which contain tetratricopeptide repeat (TPR) domains (Scheufler et al. 2000). As a result, the Hsp90 system is of considerable complexity and the mechanisms by which this machinery mediates client protein conformational changes are not yet fully understood (Wandinger et al. 2008). However, recent crystal structures of full-length Hsp90s provided long awaited new information (Ali et al. 2006; Shiau et al. 2006; Dollins et al. 2007) (Fig. 4B, left panel). Hsp90 functions as a dimer. Similar to other chaperones, the Hsp90 cycle is ATP-driven and is accompanied by considerable structural rearrangements (Hessling et al. 2009) (Fig. 4B, right panel). How these changes translate into structural work on a bound client protein is not yet clear, however. A number of cofactors modulate the reaction cycle. Some of these, such as the TPR-protein Hop, form a link between Hsp90 and Hsp70, facilitating substrate transfer (Scheufler et al. 2000). Hsp90 appears to have several substrate interaction regions and the interaction strength seems to be strongly influenced by a substrate’s structural flexibility. For example, the exchange of only a few amino acids in the viral v-Src renders this kinase strongly dependent on Hsp90 as compared with the cellular c-Src variant (Xu and Lindquist 1993; Falsone et al. 2004). This observation is in line with the proposed role of Hsp90 as an evolutionary capacitor in protecting mutated and thus destabilized forms of proteins from degradation (Rutherford and Lindquist 1998; Queitsch et al. 2002; Cowen and Lindquist 2005). When displaced from Hsp90 by other denatured proteins under stress conditions, these mutant proteins may provide a critical advantage to the organism, as shown in Drosophila and plants. The regulatory role of Hsp90 extends to a large variety of cellular processes, including vesicular transport, the cell cycle, and cell division (McClellan et al. 2007). Because a number of Hsp90 substrates are kinases with well-documented roles in tumor development, inhibition of Hsp90 with drugs like geldanamycin has emerged as a new therapeutic strategy for certain cancers (Neckers 2007).
CYTOSOLIC STRESS
The concept of stress as a disturbance of homeostasis is of fundamental importance in biology. Although initially formulated in physiological and clinical contexts, we are now beginning to understand cell stress at the level of precise biochemical mechanisms and networks. Of central concern are environmental insults that affect proteostasis, the stability of the proteome (Balch et al. 2008), as its acute or chronic disturbance is increasingly recognized as the cause of disease. Except perhaps for specific chemical noxes, stress generally affects the cell as a system. The classical physical stressors include heat shock and cold shock, osmolality changes, and pH fluctuations. Aging may be considered as a chronic form of proteome stress (Cohen et al. 2006). Heat shock is the best understood stressor in regard to its physico-chemical consequences for the proteome. Proteins are dynamic polymers that can leave their native conformation, fluctuating into and out of aggregation-prone, near-native states (Chiti and Dobson 2009), and additional input of thermal energy can critically shift the conformational equilibrium of a subset of proteins to more unfolded states. As a result, the danger of proteins aggregating via exposed hydrophobic surfaces increases sharply, explaining the need for the rapid up-regulation of molecular chaperones (stress proteins) under these conditions. In a similar way cells respond to various covalent modifications of proteins, including prominently those elicited by reactive oxygen species. Generally, cells seem to be better equipped to respond to acute proteome stress than to chronic forms, as it may occur during aging (Morimoto 2008).
STRESS RESPONSE—THE HSF AXIS
Molecular chaperones that are transcriptionally up-regulated under stress participate in the restoration of proteostasis in various ways. As in de novo folding, their primary role is to prevent protein aggregation by shielding the hydrophobic surfaces exposed by partially unfolded proteins. They may facilitate refolding when more favorable conditions return. Moreover, these chaperones cooperate with various proteolytic systems, primarily the ubiquitin proteasome system (UPS) and autophagy (Chiang et al. 1989; Dice 1990; Arndt et al. 2007; Kon and Cuervo 2009), in clearing terminally misfolded proteins. As suggested more recently, chaperones may also have a role in mediating the temporary deposition of misfolded proteins as large aggregates for later resolubilization (Weibezahn et al. 2005; Kaganovich et al. 2008). This may be beneficial, because large aggregates expose less potentially dangerous surfaces than the monomeric or oligomeric states of non-native chains.
In E. coli, the transcriptional up-regulation of the heat shock genes on stress is mainly regulated by σ32, a transcriptional activator of RNA polymerase with specificity for heat shock genes (Grossman et al. 1984; Landick et al. 1984; Cowing et al. 1985; Bukau 1993). A dramatic up-regulation in chaperone capacity is observed on shift to high temperatures, such as 46°C, with DnaK and GroEL accounting for as much as 20% of cellular protein (Herendeen et al. 1979; Bukau 1993). The rapid synthesis of co-chaperones, such as DnaJ and GrpE, as well as of the La and Clp proteases is also observed under these conditions.
The transcriptional induction of the stress response in higher eukaryotes is a highly regulated multistep event. Activation of heat shock transcription factors (HSFs) is central to this process. Four different HSFs (HSF1-4) are known in vertebrates (Akerfelt et al. 2007), with HSF1 being considered the master regulator for the induced transcription of hsp genes during most stress conditions (Morimoto 1998; Akerfelt et al. 2007). Simultaneous activation of HSF1 and HSF2 has been described in stress conditions triggered by proteosomal malfunction (Ostling et al. 2007). HSF1 and HSF2 are also involved, as a heterotrimer, in regulating the transcription of satellite III DNA at the nuclear stress bodies, although the consequence of this process remains elusive (Sandqvist et al. 2009). HSF3 is mainly present in avian species, except for the recent report of a novel functional HSF3 in mouse (Fujimoto et al. 2009). There is as yet no evidence for the involvement of HSF4 in the stress response (Akerfelt et al. 2007).
Under normal physiological conditions, HSF1 is present as an inert monomer, mostly bound to chaperones, either in the cytoplasm or nucleus, or both (Morimoto 1998; Vujanac et al. 2005; Whitesell and Lindquist 2009) (Fig. 5). Displacement from chaperones by stress-denatured proteins, as well as various posttranslational modifications, including phosphorylation/dephosphorylation, acetylation/deacetylation, and sumolyation during stress play a role in shifting the monomeric HSF1 to an active trimeric form, which is only present in the nucleus (Hong et al. 2001; Guettouche et al. 2005; Westerheide et al. 2009). This trimeric form of HSF1 is competent for DNA binding at the conserved promoter sequence of the 5′ flanking regions of hsp genes, the so called heat shock element (HSE) (Lis and Wu 1993). Various transcriptional cofactors may regulate this step (Gomez et al. 2008; Xu et al. 2008). HSF1 binding to the HSE promotes the transactivation function of HSF1 through interaction with various regulatory factors downstream of the HSE. This sequence of events culminates in the activation of RNA polymerase II, thus inducing transcription (Lis and Wu 1993; Shamovsky and Nudler 2008; Whitesell and Lindquist 2009). According to a recent report, HSF1 is not only self-regulated by various posttranslational modifications but also specifically interacts with histone deacetylases, HDAC1 and HDAC2, to regulate gene expression during heat shock through large-scale epigenetic remodeling by histone deacetylation (Fritah et al. 2009).
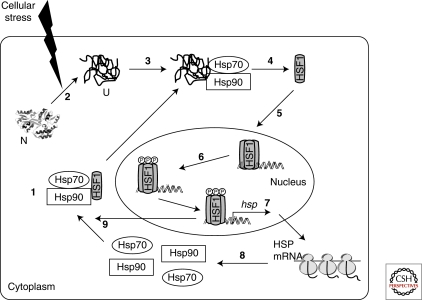
Figure 5.
Transcriptional regulation of heat shock proteins by HSF1. (1) Under normal conditions HSF1 exists as an inert monomer in a complex with Hsp90 and Hsp70. (2) Cellular stress increases the amount of denatured proteins in the cytoplasm. (3) Denatured proteins bind to Hsp70 and Hsp90, resulting in the displacement of HSF1 (4). (5) HSF1 then trimerizes and translocates to the nucleus where it undergoes a series of posttranslational modifications, including phosphorylation (6). (7) Activated HSF1 trimer induces the transcription of a number of hsp genes, resulting in the translation of HSPs, including Hsp90 and Hsp70 (8). (9) The increased cellular concentration of these chaperones in turn inactivates HSF1 by binding to monomeric or trimeric forms of HSF1. N, native protein; U, unfolded protein. (Powers and Workman 2007.)
Chaperones have a critical role in the stress response as negative regulators of HSF1 (Fig. 5). In particular, the Hsp90 multichaperone complex is important in this regard. Both, in vitro and in vivo experiments have shown that Hsp90 binds HSF1 in the inactive, monomeric form (Zou et al. 1998; Guo et al. 2001). The application of geldanamycin and other ansamycins to mammalian cells, which inhibit Hsp90 by blocking its ATP binding pocket (Prodromou et al. 1997; Stebbins et al. 1997), disrupts the Hsp90-HSF1 complex and induces HSF1 activation (Murakami et al. 1991; Grenert et al. 1997; Prodromou et al. 1997; Zou et al. 1998). As shown in an in vitro system derived from HeLa cells, immunodepletion of Hsp90 induces HSF1 trimerization and HSE binding activity (Zou et al. 1998). Because Hsp90 has an important role in stabilizing the non-native states of proteins in the cytosol, the increased concentration of unfolded proteins under stress conditions causes the displacement of HSF1 from Hsp90, triggering its trimer formation and activation for transcription by phosphorylation (Morimoto 1998; Voellmy and Boellmann 2007) (Fig. 5). Hsp70 and Hsp40 are associated with Hsp90 multichaperone complexes under various conditions (Shi et al. 1998) and a direct interaction of Hsp70 with HSF1 trimers has been observed during the attenuation of the stress response (Abravaya et al. 1992; Baler et al. 1992; Shi et al. 1998). Overexpression of Hsp70 or Hsp40 can repress HSF1 transcriptional activity (Shi et al. 1998), further suggesting a role of these chaperones in HSF1 regulation, either as part of the Hsp90 multichaperone complex or independently. During the attenuation of the stress response, as the level of non-native proteins decreases, HSF1 shifts back to its inactive, Hsp90-bound state (Fig. 5).
IMPLICATIONS IN AGING AND DISEASE
An important insight from studies in recent years is that the aging process goes along with a decline in the cellular capacity to maintain proteostasis (Balch et al. 2008; Morimoto 2008). This decline of folding capacity has been shown in Caenorhabditis elegans by a semiquantitative phenotypic analysis of temperature sensitive mutants of different metastable proteins (Ben-Zvi et al. 2009). Although the mechanisms underlying this phenomenon are not yet fully understood, a decrease in the production of molecular chaperones and other components of the proteostasis network is frequently observed during aging in various systems (Rattan 1995; Rattan 1998; Finkel and Holbrook 2000). Interestingly, the level of HSF1 itself appears not to be affected, rather its activation is impaired, presumably as a result of a deregulation in the signal transduction pathways involved (Heydari et al. 2000; Soti and Csermely 2000; Morimoto 2008). As a consequence, aging renders cells increasingly sensitive toward protein misfolding, providing a plausible explanation for why age is the major risk factor for neurodegenerative diseases and other disorders associated with amyloid-like protein aggregation.
Although the toxic principle operating in the age-of-onset diseases is far from being understood, a consensus is emerging that oligomeric, soluble states of the respective disease protein are the primary cytotoxic species. These aggregation intermediates are thought to interact aberrantly with other proteins or membranes, altering their functional properties (Barral et al. 2004). Interestingly, molecular chaperones of several classes, most prominently the Hsp70s, have been shown to inhibit the formation of such oligomers and to deviate the aggregation pathway from the amyloidogenic route toward the formation of amorphous aggregates (Muchowski et al. 2000; Schaffar et al. 2004; Behrends et al. 2006; Kitamura et al. 2006; Tam et al. 2006). In the case of Huntington’s disease and related disorders, Hsp70 may cooperate with the cytosolic chaperonin TRiC/CCT in modulating aggregation and facilitating the formation of benign oligomers of the disease protein (Behrends et al. 2006; Kitamura et al. 2006; Tam et al. 2006).
CONCLUDING REMARKS
The discovery and detailed analysis of the cellular machinery of protein folding has led to unexpected insights of tremendous medical relevance. We are only just beginning to comprehend the enormous implications of molecular chaperone functions and the stress response in preventing disease and possibly in delaying aging. Searching for ways to re-establish the normal balance between chaperone capacity and the production of misfolded proteins may offer an effective therapeutic strategy, as suggested by several proof-of-principle experiments. For example, induction of the HSF1-mediated stress response by small molecules can result in a powerful up-regulation of the chaperone network sufficient to prevent the aggregation and toxicity of huntingtin in cell culture models (Sittler et al. 2001; Westerheide and Morimoto 2005). In contrast, the well documented dependence of cancer cells on molecular chaperones, such as Hsp70 and Hsp90, has added chaperone inhibitors to the arsenal of anticancer drugs (de Billy et al. 2009; Whitesell and Lindquist 2009). Hsp90 inhibitors are already in clinical trials, with promising results (Taldone et al. 2008; Hwang et al. 2009), and Hsp70 inhibitors are in the pipeline (Galluzzi et al. 2009; Leu et al. 2009). Taken together, the chaperone field is an excellent example for how curiosity-driven research can open up rich opportunities in improving human health.
Footnotes
Editors: Richard Morimoto, Jeff Kelly, and Dennis Selkoe
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Abravaya K, Myers MP, Murphy SP, Morimoto RI 1992. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev 6: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM 2004. Function of trigger factor and DnaK in multidomain protein folding: Increase in yield at the expense of folding speed. Cell 117: 199–209 [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Trouillet D, Mezger V, Sistonen L 2007. Heat shock factors at a crossroad between stress and development. Ann N Y Acad Sci 1113: 15–27 [DOI] [PubMed] [Google Scholar]
- Albanese V, Yam AYW, Baughman J, Parnot C, Frydman J 2006. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124: 75–88 [DOI] [PubMed] [Google Scholar]
- Ali MMU, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH 2006. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440: 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB 1973. Principles that govern the folding of protein chains. Science 181: 223–230 [DOI] [PubMed] [Google Scholar]
- Apetri AC, Horwich AL 2008. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc Natl Acad Sci 105: 17351–17355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V, Rogon C, Hohfeld J 2007. To be, or not to be—molecular chaperones in protein degradation. Cell Mol Life Sci 64: 2525–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW 2008. Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Baler R, Welch WJ, Voellmy R 1992. Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J Cell Biol 117: 1151–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Baram D, Pyetan E, Sittner A, Auerbach-Nevo T, Bashan A, Yonath A 2005. Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action. Proc Natl Acad Sci 102: 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Broadley SA, Schaffar G, Hartl FU 2004. Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol 15: 17–29 [DOI] [PubMed] [Google Scholar]
- Bartlett AI, Radford SE 2009. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat Struct Mol Biol 16: 582–588 [DOI] [PubMed] [Google Scholar]
- Baumketner A, Jewett A, Shea JE 2003. Effects of confinement in chaperonin assisted protein folding: Rate enhancement by decreasing the roughness of the folding energy landscape. J Mol Biol 332: 701–713 [DOI] [PubMed] [Google Scholar]
- Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, et al. 2006. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell 23: 887–897 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci 106: 14914–14919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W 2009. The native 3D organization of bacterial polysomes. Cell 136: 261–271 [DOI] [PubMed] [Google Scholar]
- Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M 2001. Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107: 223–233 [DOI] [PubMed] [Google Scholar]
- Brockwell DJ, Radford SE 2007. Intermediates: Ubiquitous species on folding energy landscapes? Curr Opin Struct Biol 17: 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B 1993. Regulation of the Escherichia coli heat-shock response. Mol Microbiol 9: 671–680 [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M 2010. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell 142: 112–122 [DOI] [PubMed] [Google Scholar]
- Chang HC, Tang YC, Hayer-Hartl M, Hartl FU 2007. SnapShot: Molecular chaperones, Part I. Cell 128: 212. [DOI] [PubMed] [Google Scholar]
- Chaudhuri TK, Farr GW, Fenton WA, Rospert S, Horwich AL 2001. GroEL/GroES-mediated folding of a protein too large to be encapsulated. Cell 107: 235–246 [DOI] [PubMed] [Google Scholar]
- Chiang H-L, Terlecky SR, Plant CP, Dice JF 1989. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246: 382–385 [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM 2009. Amyloid formation by globular proteins under native conditions. Nat Chem Biol 5: 15–22 [DOI] [PubMed] [Google Scholar]
- Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR 2009. Chaperonin complex with a newly folded protein encapsulated in the folding chamber. Nature 457: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A 2006. Opposing activities protect against age-onset proteotoxicity. Science 313: 1604–1610 [DOI] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S 2005. Hsp90 potentiates the rapid evolution of new traits: Drug resistance in diverse fungi. Science 309: 2185–2189 [DOI] [PubMed] [Google Scholar]
- Cowing DW, Bardwell JC, Craig EA, Woolford C, Hendrix RW, Gross CA 1985. Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci 82: 2679–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar J, Martin-Benito J, Scheres SH, Sousa R, Moro F, Lopez-Vinas E, Gomez-Puertas P, Muga A, Carrascosa JL, Valpuesta JM 2008. The structure of CCT-Hsc70 NBD suggests a mechanism for Hsp70 delivery of substrates to the chaperonin. Nat Struct Mol Biol 15: 858–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Billy E, Powers MV, Smith JR, Workman P 2009. Drugging the heat shock factor 1 pathway Exploitation of the critical cancer cell dependence on the guardian of the proteome. Cell Cycle 8: 3806–3808 [DOI] [PubMed] [Google Scholar]
- Dice JF 1990. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci 15: 305–309 [DOI] [PubMed] [Google Scholar]
- Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S 1998. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 93: 125–138 [DOI] [PubMed] [Google Scholar]
- Dobson CM 2003. Protein folding and misfolding. Nature 426: 884–890 [DOI] [PubMed] [Google Scholar]
- Dollins DE, Warren JJ, Immormino RM, Gewirth DT 2007. Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol Cell 28: 41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcock AH 2006. Molecular simulations of cotranslational protein folding: Fragment stabilities, folding cooperativity, and trapping in the ribosome. PLoS Comput Biol 2: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, Minton AP 2006. Protein aggregation in crowded environments. Biol Chem 387: 485–497 [DOI] [PubMed] [Google Scholar]
- England JL, Lucent D, Pande VS 2008. A role for confined water in chaperonin function. J Am Chem Soc 130: 11838–11839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells SA, Meyer AS, Yam AY, Roobol A, Miao YW, Shao YL, Carden MJ, Skach WR, Frydman J, Johnson AE 2005. The cotranslational contacts between ribosome-bound nascent polypeptides and the subunits of the hetero-oligomeric chaperonin TRiC probed by photocross-linking. J Biol Chem 280: 28118–28126 [DOI] [PubMed] [Google Scholar]
- Falsone SF, Leptihn S, Osterauer A, Haslbeck M, Buchner J 2004. Oncogenic mutations reduce the stability of SRC kinase. J Mol Biol 344: 281–291 [DOI] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431: 590–596 [DOI] [PubMed] [Google Scholar]
- Fersht AR 2008. From the first protein structures to our current knowledge of protein folding: Delights and scepticisms. Nat Rev Mol Cell Biol 9: 650–654 [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247 [DOI] [PubMed] [Google Scholar]
- Fritah S, Col E, Boyault C, Govin J, Sadoul K, Chiocca S, Christians E, Khochbin S, Jolly C, Vourc’h C 2009. Heat-shock factor 1 controls genome-wide acetylation in heat-shocked cells. Mol Biol Cell 20: 4976–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J 2001. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU 1994. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature 370: 111–117 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Hayashida N, Katoh T, Oshima K, Shinkawa T, Prakasam R, Tan K, Inouye S, Takii R, Akira Nakai 2009. A novel mouse HSF3 has the potential to activate non-classical heat shock genes during heat shock. Mol Biol Cell DOI: 10.1091/mbc.E1009-1007-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Giordanetto F, Kroemer G 2009. Targeting HSP70 for cancer therapy. Mol Cell 36: 176–177 [DOI] [PubMed] [Google Scholar]
- Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ 1992. A cytoplasmic chaperonin that catalyzes β-actin folding. Cell 69: 1043–1050 [DOI] [PubMed] [Google Scholar]
- Gavin A-C, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, Cruciat C-M, et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E 1998. A novel protein complex promoting formation of functional α- and γ-tubulin. EMBO J 17: 952–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez AV, Galleguillos D, Maass JC, Battaglioli E, Kukuljan M, Andres ME 2008. CoREST represses the heat shock response mediated by HSF1. Mol Cell 31: 222–231 [DOI] [PubMed] [Google Scholar]
- Grenert JP, Sullivan WP, Fadden P, Haystead TA, Clark J, Mimnaugh E, Krutzsch H, Ochel H-J, Schulte TW, Sausville E, et al. 1997. The amino-terminal domain of heat shock protein 90 (Hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates Hsp90 conformation. J Biol Chem 272: 23843–23850 [DOI] [PubMed] [Google Scholar]
- Gromiha MM, Selvaraj S 2004. Inter-residue interactions in protein folding and stability. Prog Biophys Mol Biol 86: 235–277 [DOI] [PubMed] [Google Scholar]
- Grossman AD, Erickson JW, Gross CA 1984. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell 38: 383–390 [DOI] [PubMed] [Google Scholar]
- Guettouche T, Boellmann F, Lane WS, Voellmy R 2005. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Guettouche T, Fenna M, Boellmann F, Pratt WB, Toft DO, Smith DF, Voellmy R 2001. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem 276: 45791–45799 [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ 2007. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat Rev Mol Cell Biol 8: 101–112 [DOI] [PubMed] [Google Scholar]
- Hartl FU 1996. Molecular chaperones in cellular protein folding. Nature 381: 571–580 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M 2002. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M 2009. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Hayer-Hartl M, Minton AP 2006. A simple semiempirical model for the effect of molecular confinement on the rate of protein folding. Biochemistry 45: 13356–13360 [DOI] [PubMed] [Google Scholar]
- Herendeen SL, VanBogelen RA, Neidhardt FC 1979. Levels of major proteins of Escherichia coli during growth at different temperatures. J Bacteriol 139: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessling M, Richter K, Buchner J 2009. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90.[see comment]. Nat Struct Mol Biol 16: 287–293 [DOI] [PubMed] [Google Scholar]
- Heydari AR, You S, Takahashi R, Gutsmann-Conrad A, Sarge KD, Richardson A 2000. Age-related alterations in the activation of heat shock transcription factor 1 in rat hepatocytes. Exp Cell Res 256: 83–93 [DOI] [PubMed] [Google Scholar]
- Hillger F, Hanni D, Nettels D, Geister S, Grandin M, Textor M, Schuler B 2008. Probing protein-chaperone interactions with single-molecule fluorescence spectroscopy. Angew Chem Int Ed Engl 47: 6184–6188 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification.[erratum appears in J Biol Chem 2002 Jul 19;277(29):26708]. J Biol Chem 276: 40263–40267 [DOI] [PubMed] [Google Scholar]
- Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wuthrich K 2005. Direct NMR observation of a substrate protein bound to the chaperonin GroEL. Proc Natl Acad Sci 102: 12748–12753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Chapman E, Farr GW 2007. Two families of chaperonin: Physiology and mechanism. Annu Rev Cell Dev Biol 23: 115–145 [DOI] [PubMed] [Google Scholar]
- Hundley HA, Walter W, Bairstow S, Craig EA 2005. Human Mpp11 J protein: Ribosome-tethered molecular chaperones are ubiquitous. Science 308: 1032–1034 [DOI] [PubMed] [Google Scholar]
- Hwang M, Moretti L, Lu B 2009. HSP90 inhibitors: Multi-targeted antitumor effects and novel combinatorial therapeutic approaches in cancer therapy. Curr Med Chem 16: 3081–3092 [DOI] [PubMed] [Google Scholar]
- Jahn TR, Radford SE 2005. The Yin and Yang of protein folding. FEBS J 272: 5962–5970 [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM 2006. Real-time observation of trigger factor function on translating ribosomes. Nature 444: 455–460 [DOI] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122: 209–220 [DOI] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K 2006. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol 8: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Kon M, Cuervo AM 2009. Chaperone-mediated autophagy in health and disease. FEBS Lett DOI: 10.1016/j.febslet.2009.1012.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16: 589–597 [DOI] [PubMed] [Google Scholar]
- Lakshmipathy SK, Tomic S, Kaiser CM, Chang HC, Genevaux P, Georgopoulos C, Barral JM, Johnson AE, Hartl FU, Etchells SA 2007. Identification of nascent chain interaction sites on trigger factor. J Biol Chem 282: 12186–12193 [DOI] [PubMed] [Google Scholar]
- Landick R, Vaughn V, Lau ET, VanBogelen RA, Erickson JW, Neidhardt FC 1984. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell 38: 175–182 [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU 1992. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683–689 [DOI] [PubMed] [Google Scholar]
- Leroux MR, Fandrich M, Klunker D, Siegers K, Lupas AN, Brown JR, Schiebel E, Dobson CM, Hartl FU 1999. MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J 18: 6730–6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JIJ, Pimkina J, Frank A, Murphy ME, George DL 2009. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell 36: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinthal C 1968. Are there pathways for protein folding? J Chimie Phys Physico-Chimie Biol 65: 44–45 [Google Scholar]
- Lin Z, Madan D, Rye HS 2008. GroEL stimulates protein folding through forced unfolding. Nat Struct Mol Biol 15: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg MO, Oliveberg M 2007. Malleability of protein folding pathways: A simple reason for complex behaviour. Curr Opin Struct Biol 17: 21–29 [DOI] [PubMed] [Google Scholar]
- Lis J, Wu C 1993. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell 74: 1–4 [DOI] [PubMed] [Google Scholar]
- Lu JL, Deutsch C 2005. Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol 12: 1123–1129 [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S, Roth JR, Fredriksson A, Nystrom T, Berg OG, Andersson DI 2005. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nature Genet 37: 1376–1379 [DOI] [PubMed] [Google Scholar]
- Martin-Benito J, Boskovic J, Gomez-Puertas P, Carrascosa JL, Simons CT, Lewis SA, Bartolini F, Cowan NJ, Valpuesta JM 2002. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J 21: 6377–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hackert E, Hendrickson WA 2009. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Rudiger S, Bukau B 2000. Molecular basis for interactions of the DnaK chaperone with substrates. Biol Chem 381: 877–885 [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J 2007. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131: 121–135 [DOI] [PubMed] [Google Scholar]
- Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, et al. 2008. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J 27: 1622–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz F, Hoffmann A, Rutkowska A, Zachmann-Brand B, Bukau B, Deuerling E 2006. The C-terminal domain of Escherichia coli trigger factor represents the central module of its chaperone activity. J Biol Chem 281: 31963–31971 [DOI] [PubMed] [Google Scholar]
- Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J 2003. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 113: 369–381 [DOI] [PubMed] [Google Scholar]
- Morimoto RI 1998. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Morimoto RI 2008. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev 22: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU 2000. Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci 97: 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Uehara Y, Yamamoto C, Fukazawa H, Mizuno S 1991. Induction of Hsp 72/73 by herbimycin A, an inhibitor of transformation by tyrosine kinase oncogenes. Exp Cell Res 195: 338–344 [DOI] [PubMed] [Google Scholar]
- Neckers L 2007. Heat shock protein 90: The cancer chaperone. J Biosci 32: 517–530 [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU 1997. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388: 343–349 [DOI] [PubMed] [Google Scholar]
- Onuchic JN, Wolynes PG 2004. Theory of protein folding. Curr Opin Struct Biol 14: 70–75 [DOI] [PubMed] [Google Scholar]
- Ostling P, Bjork JK, Roos-Mattjus P, Mezger V, Sistonen L 2007. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem 282: 7077–7086 [DOI] [PubMed] [Google Scholar]
- Otto H, Conz C, Maier P, Wolfle T, Suzuki CK, Jeno P, Rucknagel P, Stahl J, Rospert S 2005. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci 102: 10064–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C 2006. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem 75: 271–294 [DOI] [PubMed] [Google Scholar]
- Pellecchia M, Montgomery DL, Stevens SY, Vander Kooi CW, Feng HP, Gierasch LM, Zuiderweg ERP 2000. Structural insights into substrate binding by the molecular chaperone DnaK. Nat Struct Biol 7: 298–303 [DOI] [PubMed] [Google Scholar]
- Powers MV, Workman P 2007. Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett 581: 3758–3769 [DOI] [PubMed] [Google Scholar]
- Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH 1997. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90: 65–75 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Rattan SI 1995. Ageing–a biological perspective. Mol Aspects Med 16: 439–508 [DOI] [PubMed] [Google Scholar]
- Rattan SI 1998. Repeated mild heat shock delays ageing in cultured human skin fibroblasts. Biochem Mol Biol Int 45: 753–759 [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Buchberger A, Bukau B 1997. Interaction of Hsp70 chaperones with substrates. Nat Struct Biol 4: 342–349 [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S 1998. Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342 [DOI] [PubMed] [Google Scholar]
- Sandqvist A, Bjork JK, Akerfelt M, Chitikova Z, Grichine A, Vourch C, Jolly C, Salminen TA, Nymalm Y, Sistonen L 2009. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol Biol Cell 20: 1340–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, Hartl FU 2004. Cellular toxicity of polyglutamine expansion proteins: Mechanism of transcription factor deactivation. Mol Cell 15: 95–105 [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I 2000. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101: 199–210 [DOI] [PubMed] [Google Scholar]
- Schlunzen F, Wilson DN, Tian PS, Harms JM, McInnes SJ, Hansen HAS, Albrecht R, Buerger J, Wilbanks SM, Fucini P 2005. The binding mode of the trigger factor on the ribosome: Implications for protein folding and SRP interaction. Structure 13: 1685–1694 [DOI] [PubMed] [Google Scholar]
- Shamovsky I, Nudler E 2008. New insights into the mechanism of heat shock response activation. Cell Mol Life Sci 65: 855–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Chakraborty K, Mueller BK, Astola N, Tang YC, Lamb DC, Hayer-Hartl M, Hartl FU 2008. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133: 142–153 [DOI] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI 1998. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev 12: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA 2006. Structural analysis of E-coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 127: 329–340 [DOI] [PubMed] [Google Scholar]
- Shimon L, Hynes GM, McCormack EA, Willison KR, Horovitz A 2008. ATP-induced allostery in the eukaryotic chaperonin CCT is abolished by the mutation G345D in CCT4 that renders yeast temperature-sensitive for growth. J Mol Biol 377: 469–477 [DOI] [PubMed] [Google Scholar]
- Siegers K, Waldmann T, Leroux MR, Grein K, Shevchenko A, Schiebel E, Hartl FU 1999. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J 18: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I 2000. Structure of the molecular chaperone prefoldin: Unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell 103: 621–632 [DOI] [PubMed] [Google Scholar]
- Sittler A, Lurz R, Lueder G, Priller J, Hayer-Hartl MK, Hartl FU, Lehrach H, Wanker EE 2001. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Hum Mol Genet 10: 1307–1315 [DOI] [PubMed] [Google Scholar]
- Soti C, Csermely P 2000. Molecular chaperones and the aging process. Biogerontology 1: 225–233 [DOI] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J 2004. Mechanism of the eukaryotic chaperonin: Protein folding in the chamber of secrets. Trends Cell Biol 14: 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP 1997. Crystal structure of an Hsp90-geldanamycin complex: Targeting of a protein chaperone by an antitumor agent. Cell 89: 239–250 [DOI] [PubMed] [Google Scholar]
- Taldone T, Gozman A, Maharaj R, Chiosis G 2008. Targeting Hsp90: small-molecule inhibitors and their clinical development. Curr Opin Pharmacol 8: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J 2006. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol 8: 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Chakraborty K, Hartl FU, Hayer-Hartl M 2008. Essential role of the chaperonin folding compartment in vivo. EMBO J 27: 1458–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Hayer-Hartl M, Hartl FU 2007. SnapShot: molecular chaperones, Part II. Cell 128: 412. [DOI] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M 2006. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125: 903–914 [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU 1999. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97: 755–765 [DOI] [PubMed] [Google Scholar]
- Thirumalai D, Lorimer GH 2001. Chaperonin-mediated protein folding. Annu Rev Biophys Biomolec Struct 30: 245–269 [DOI] [PubMed] [Google Scholar]
- Thulasiraman V, Yang CF, Frydman J 1999. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J 18: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS 2009. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459: 668–671 [DOI] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ 1998. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93: 863–873 [DOI] [PubMed] [Google Scholar]
- Valpuesta JM, Martin-Benito J, Gomez-Puertas P, Carrascosa JL, Willison KR 2002. Structure and function of a protein folding machine: The eukaryotic cytosolic chaperonin CCT. FEBS Lett 529: 11–16 [DOI] [PubMed] [Google Scholar]
- Voellmy R, Boellmann F 2007. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol 594: 89–99 [DOI] [PubMed] [Google Scholar]
- Vujanac M, Fenaroli A, Zimarino V 2005. Constitutive nuclear import and stress-regulated nucleocytoplasmic shuttling of mammalian heat-shock factor 1. Traffic 6: 214–229 [DOI] [PubMed] [Google Scholar]
- Wandinger SK, Richter K, Buchner J 2008. The Hsp90 chaperone machinery. J Biol Chem 283: 18473–18477 [DOI] [PubMed] [Google Scholar]
- Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS 2002. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell 111: 1027–1039 [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Tessarz P, Mogk A, Bukau B 2005. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol Chem 386: 739–744 [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323: 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide SD, Morimoto RI 2005. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem 280: 33097–33100 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist S 2009. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Targets 13: 469–478 [DOI] [PubMed] [Google Scholar]
- Woolhead CA, McCormick PJ, Johnson AE 2004. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116: 725–736 [DOI] [PubMed] [Google Scholar]
- Wright CF, Teichmann SA, Clarke J, Dobson CM 2005. The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438: 878–881 [DOI] [PubMed] [Google Scholar]
- Xu Y, Lindquist S 1993. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci 90: 7074–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZH, Horwich AL, Sigler PB 1997. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388: 741–749 [DOI] [PubMed] [Google Scholar]
- Xu D, Zalmas LP, La Thangue NB 2008. A transcription cofactor required for the heat-shock response. EMBO Rep 9: 662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H 1992. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature 358: 245–248 [DOI] [PubMed] [Google Scholar]
- Young JC, Barral JM, Hartl FU 2003. More than folding: Localized functions of cytosolic chaperones. Trends Biochem Sci 28: 541–547 [DOI] [PubMed] [Google Scholar]
- Zhao R, Houry WA 2007. Molecular interaction network of the Hsp90 chaperone system. Adv Exp Med Biol 594: 27–36 [DOI] [PubMed] [Google Scholar]
- Zhu XT, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272: 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SB, Trach SO 1991. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol 222: 599–620 [DOI] [PubMed] [Google Scholar]
- Zou JY, Guo YL, Guettouche T, Smith DF, Voellmy R 1998. Repression of heat shock transcription factor Hsf1 activation by Hsp90 (Hsp90 Complex) that forms a stress-sensitive complex with Hsf1. Cell 94: 471–480 [DOI] [PubMed] [Google Scholar]