Abstract
Proteomics has evolved into an invaluable tool for biomedical research and for research on renal diseases. A central player in the proteomic revolution is the mass spectrometer and its application to analyze biological samples. Our need to understand both the identity of proteins and their abundance has led to improvements in mass spectrometers and their ability to analyze complex tryptic peptide mixtures with high sensitivity and high mass accuracy in a high throughput fashion (such as the LTQ-Orbitrap). It should not be surprising that this occurred coincident with dramatic improvements in our understanding chronic kidney disease (CKD), the mechanisms through which CKD progresses and the development of candidate CKD biomarkers. This review attempts to present a basic framework for the operational components of mass spectrometers, basic insight into how they are used in renal research and a discussion of CKD research that was driven by mass spectrometry.
Keywords: urine, serum, plasma, peptidome, proteome, targeted proteomics
Introduction
Chronic kidney disease (CKD) is not an easily diagnosed and easily treated disease entity. CKD has been defined as a state of reduced renal function that lasts three or more months.(1) The reduced renal function may be characterized by one or more of the following: renal damage with or without reduced glomerular filtration rate (GFR), renal ultra-structural changes, and/or proteinuria. It is often undiagnosed until it has progressed to a moderate stage. The etiology of CKD is diverse and the diagnosis of CKD can be associated with or causally linked to diabetes, hypertension, glomerulonephritis, chronic interstitial nephritis, hereditary diseases such as cystic disease, and secondary glomerulonephritis such as vasculitis.(2) The prevalence of CKD as a disease entity has increased from 10% in 1994 to 16% in 2007.(3–4) It is unclear whether the increased prevalence arises from a sustained increase in the disease incidence or a result of people living longer with increased prevalence of the metabolic syndrome; approximately 70% of CKD has been associated with diabetes and hypertension.(4) An estimated two percent (2%) of patients with CKD will experience an unremitting loss of renal function that leads to end stage renal disease requiring renal replacement therapy (dialysis or renal transplant).(5) Medicare expenditures in 2007 for patients with CKD reached almost $58 billion, accounting for nearly 28% of Medicare spending.(4) The unremitting development of interstitial fibrosis is pathognomonic for progressive renal function loss in CKD patients.(6) The pathology of interstitial fibrosis is complex, variable and a product of hyper-filtration and/or persistent proteinuria that contributes to cellular inflammation and sustained, detrimental paracrine cytokine effects. These data highlighting multiple routes toward the development and the progression of CKD suggest there is a great need for a better understanding of CKD pathogenesis and more sensitive methods to elucidate, detect, and diagnose both its presence and likely outcome to treatment. Several approaches being used to address the study of CKD have developed out of revolutions in mass spectrometric methods to analyze complex protein solutions.
Proteomics is a field that addresses both the qualitative and quantitative analyses of complex protein mixtures. Watershed events in mass spectrometry (MS) and computation methods to analyze large data sets are largely responsible for the successes of large scale proteomic studies. These events stem from developments in a set of core technologies including separation of complex mixtures of proteins and peptides,(7) soft ionization approaches used to characterize biological molecules by mass spectrometry.(8) methods to quantitatively measure the mass of a target analyte with high mass accuracy,(8–13) and advanced computer-assisted data analysis approaches capable of handling complex data sets.(14) Proteomics has evolved into a high-throughput, analytical discipline largely reliant on highly sensitive mass spectrometric methods to analyze complex biological sample sets. These open-ended, hypothesis generating approaches, when appropriately designed and interpreted, are well suited to the study of the cellular mechanisms leading to and propagating CKD. In support of this position, we will review here some basic elements of mass spectrometers and selected cases highlighting their contribution to our understanding of the natural history of CKD. These methods will be discussed to address how improvements in mass spectrometry methods have enabled the study of events that contribute to the development and progress of CKD as well as diagnosis of CKD.
Mass spectrometric methods used to advance the study of CKD
Large scale studies of nucleic acids, proteins, and metabolites of intermediary metabolism using high throughput methods have become referenced as “omics” studies. Collectively these omics studies are assisting with significant advances in our larger understanding of CKD and its treatment.(15–17) Given the prominent role of proteins as regulators of cellular responses, those methods such as proteomic methods that provide unbiased sources of qualitative and quantitative protein-centric information have become a central feature of current studies into CKD. MS-based proteomic methods have been applied to the study of CKD to great success. As highlighted by case studies presented later it is apparent that no one MS-based method occupies an exclusive license for CKD research. The caveat here is the understanding of the limitations of MS data acquired with instruments achieving lower and higher resolutions and mass accuracies. To frame out this understanding we will first address some of the fundamental characteristics of the MS approaches used by various laboratories. We will then address case studies that highlight the recent contributions of liquid chromatography (LC)-MS or LCMS, including LC-electrospray (ESI)-MS or LC- ESI-MS (15–18) and matrix assisted laser desorption ionization (MALDI)-time of flight (TOF) MS or LC-MALDI-TOF MS,(19) and of capillary electrophoresis (CE)-MS or CEMS methods toward advancing our understanding of early features of renal diseases(20–22). These features include the initiating mechanistic processes that lead to CKD, understanding the differences of renal diseases leading to later stages of CKD, and features of renal physiology that lend toward successful transplantation outcomes.
Fundamentals of mass spectrometric methods used in CKD Research
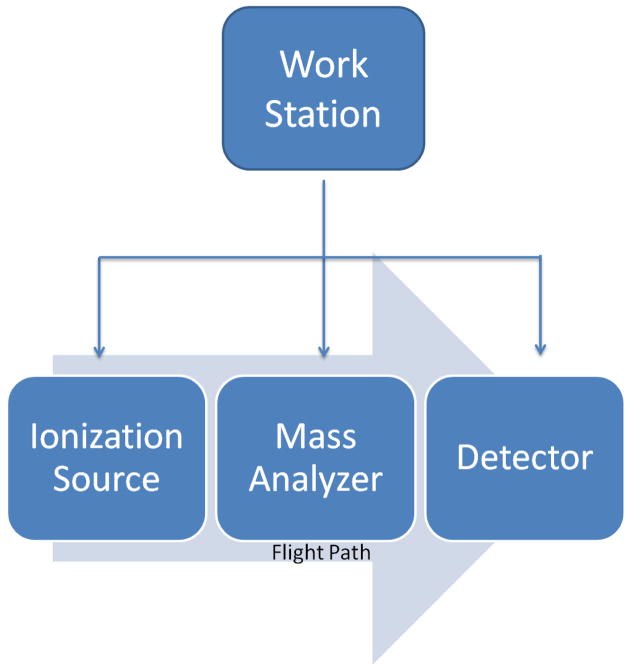
MS is a technique whereby the mass to charge ratio (m/z) of a gas phase ion is determined; essentially making it a particle counting method that yields knowledge of the particle s mass and charge. The mass spectrometer is functionally defined by the sample ionization source, the mass analyzer, and the detector. (Figure 1) The type and quality of the MS data acquired are affected by the quality of the sample as well as the individual components of the mass spectrometer being used in the study.
Figure 1.
The operational components of a mass spectrometer.
A fundamental requirement for MS experiments is that the biomolecules to be analyzed must have a charge and must be present in the gas phase. The overarching problem for the analysis of proteins and peptides by MS is their lack of volatility. The energy required for vaporization of most proteins (under normal laboratory conditions) is more than sufficient for combustion (e.g., formation of carbon dioxide) to occur. The principle event that enabled the analysis of biomolecules with MS was the development of soft (non-destructive) methods for protein ionization (converting to the gas phase). The two ionization methods known as matrix assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) have emerged to play important roles in biomedical sciences.(23) The impact of these ionization techniques cannot be understated as evidenced by the awarding of a portion of the 2002 Nobel Prize in Chemistry for their development. The major revolutions in mass spectrometry have since occurred at the level of the mass analyzer.(8, 24–25) The efforts in this revolution have been directed to improve the limits of detection (sensitivity), to improve the ability to assign the m/z, mass for the +1 charge state, with great accuracy and to increase the resolution of the observed signals such that we can reliably distinguish unique peptides that differ by as little as 0.05 Daltons.
Sample ionization source
ESI is a soft ionization technique that is compatible with peptides or proteins dissolved in solution at typical concentration ranges of 10−15 to 10−6 M.(26) The sample forms an aerosol as it is finely sprayed out a capillary (μL/min flow rate) into a concentric sheath of warm inert carrier gas. A high electrical potential is applied between the capillary tip and the counter electrode. Electrostatic forces produce a charge-repulsion at the liquid-gas interface resulting in the formation of micrometer sized droplets. These droplets are attracted to the counter-electrode and during the migration of the droplet toward the counter-electrode, the solvent is evaporated. The proteins and peptides achieve a final protonated (charge) state. The ESI spectrum of a single peptide can thus contain many species of one peptide solely varying by the final number charged groups. Often, the solution is chosen to be acidic, thus the possible ionic species would be the sum of the side chains for histidine, lysine, arginine and free amino termini, which can easily be dozens of charges. A protein with a molecular weight of 60,000 Da and having a range of total charge from +20 to +30 results in an ESI measured m/z of 3,000 to 2,000. The operational range with highest resolution on most mass spectrometers is from 0 m/z to 4,000 m/z. Thus, larger proteins may be accurately analyzed by ESI-MS.
MALDI is a technique for the analysis of biomolecules wherein the analyte must be co-crystallized with a low molecular weight, organic acid of relatively high volatility.(27) The organic acid is typically aromatic and strongly absorbs ultraviolet light in the output range of the laser employed. Following excitation, the organic acid dissipates energy by protonization of analyte and volatilization of the mixture. As opposed to ESI-MS where the sample is continuously introduced into the spectrometer, MALDI analysis is a pulsed technique. Sample introduction is controlled by the rate at which the organic matrix is volatilized which is in turn controlled by the rate at which a UV laser is fired at the sample-matrix co-crystal. Analyte is carried into the plasma plume and volatilized. Unlike ESI, the MALDI process typically produces singly charged ions. The m/z value measured for the ion equals the mass value for the ion observed. Since the firing of the UV laser controls the rate of introduction of ions into the mass analyzer, very little sample is wasted. High sensitivities (sub-femtomole) are achieved via MALDI sample introduction. As compare to ESI, MALDI is more tolerant of salt and buffers. Perhaps the greatest drawback of the MALDI process is that the ionization process is typically only connected to a time-of-flight mass analyzer. But more recently, this sample source has been integrated onto linear ion trap instruments such as the LTQ-Orbitrap (Thermo Fisher, Inc).
Two characteristics obtained from MS experiments that are used in subsequent data analyses include the m/z value and some quantitative measure of peptide abundance; either the number of times a peak or spectrum was observed or the area under the curve for an extracted ion chromatogram. Two key parameters that contribute to the level of confidence in the assigned m/z value are the accuracy of the assigned m/z value and the signal resolution achieved during the survey scan.(28) The mass accuracy itself is highly dependent on the signal resolution achieved. Accuracy is a value that provides a measure of the error in the determination of the m/z value for any given ion. Unit listing for accuracy can be on a fixed scale (Dalton; Da) or on a proportional scale (parts per million; ppm). A low numerical value for error (e.g. ±25ppm or ±0.025Da at 1,000 m/z) implies a high degree of confidence in the value measured for an ion. The ability to provide the database search algorithms and bioinformatics software with m/z values determined with high accuracy is paramount for success. The ability to resolve or disperse the MS signals of multiple ions of near-like m/z values (so called isobaric species) allows for the accurate determination of the m/z value for the monoisotopic ion. The signal resolution for the mass spectrum is calculated for each ion peak using the formula m/Δ m. In this formula, m is the integer mass of the ion and Δ m is the mass difference between the two ions where the resolution is being calculated. The m/z value for the monoisotopic ion and each of the daughter or grand-daughter fragments are also used to establish the ion lists and interpreted to assign identity.
Mass analyzers
Essentially, there are four fundamental types of mass analyzers in use for proteomics research today. Two of the mass analyzers (time of flight (TOF) and quadrupoles (q)) are beam analyzers while two types of mass analyzers (ion traps, including quadrupole ion traps (Qit) and linear ion traps (LIT or LTQ), and Fourier-transform ion cyclotron resonance (FTICR)) are trapping analyzers. Mass spectrometers built from a composite of analyzers such as quadrupole TOF (qTOF) or LTQ-FTICR are considered “hybrid” instruments. Perhaps one of the most striking differences between these two categories of mass analyzers is their use in various methods to fragment or dissociate analytes (i.e., peptide ions). Beam analyzers have the potential to be used in single MS experiments or in single ion-fragmentation experiments (MS-MS; e.g., MALDI TOF-TOF). Trapping analyzers have the ability to trap an analyte in a magnetic field and through sequential collision and daughter fragment trapping conduct MSn experiments (e.g., MS-MS-MS).
TOF mass analyzers simply measure the time of flight of individual ions and a practical benefit of the TOF mass analyzer is that the size of the ion (m/z value) is theoretically limitless. The quadrupole is a common mass analyzer that is typically used in mass detectors for chromatographic instruments (gas or liquid chromatography and capillary electrophoresis). The mass values measured with a quadrupole vary from 300 to 4,000 with a mass accuracy in the ppm range. The quadrupole ion trap (Qit) functions as a modified quadrupole in it can control the electric field in all co-ordinate space (x, y and z). Instead of guiding the ions down the quadrupole flight path the Qit can apply a third electric field to essentially trap an ion in the quadrupole. Qit instruments have resolution and accuracy similar to quadrupoles but their resolution can be enhanced by application of varying rf voltages. One additional difference is that the mass range is up to about 70,000 m/z. FTICR mass analyzers are conceptually related to sector and to trap analyzers. Both analyzers use a magnetic field to determine the ion m/z value but the FTICR operates at lower kinetic energy ranges. The notable point here is that at such low kinetic energies the ions do not pass through the sector fields but are trapped within the sector field. The ions are trapped and oscillate within a magnetic field with a frequency that is inversely proportional to their m/z value. The resolution of FTICR instruments is typically in the hundreds of thousands as compared to the tens of thousands for state of the art TOF instruments. Additionally, mass accuracies down to the ppb level are obtainable with FTICR instruments. A major drawback of such high resolution and mass accuracy is the cost, exceeding $1 million and requiring significant upkeep costs.
Detectors
A detector is a device that registers an electric signal when an ion strikes the detector surface. Three types of detectors are in common use with mass analyzers- a) electron multipliers, b) photomultipliers, and c) microchannel plates. The last type of detector, the microchannel plate, has a response time of less than 1ns and a very high sensitivity (single ion signal greater than 50mV). The microchannel plates are optimized to have large planar detection areas and therefore a large acceptance volume for collection of ions. Only a few microchannels out of thousands are affected by the collision of a single ion; therefore the simultaneous detection of multiple ions at the same time is possible. This fast detector response/recovery time is important in ion beams like those associated with MALDI sources (where thousands of ions can be generated in nanoseconds).
Fragmentation analysis
Additional information is gained from a protein or peptide identified in a precursor scan by tandem mass spectrometry experiments and fragmentation analysis. In tandem mass spectrometry, essentially two mass spectrometers in series, one must have an established knowledge of the specific ion one wishes to fragment. A precursor scan (the initial MS of the tandem MS) is first performed. The ion of interest is selected and the ion is then isolated from the other ions. The identity of the ions is changed by a chemical reaction; either the m is changed or the z is changed. The internal energy of the ion is increased such that activation barriers are crossed and the ion can fragment. In complex biomolecules, numerous fragmentation pathways exist. The fragmentation products are analyzed with the second survey scan in the mass analyzer; essentially a second dimension beyond the first dimension (the survey scan). An important feature of the tandem mass spectrometer is that the second or n-dimension has better signal to noise because all of the ions originate from the fragmentation of the precursor ion.
Tandem MS is the most basic multidimensional MS experiment possible. Ion-trap mass analyzers in theory can perform MSn experiments where n is the number of times a survey scan is conducted with data acquired and (n-1) is the number of times fragmentation is induced. Tandem MS is a powerful tool and when used as a detector for liquid chromatography the utility is twofold. The detector can be utilized to use the precursor or MS scans as a dependent variable or independent variable (likewise for the MS-MS scan). For example, the MS can be used to sample the eluent from a chromatography experiment. The eluent is continuously sampled and the mass spectrum acquired. If an ion of interest is sought in the LC experiment, individual ion chromatograms can be plotted and used to show elution of specific identities from the column. Conversely, the first MS scan can be used to define the independent variable and the MS-MS spectrum the dependent variable. The MS-MS spectrum can be evaluated for mass losses corresponding to those characteristic of, for example, the loss of phosphate, myristoyl groups, or carbohydrates. Therefore, an ion chromatogram can be constructed to identify peptides or proteins eluting from the LC that contain phosphoryl groups. Those peptides containing specific post-translational-modifications (PTM s) can be selected for amino acid sequence analysis and potentially amino acid sequence tagging. When beam analyzers (TOF or quadrupole) are used for tandem mass spectrometers, the ion fragmentation is tandem-in-space. Tandem-in-space requires a distinct analyzer or a distinct ionization point for the two stages of ion formation. These beam analyzers have an advantage over the trap analyzers in that they are able to perform parent-ion scans or constant neutral loss scans. Trap analyzers are tandem-in-time analyzers and examples of these are the quadrupole ion traps and the FTICR s. Tandem-in-time instruments have the advantage of being able to perform MSn where n ≥ 2 and all using a single mass analyzer. Tandem-in-time analyzers generally have better or higher MS-MS efficiencies since the ability to contain/trap the precursor ions yields more fragment ions.
Hybrid instruments
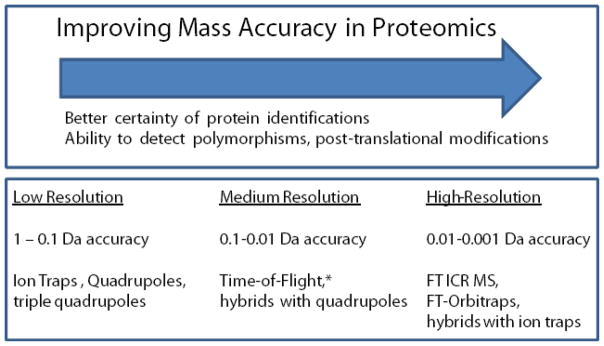
A new set of instruments have emerged to play the lead role in high sensitivity, high mass accuracy proteomic experiments. These instruments are hybrid instruments that merge components providing high sensitivity (such as the linear ion trap) with high mass accuracy (such as the FTICR or the Thermo Orbitrap) and augment traditional approaches to fragmentation (collision induced dissociation, CID) with new approaches (such as electron-capture dissociation (ECD) (29–33) and electron-transfer dissociation (ETD))(34–37) that provide richer fragmentation spectra and preserve post-translational modifications. Unlike the lower mass accuracies of the triple quadrupole and the three dimensional ion traps, these newer instruments are associated with improved peptide identifications and elucidations of post-translational modifications. (Figure 2) Due to its reduced upfront cost and upkeep the LTQ-Orbitrap has evolved into a preferred tool for high throughput proteomics experiments. The basis for the Orbitrap s success relies on its ability to trap ions, confine them to orbit around a central spindle-like electrode and oscillate harmonically along the electrodes axis with a frequency that is characteristic of their m/z values. This oscillation at a characteristic frequency can be Fourier transformed with very high signal resolution and compiled into mass spectra. The LTQ-Orbitrap has been successfully used for bottom up, middle down and top down shotgun proteomics experiments.
Figure 2.
A general overview of the mass accuracies association with modern MS mass analyzers and the general effect it has on the identification of peptide sequence and post translational modifications. (Reproduced with permission from (28).)
Quantitative LCMS approaches
Early in the twenty first century two quantitative mass spectrometric approaches competed to become the optimal method used to gain information on protein expression levels. One approach (labeling) requires the incorporation of a chemical label into the proteome (such as the SILAC method to incorporate heavy isotopes of carbon and or nitrogen into cells in culture) (13) or derivatized onto the trypsinized proteome (such as with isobaric labeling reagents called iTRAQ reagents).(38) The second approach (label-free) is a spectral counting approach that seeks to compare the numbers of observations for a particular peptide from experiment to experiment.(39) The SILAC method has emerged to play a prominent role for the analysis of eukaryotic cells in culture and the study of how they respond to external stimuli. This method is easy and straightforward. The most basic approach is the culture of two populations of cells (one normal and one treated) where one population is cultured in a medium that contains a 'light' amino acid (normal isotopic abundances of carbon, nitrogen and oxygen) and the other in a medium that contains a selected 'heavy' amino acid (the amino acid contains a group of atoms enriched for heavier isotopes such as 13C instead of 12C, or 15N instead of 14N). At the end of the treatment period, cells from the control and treated cultures are trypsinized, mixed 1:1, lysed and used for a shotgun type experiment. Incorporation of the heavy amino acid into a tryptic peptide introduces a known mass shift as compared with the peptide containing the light amino acid. A commonly selected amino acid for these experiments is arginine enriched for six 13C or six 13C plus four 15N (leading to a mass shift of +6Da or +10Da). The integrated signal area for each peptide is converted to a ratio and all ratios for peptides from a common protein are averaged to determine the expressional change with the treatment.
With the broad implementation of hybrid instruments such as the LTQ-Orbitrap and accurate mass measurements, the spectral counting method has developed into a routine method to perform proteome profiling and biomarker discovery work. Early studies into the observed signal response of peptides in complex mixtures demonstrated a linear correlation of MS/MS spectral values with peptide or protein concentration.(40–41) These studies have been used to establish the framework for effective quantitative analysis of LC-MS/MS data. Spectral counting uses the number of unique or total MS/MS spectra that match to each identified protein in a selected database. Using a spectral counting approach, an estimate for protein abundance termed the protein abundance factor (PAF) has been derived. Here the PAF is defined as the total number of non-redundant spectra (spectral counts) that correlate significantly to each respective candidate protein normalized to the protein s MW (×104). By using the non-redundant spectra (inclusive of charge state), peptides with unusual chromatographic properties such as peptides eluting with poor chromatographic resolution are prevented from biasing the PAF value. Thus biases arising from increased observation of peptides derived from proteins with larger MW are eliminated. By normalizing the spectral counts to the cognate protein s MW, PAF scoring can be used to index the relative abundance of unrelated proteins in the same sample and across multiple samples.
The triple quadrupole (triple quad) played a significant role in the early days of biomolecular MS. That role has since been supplanted by higher mass accuracy TOF instruments and MSn trapping instruments. Recently, the interest in the triple quad has been renewed due to the ability to selectively fragment targeted peptide masses and to gain quantitative peptide information.(42–44) These experiments are referred to as selective reaction monitoring (SRM) or multiple reaction monitoring (MRM) MS experiments. In SRM, the triple quad is programmed to selected targeted m/z values for fragmentation. Based on prior studies of synthetic peptides or empirical studies of tryptic digests of purified proteins, the fragmentation spectrum for each target is established (the fragment m/z values). The selectivity of the triple quad can be used to allow only ions of a specific mass to pass from the first quadrupole to the second quadrupole. The second quadrupole acts as a collision cell to generate fragments of that ion. The third quadrupole is used to survey the resulting fragmentation ions. Effectively the triple quad monitors for the parent, fragments the parent and quantitatively observes for the daughter fragments. Peptides with similar masses to the target (isobaric species) can also be gaited into the triple quad fragmentation chamber, but the lack of the correct daughter fragment masses aids in their exclusion from the quantitative experiment.
For purposes of experiments aimed at confirming candidate biomarkers, the targeted peptide masses can be commercially synthesized as unlabeled (light) and labeled (heavy) peptides. A heavy peptide is defined as a peptide wherein one amino acid is enriched in stable heavy isotopes of carbon (13C) and nitrogen (15N). These peptides can be used to identify and establish a rank ordered list of daughter fragment ions having the highest stability and detection sensitivity. The quantification of the parent peptide is typically derived from multiple fragments (e.g., the three most stable and easily detected fragment ions) of the parent peptide. The quantification for the individual fragment ions is based on the mean for the measured areas under the curve (AUCs) for the detector response to the multiple fragments. Calibration curve can be constructed using solutions composed of combinations of the heavy and light candidate biomarker peptides. The heavy peptide concentration is held constant and the light peptide concentration is varied from low to high concentration. The ordinate (y- axis value) is defined by the ratio of heavy to light ion fragment. The abscissa (x-axis value) is defined by the light peptide concentration. The heavy peptide is be spiked into the patient samples (unknowns) at a fixed concentration and the ratio of the AUC for the spiked (heavy) and endogenous (light) peptide fragments quantified. The concentration of the endogenous peptide can then be based on the average of the measured values for all detected transitions.
High mass accuracy shotgun proteomic experiments to understand transplant rejection
Renal replacement therapy including dialysis and renal transplant are the most common treatments for patients with end stage renal disease (ESRD) and can offer substantial improvements in the quality of life.(5, 45) As of 2007 approximately 368,500 ESRD patients received dialysis (4) and 16,600 patients received renal transplant operations.1 Chronic allograft nephropathy (CAN) is the most prevalent cause of renal transplant failure in the first 10 years post-transplant.(46) While much has been speculated, no single mechanism has evolved to explain CAN. Instead a set of related processes including immune mediated and non-immune mediated processes contribute to the progressive loss of graft function. Several efforts using high throughput proteomic and genomic methods have recently been used to enhance our understanding of why CAN may occur and how we might be able to predict that CAN will occur. The success of these studies can be equally attributed to the assembled teams of experts, the assembled sets of tissue and blood samples, cutting edge mass spectrometric approaches used to acquire data, and the Banff classification system used to stratify patient samples. The Banff classification system, established as a working classification of kidney transplant pathology was developed to aid in the accurate and consistent diagnosis of CAN by renal pathologists.(47) The Banff classification parameterizes CAN in quantitative histological terms of interstitial fibrosis and tubular atrophy (IFTA). These conditions are based on the observation that the incidence of IFTA approaches 60% in first year kidney transplants post-transplantation and increases to greater than 90% beyond 10 years post-transplant and strongly correlates with progressive renal function decline.(48) These data suggest that significant insight could be gained into the mechanism of progressive CAN if protein expression patterns (at the transcriptional or translational level) can be paired and correlated to Banff scores of specific renal biopsies.
To help elucidate the mechanisms that might contribute to the progressive nature of CAN a quantitative proteogenomics study of renal transplant biopsies was undertaken. The study was a discovery-driven analysis of protein expression in biopsies collected from transplanted kidneys of patients with mild (b0 and b1) to moderate/severe stages of IFTA (b2,3) and compared to expression patterns of proteins extracted from biopsies of patients with normal renal function and biopsy histology.(17) The b0 samples showing little to no histological evidence of IFTA provided a control or normal kidney transplant biopsy. The b1 samples represented mild IFTA in early stages of progressive tissue injury and IFTA. The b2 represented moderate to severe IFTA with progressive injury and the b3 group represented those with a high risk for graft loss. The grouping of b2 and b3 samples might serve to reduce any subjective influence of assigning samples borderline b2 and b3 samples into increase the sample size for the moderate to severe IFTA affected samples.
A systematic series of shotgun LCMS experiments using the hybrid LTQ-Orbitrap instrument were conducted as the proteomics arm to identify and correlate specific proteins to functional molecular pathways using pathways analysis software. The goal was to identify groups of proteins that would provide diagnostic information (such as activated signaling pathways) and also proteins that might serve as metrics for disease progression. Shotgun experiments are bottom-up proteomics experiments that involve the analysis of very complex peptide mixtures.(49) Typically this approach results in the analysis of tens of thousands of tryptic peptides using multiple chromatographic runs and analysis of aggregated or concatenated MS data sets. One problem with this approach is the large dynamic range for the observed peptides (the abundance of individual peptides) and sensitivity for detection of low abundance peptides. Improvements in chromatographic methods used to separate complex peptide mixtures have improved the depth into the low abundance proteins that can be analyzed.(7) (Note that abundance is a reflection of expression levels of the parent protein and also the natural susceptibility of a peptide to achieve ionization in the mass spectrometer (heavily influenced by amino acid sequence)). Early shotgun experiments were conducted using MS instruments (ion traps) with lower resolving power. Hence, many co-eluting peptides were assigned overlapping m/z values and resulted in an increased number of false positive protein identifications. With the newer hybrid instruments such as the LTQ Orbitrap the greater dispersive capacity of the Orbitrap results in much higher signal resolutions and more accurate mass assignments.(8, 24–25, 34)
A common problem with proteomic experiments especially those that identify thousands of proteins within each data set is simply data overload. One method to address this data congestion in a statistically significant manner is the application of unbiased methods to filter the data. A method that achieves such a goal is a natural language tool that looks for correlation of proteins within a list with known associations within known databases. To address the extensive list of identified proteins within the three groups (in b0, b1, and b2,3), including data reflecting both the expression level and/or presence, the data were analyzed using Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Mountain View, CA). Using the IPA software, lists of submitted protein (or gene) accession numbers submitted for analysis. For each accession number all known functional annotations for each protein are used to assign it to either a biologic function or canonical pathways and hierarchically clustered. The assignments are used to establish a) groups of proteins that are specific components of discrete signaling pathways, b) groups of proteins that are loosely associated with known diseases, or c) groups of proteins that act as specific components of canonical signaling pathways such as the acute phase response. The analysis software predicts hypothetical protein networks based on large databases gathered from curated biologic literature, including physical binding reactions, cis-trans interactions in transcriptional regulation and enzyme-substrate relationships. This analysis software uses computational algorithms to identify local networks consisting of identified proteins and their interactions with each other and with other proteins found in the Ingenuity Systems Knowledge Base. The association scoring is probability based (negative log of the P value) and is calculated for each network according to the fit of the network against the set of identified proteins. For example, a scoring of 12 is equivalent to (−log (1/(1 × 10P))) where P equals 12. Therefore a scoring of 12 suggests that the probability that a group of identified proteins being randomly identified and associated together in the identified network would be 1 out of 1 × 1012 or 1 out of one trillion. In addition to defining networks, the software was used to define global functions and canonical pathways in which the identified proteins might function.
Nakorchevsky et al identified proteins with absolute expression differences (492 unique between b0, b1, and b2,3) and those with relative expression differences (904 differentially expressed between b0, b1, and b2,3).(17) Proteins up-regulated or unique to b0 were enriched for amino acid metabolism, small-molecule biochemistry, and lipid metabolism. This observation may be more a reflection of decreased expression in the b1 and b2,3 samples rather than more in the b0 samples. Those proteins with increased expression in b1 and b2,3 tissue digests reflected cellular distress/tissue injury and represented general biologic functions such as cancer and gastrointestinal, immunologic, and metabolic disease. The association of these proteins into canonical metabolic or signaling pathways for b0 represented typical pathways of intermediary metabolism. But in the analysis of b1 and b2,3 expression profiles, the pathways that were assembled were enriched for proteins belonging to phosphatidylinositol-3 kinase (PI3K)/AKT signaling, acute-phase response signaling, complement system, actin cytoskeleton signaling, fibrosis, and integrin signaling. Moreover, the cluster analysis of these pathways suggested that the b1 and b2,3 clusters were enriched for activation of renal apoptosis, necrosis, fibrosis, and cell death. Collectively the data supported a significant role for fibrosis and tissue remodeling in late-stage IFTA. In considering the result of the IPA analysis, the important pathways activated in b2,3 are composed of differentially abundant proteins of the actin cytoskeleton pathway including multiple actins, fibronectin, mitogen-activated protein kinases (MAPK1 and MAP2K1), Rac2, Ras (RHOA and RHOC), vasodilator-stimulated protein, complements 7 and 8, and complement factors B and I. In addition to the collection of proteomic data sets, the authors collected total RNA for the sample samples. These samples were reserved from the original Triazol sample preparation step that yielded the protein used for proteomic analysis. The microarray expression data was correlated with protein expression data using a Pearson correlation analysis. This analysis yielded a correlation coefficient of 0.13. The low or poor correlation of the two data sets was attributed to posttranslational regulation and regulatory degradation of RNA in IFTA.
A separate quantitative LCMS method was employed to validate these protein expression findings. The goal was to identify proteins whose expression was unique to b0 and b2,3 samples. This goal was achieved and 86 proteins unique to b0 samples and 333 unique to b2,3 samples were identified. But for validation studies using SRM methods, a total of 21 abundant b0 proteins and 23 abundant unique b2,3 proteins were selected. The authors used exact criteria to select easily observed peptides whose sequences were uniquely associated with the parent protein. A total of 96 peptides for the 23 b2,3 proteins and 92 peptides for the 21 b0 proteins were synthesized and used for the SRM approach described earlier. In this case, the expression patterns of all b23 proteins and 81% of the b0 proteins were confirmed..
The discovery phase of this work relied on unbiased shotgun style proteomic experiments. In this case many of the unique proteins may exist in other samples but at a concentration low enough that any tryptic peptides were never selected for tandem MS experiments and hence never identified. Conversely, the LCMS methods used in the validation phase represents a biased approach of the SRM method to observe and quantify a peptide in spite of its relative abundance to other proteins. Retrospective analyses of the SRM data were conducted to establish the presence of the unique b2,3 proteins in b0 samples and vice versa. These analyses demonstrated 13 b0 target proteins in b2,3 samples and one b2,3 protein in b0 samples. This highlights a key point that the SRM validation experiment has a 2–3 log orders higher sensitivity than that of the large-scale shotgun method.(44)
While kidney transplants offer a significant improvement in the patient s quality of life, beyond the use of percutaneous biopsies the ability to predict those patients who will suffer progressive allograft dysfunction is problematic. Therefore, the identification of CAN associated biomarkers would be invaluable to the practicing clinician as well as to the biomedical research interested in understanding the causal factors behind the rejections. To achieve a greater understanding of this problem, Kurian et al (16) used DNA microarrays, high sensitivity, high mass accuracy MS methods (LTQ-Orbitrap XL) and bioinformatics to identify genomic and proteomic markers of mild and moderate/severe CAN in peripheral blood cell proteome with the goal of establishing biomarkers of CAN. Two distinct cohorts of kidney transplant patients with biopsy-documented histology were studied. Test Set 1 comprised 42 kidney transplant patients. Test Set 2 comprised 35 kidney transplant patients. A fundamental difference between these samples sets arise from the method of blood collection. Test Set 1 whole blood was collected and peripheral blood lymphocytes (PBL) were isolated using density gradient purification. With Test Set 2, blood was collected in PaxGene tubes. PaxGene tubes are based on the BD Vacutainer tube design and are a self contained method for isolating RNA but not protein. Only samples from Test Set 1 were available for proteomic analysis.
Using methods similar to those of Nakorchevsky (17) Kurian et al used a spectral counting approach to compare shotgun proteomic datasets of PBL protein digests. Similar to Nakorchevsky, the samples from patients with Banff 2 and Banff 3 scored renal biopsies were combined into a b2,3 dataset and iteratively compared with the PBL digests of patients with b0 and b1 scored biopsies. Several observations were drawn from data including 94 proteins were differentially expressed only in b1 samples and 168 proteins differentially expressed only in b2,3 samples. Ninety five proteins were differentially up-regulated in the ITFA positive CAN samples (samples collected from patients with b1 or b2,3 scored biopsies). Bioinformatically these b1 proteins were linked to these were linked to cell death, cell signaling, and post-translational protein modifications. Differentially expressed b2,3 proteins were also linked to cell death, cell signaling, and post-translational protein modifications and linked to cellular morphology, growth and proliferation and signaling via ERK map kinases, acute phase responses, insulin like growth factor 1 signaling pathways, and peroxisome proliferator- activated receptor A/retinoid X receptor activated transcription factor pathways. Proteins that were uniquely upregulated in IFTA samples were associated with immune and inflammatory pathways (signaling via T and B cell receptors, IL4 and JAK/STAT). These proteins were posited to represent biomarkers with 100% sensitivity and specificity for CAN. The question that could not yet be answered was how many biomarkers are necessary to insure a robust diagnostic test. In consideration of both the transcript and protein profiling experiments, over 80 protein/transcript matches for CAN were coordinately observed, thus providing candidate validation based on two independent technologies. The incorporation of the differentially expressed protein and transcript matches was reported to not significantly improve the classifications obtained with the consensus gene expression set alone. While these data are very encouraging that a blood-based biomarker assay for CAN could be developed, the next critical step would be a prospective clinical trial in kidney transplantation with serial blood-monitoring and genome-wide gene expression and proteomic profiling. The questions to be addressed by this study would be to determine if these biomarker panels are as predictive as they are now diagnostic and also to determine if these protein and gene expression signatures could be used to guide management of immuno-suppression regimes for CAN treatment.
LC-MALDI-TOF MS and Biomarker Discovery for Diabetic Kidney Disease and Progressive Renal Function Decline
Biomarkers have potential value to be used diagnostically as a marker for disease or disease progression and also to provide insight into the mechanism of the disease. We were interested in identifying urinary markers of progression of renal function decline in type-1 diabetics with microalbuminuria. As with the successes of previously mentioned studies, the characterization or curation of the clinical samples was paramount. This goal was been greatly aided by the use of well characterized, clinically curated urine samples collected in the First Joslin Study on the Natural History of Microalbuminuria and Type-1 Diabetes. The onset and levels of microalbuminuria as well as the albumin excretion rate was determined in patients who were followed from 1991 to 2007. In 943 patients with normoalbuminuria, new onset microalbuminuria developed in 109 of the patients within four years after the initial evaluation. Sixty one of 86 patients followed until 2007 were included in the analysis based on the following criteria: 1) greater than 8 years of biennial follow-up examinations after the onset of microalbuminuria until 2007 in order to measure serial estimates of the GFR; 2) sufficient stored urine specimens for analysis of peptide components taken within five years of the onset of microalbuminuria. The critical sample selected for analysis was identified as the earliest available urine sample after the documentation of microalbuminuria onset. This study utilized LC-MALDI-TOF MS to study endogenous urinary peptides in a middle-down approach. The strengths of our approach were that a) we were not required to proteolyze our sample, b) the chromatographic separations of our samples were collected on archival MALDI target plates, c) the high throughput nature of the MALDI-TOF MS survey scans, and d) lack of the need to immediately conduct tandem MS experiments following each MS survey scan. The weaknesses of our approach were the moderate sensitivity due to no ion trapping capacity and the moderate mass spectral resolution/mass accuracy of the TOF analyzer as compared to the substantially more accurate mass analyzer of the Orbitrap or FTICR instruments highlighted in the other case studies.
In these studies the low molecular weight proteins found in the urine of type-1 diabetics were isolated using size filtration and solid phase extraction, characterized by LC-MALDI-TOF MS, and the abundance correlated within two patient cohorts: T1DM patients with microalbuminuria who demonstrated stable or age-equivalent loss of renal function (referred to as controls or non-decliners) and patients who demonstrated early progressive loss of renal function (referred to as cases or decliners).(19) The cohort assignment was based on serum cystatin C estimates of the GFR. The GFR in ml/min was approximated numerically by the reciprocal of cystatin C (in mg/L) multiplied by 100 (cC-GFR) and a regression slope fitted to serial measurements of cC-GFR over several years was used to accurately track the trend in renal function over that time. Data available from the Baltimore Aging Study was used to establish the reference distribution for evaluating whether a negative slope or trend in renal function qualified as an abnormal rate of decline (designated early renal function decline or ERFD ). Nineteen subjects (cases) had renal function loss of 3.3% or more per year (slopes ranging from −3.3 to −16.1% per year). The remaining 42 patients without such rapid renal function loss (slopes ranging from +1.9 and −3.2% per year) were designated as controls.
The statistical analysis of the urinary peptide data sets collected from LC-MALDI-TOF MS experiments presented several challenges. To focus effort on the most promising peptides, we imposed several stringent criteria for selecting peptides for further analysis. The first criterion was aimed at reducing the number of peptides observed in a few samples; we eliminated 3364 peptides that were detected in less than 20% of the specimens. While this data reduction was at the expense of eliminating potential true positive biomarkers, we also decreased the number of false positive associations between peptide expressions and early renal function decline. Next we required that there be at least a 50% difference in the frequency of a peptide between case patients and control subjects and that this difference be statistically significant. This had the effect of reducing the curated peptidome from 825 to six. For these six peptides, we compared their urinary abundances using the peptide peak s characteristics. The peak characteristics were defined from the integrated signal area for the peptide isotopic series. Three peptides were present more frequently in urine of case patients in comparison with urine of control subjects, and three were present less frequently in urine of case patients in comparison with urine of control subjects. Interesting, the abundance and detection frequency of two peptides 983.534 and 1190.638 m/z were completely correlated whereas the others were less correlated or not correlated at all among themselves. The association of these peptides with early renal function decline was studied further using logistic regression analysis controlling for the effects of other covariates such as HbA1c and albumin excretion rates. Urinary presence of peptides 983.534, 1190.638, and 1838.851 m/z was strongly and independently associated with presence of ERFD. The adjusted odds ratios (ORs) varied from 4.4 to 4.9 (95% confidence interval [CI] 1.2 to 20.0). Conversely, urinary presence of peptides (i.e., 1841.811, 2195.965, and 2315.018 m/z) was protective against ERFD. Adjusted ORs varied from 0.2 to 0.4. 95% CI for the 1841.811 m/z was less than 1.0 but for the two others slightly above 1, indicating that after adjustment for other covariates, the negative association of these peptides with ERFD had only borderline significance. Analysis of contemporaneous plasma samples from the same patients by similar methods established that the observed differences in these peptides were specific to the kidney and not derived from filtered, differentially abundant plasma peptides. Therefore these peptides were now considered to be candidate biomarkers for early renal function decline in T1DM patients with microalbuminuria.
To increase the value of these biomarkers beyond that of a m/z value whose abundance appeared to be different between clinical cohorts, we conducted tandem MS experiments to gain amino acid sequence information for the six peptides. Full or partial amino acid sequence information would allow identification of the parent protein from which the peptide was proteolyzed and thereby an avenue to explore a role of the candidate biomarker in the pathophysiology of early progressive renal function decline. The three more abundant peptides were fragments of the cadherin-like protein FAT tumor suppressor 2, zona occludins-3 (ZO-3), and inositol pentakisphosphate-2 kinase (IPP2K). The three peptides decreased in the early renal function decline specimens were fragments of extracellular matrix proteins- tenascin- X, α-I (IV) collagen, and α-I (V) collagen. The analysis of the MS/MS data for the 1838.851 m/z peptide, assigned to IPP2K, was consistent with a glycyl-glycyl posttranslational modification to the epsilon amino group of the internal lysine, which would be presumed to result from ubiquitination of the parent protein IPP2K. Immunohistochemical analysis of renal biopsies of diabetics with early and late nephropathy suggested differences in the expression of ZO-3 and IPP2K.
This study achieved the goals of identifying urinary peptides that predict progressive early renal function decline and establishing an association of the observed urinary peptides with protein abundance changes in the renal parenchyma. These peptides reflect changes in both tubular and glomerular protein expression that are associated with the formation of stress granules and may define a new cellular mechanism by which DN is initiated or progresses. Before these biomarkers can have meaningful value beyond discrete observations, the usefulness of these discriminating peptides as biomarkers of diabetes-associated renal function decline must be determined in additional rigorous studies in a larger patient population. These results provide the hope that candidate biomarkers can provide insight into the mechanisms of diabetic kidney disease and progressive CKD.
CE-MS and Biomarker Discovery for CKD
As of 2007, the United States Renal Data System reports 111,000 incident cases of end-stage renal disease primarily resulting from diabetes, hypertension and glomerular nephritis.(50) Routinely, renal damage is indicated either by increased levels of serum protein in the urine (proteinuria or albuminuria) or significant increases in serum creatinine concentration (an estimation of GFR, eGFR) or both. These two methods for diagnosis of renal disease are general observations that do not covey disease specific information. Additionally increase serum creatinine and or proteinuria are late manifestations of renal damage and are inadequate for assisting in the identification of early markers of renal function loss. Percutaneous renal biopsies remain the gold standard for pathologic phenotyping of these renal diseases; despite the invasive nature of a biopsy and the complication frequency, including life threatening complications.(51) Therefore the identification of biomarkers (as defined by the NIH Biomarker Definitions Working Group) that can function as surrogate biomarkers for renal biopsies and disease diagnosis are of great interest.(52) To this end, several groups have collectively pursued the application of CE-MS toward the identification of urinary biomarkers of renal disease.(20–22) CE as a separation modality coupled to a mass spectrometer (CE-MS) is being recognized as a robust analytic platform capable of resolving several thousand different peptides per sample in less than a one hour run (53) with low interference from salt and other interfering species. The CE-MS systems used in the following case studies have been demonstrated to be a urinary protein profiling method with detection limits of ~1 fmol and achieving moderate monoisotopic resolution characteristics; below 25ppm for z +6 or less and below 100ppm for charges greater than +6. The impressive results of this collaboration are as much a result of standardized collection of samples, the depth and encompassing nature of the range of CKD samples collected, the CE-MS method to characterize and select masses for further MS analysis, and the MS-MS methods used to assign amino acid sequences to the selected peptides.
As stated previously, the incidence of CKD and ESRD is increasing perhaps due to an increased prevalence of type-2 diabetes and an aging population. Candidate biomarkers to diagnose CKD will be required to perform with sufficient specificity so as to exclude the effects of aging on the kidney and hopefully changes in renal function can be associated with genetic and environmental factors. Given these points, three recent CE-MS studies (Rossing et al (21), Zürbig et al (54), and Good et al (20)) are together made noteworthy. Zürbig utilized CE-MS methods to identify patterns of prevalent urinary polypeptides in 324 normal individuals aged 2–73. Initially, no peptide profile could be established as differentially abundant in correlation to an age. With a higher ordered analysis by ANOVA, the relative urinary abundance of 325 of more than 5000 peptides, or approximately 6% of the urinary peptidome, was seen to be regulated with age. The most striking and significant changes correlated to changes with the urinary peptidome of subjects between 11 and 18 years of age. In light of this observation, the data for 218 patients (19–73 years) was reanalyzed. Using the same statistic approach, the expression pattern of 49 peptides was correlated to the aging process in adults. A generalized trend in the data was an attenuation of peptides in the urine of the very old. A targeted analysis using tandem MS methods (LTQ-Orbitrap) identified fragments of collagen I-alpha-I, collagen III-alpha-I, fibrinogen β-chain, and psoriasis susceptibility-1 candidate gene-2 protein. Some of these peptides were observed in previous studies of urine samples profiling DN, IgA nephropathy, focal segmental glomerular sclerosis, membranous glomerular nephritis, vasculitis, and minimal change disease. These CKD peptide panels overlapped with 73.5% of aging specific peptides. Re- analysis of the original data with patients segregated into two groups (age 19–30 years old (n=96) and 51–73 years old) identified 13 peptides associated with biological renal age. Most of these urinary peptides associated with the older cohort were also associated with urinary peptides of macroalbuminuric diabetics. These data suggested that the lesions in diabetic nephropathy are similar to aging induced renal lesions.
Similarly, Rossing et al conducted biomarker discovery experiments to detect differences in the urinary peptidome of type-1 diabetics as a function of urinary protein levels. These patient groups included normoalbuminuric patients, microalbuminuric patients and proteinuric patients plus a cohort of age matched controls. Studies were conducted to determine if the urinary peptidome defined candidate biomarkers for early diabetes, early diabetic nephropathy, and diabetic nephropathy in the face of CKD. Few differences were noted for urinary peptide profiles of normal control patients, normoalbuminuric patients and microalbuminuric patients. Large differences, however, were observed in the urinary peptidome of macroalbuminuric patients as compared to the other groups. These observations supported the findings of Zürbig et al. that the urinary peptidome of macroalbuminuric patients is significantly different from that of healthier kidneys. Statistical comparisons of CE-MS data and peptide abundance were achieved using maxT testings (a statistical t-testing approach that corrects for large numbers of simultaneous comparisons). As compared to healthy controls, 40 peptides were associated with early diabetes. The comparison of microalbuminuric patient to normoalbuminuric patients indicated a large set of peptides (n=102) were significantly different between groups. An abbreviated list of peptides (n=65) was found to classify the normoalbuminuric versus macroalbuminuric type-1 diabetic patients with 93% sensitivity and 97% specificity at cross validation. Twenty four of the 102 peptides were identified as fragments of extracellular matrix proteins (collagens I and III), serum proteins (albumin, α-1 anti-trypsin, transthyretin, α-2 HS glycoprotein, serpin peptidase inhibitor, and fibrinogen β-chain), uromodulin, β 2- microglobulin, psoriasis susceptibility-1 candidate gene-2 protein, and membrane associated progesterone receptor component 1. To address candidate biomarkers of diabetic nephropathy in the face of CKD, the biomarker panel developed previously was used to evaluate urine samples from biopsy-proven IgA nephropathy, focal segmental glomerular sclerosis, membranous glomerular nephritis, and minimal change disease. More than two thirds of the CKD patients scored positively for diabetic nephropathy, suggesting that a common mechanism of progressive renal function decline may be applicable for many renal diseases. This also suggests that specific urinary peptides may indicate progressive diabetic renal disease. To address this postulate, the study correlated peptide expression between the non-diabetic urine samples and the urine samples of macroalbuminuria type- 1 diabetic patients applying similar statistical methods (support vector machine-based model, SVM- BM). A total of 17 peptides were required to correctly identify 95% of the diabetic samples and 94% of the non-diabetic renal disease samples. These data might suggest that there are common pathways of renal damage but that small differences in the urinary peptidome imply unique mechanisms in the progression of the disease.
The success of Rossing et al spawned a larger multicenter study assembled to include over 20 centers from Europe, America, and Australia. This study was designed to both assess the practicality of standardized urine collection and sample processing, the ability of CE-MS platform to reproducibly and robustly observe urinary peptides, and to validate the capacity of urinary peptides as biomarkers of CKD. In this report, Good et al uses the urine from healthy control patients (n=379) and the urine from various renal disease patients having biopsy proven disease (n=230) to identify 634 peptides whose differential abundance was statistically significant (p-value <0.05). A portion of these peptides (273 out of 634) were identified using LC MALDI-TOF/TOF MS, LC FT-ICR MS, and LC-LTQ-Orbitrap analyses. As these peptides had a known identity and might then provide some insight into the mechanism of renal function lost, they were used in further analyses as a refined set of candidate markers to test the ability to identify controls versus disease samples. The result of that analysis was 98.7% sensitivity and 100% specificity. Of note, the training set used for this classification contained samples collected at multiple centers. An additional multi-center prospective study was then conducted for confirmatory evidence of these markers to correctly classify disease and normal samples. The collection study used an independent blinded cohort of 114 individuals divided between those with known kidney disease (n=110) and those with no known kidney disease (n=34). Use of the markers to classify the samples resulted in 85.5% sensitivity and 100% specificity. Lastly, these markers were then tested against a patient dataset extracted from a urinary peptide database. All sample data extracted from the database was for patients with no evidence of renal damage (based on clinical history, serum creatinine, or urinary protein). The individuals selected for this study included type-1 and type-2 diabetic patients, untreated HIV patients, patients with benign prostatic hyperplasia, arterial hypertension, and patients with minimal change disease treated with glucocorticoids with complete remission. The goal of this study was to determine how the candidate biomarkers would classify patients with disease but no renal damage. In general these candidate peptide biomarkers, when used as classifiers of CKD, assigned 0 of 29 HIV patients, 1 of 22 type 2 diabetics, 1 of 36 type 1 diabetics, 0 of 34 benign prostatic hyperplasia patients, 1 of 13 patients with arterial hypertension and 0 of 5 patients with minimal change disease in complete remission. Therefore, the specificity of this tool to assign disease controls correctly was 97.8%.
Given the obvious value of these peptides as candidate biomarkers of CKD, the authors had great interest in the origin of the peptides. As indicated earlier, the efforts to identify these peptides involved multiple tandem MS platforms. These peptides were identified as fragments of extracellular matrix, serum and urinary proteins; prominently including fragments of collagen alpha-1 (I) and collagen alpha-1 (III), alpha-1 antitrypsin, collagen alpha-2 (I), uromodulin, albumin, fibrinogen alpha chain, polymeric immunoglobulin receptor, alpha-2 HS-glycoprotein, clusterin, collagen alpha-1 (II), membrane-associated progesterone receptor component 1, osteopontin, sodium/potassium ATPase gamma chain, and tryansthyretin. While CE-MS is a robust and high throughput platform for urinary peptide analysis it has some limitations regarding the resolution and hence the mass accuracy of the acquired data. These limitations, however, were addressed within a small validation cohort by a) use of CE-FT-ICR-MS to assist with accurate mass assignments and calibration of peptide CE migration time, and b) use of high sensitivity, high mass accuracy platforms (FT-ICR MS, LTQ Orbitrap) and moderate mass accuracy platforms (MALDI-TOF/TOF) to aid in identification of targeted peptides. These data and the approach developed holds great hope that methods to identify renal damage in lieu of the use of percutaneous biopsies exist and are within reach.
Summary Conclusion
In summary, MS is a rapidly evolving discipline and has been successfully used to aid with understanding the evolution of CKD and to aid in the identification of candidate serum and urinary biomarkers for diagnosis of CKD. In most cases, the development of meaningful data sets will require a large number of patient samples and in turn will require high throughput LCMS methods. Currently, those approaches that have seen the greatest integration utilize bottom-up or shotgun methods and hybrid instruments like the LTQ-Orbitrap; taking advantage of the Obritrap s mass accuracy and gentle methods of peptide fragmentation that preserve post translational modification. Several challenges for mass spectrometry and include improvements for the analysis of low abundant proteins and peptides in high throughput LCMS assays, for the shotgun-style identification of post- translational modifications, and for the identification of proteins without the need for proteolysis (the middle down or top down approach) in a high throughput setting. As these challenges are successfully addressed, we will see increase use of MS to increase our knowledge behind the pathogenic mechanisms of the etiologic components of CKD.
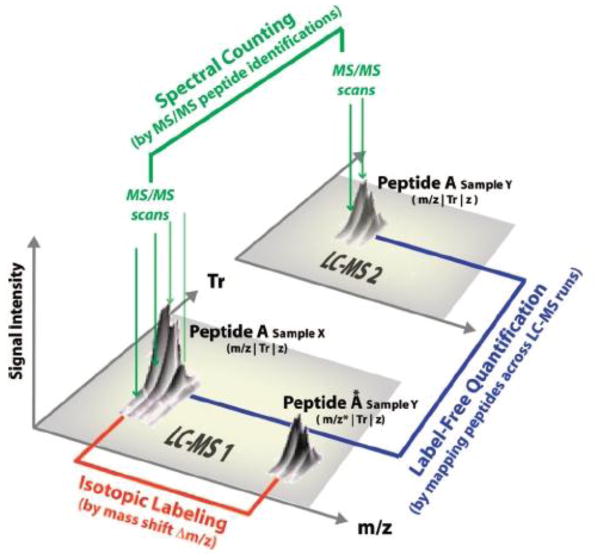
Figure 3.
A graphical representation of LC-MS data used for label and label free quantitative MS experiments. LC-MS1 and LCMS2 represent two difference LCMS experiments. Vertical green lines seen above the peaks in LCMS1 and LCMS 2 represent time dependent survey scans used to acquire MS data. Arrayed gray peaks seen in LCMS1 and LCMS2 represent the isotopic series routinely observed for all peptides. Here the largest peak represents the monoisotopic peak. The two series of grey peaks shown in LCMS1 represents the pair of light (A) and heavy (A*) peptides characterized by a mass difference (Δ m/z) that might be observed in a stable isotope labeling (e.g. SILAC) experiment. Spectral counting is a label-free quantification approach that is based on computing the number of times a peptide (characterized by m/z, MS/MS spectrum, retention time (Tr) and charge state (z)) was successfully identified from tandem MS data and comparing that across all LCMS experiments. A second label free quantification approach is to extract our signal data for peptides soley characterized by m/z, Tr, and z in the absence of MS/MS spectral data. (Reproduced with permission from (55).
Footnotes
The author has no financial and interpersonal relationships to disclose that could be viewed as presenting a potential conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002 Feb;39(2 Suppl 1):S1–266. [PubMed] [Google Scholar]
- 2.Abboud H, Henrich WL. Clinical practice. Stage IV chronic kidney disease. N Engl J Med. 2010 Jan 7;362(1):56–65. doi: 10.1056/NEJMcp0906797. [DOI] [PubMed] [Google Scholar]
- 3.Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS. Prevalence of chronic kidney disease in the United States. JAMA. 2007 Nov 7;298(17):2038–47. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 4.System USRD. USRDS 2009 Annual Data Report: Atlas of End-Stage Renal Disease in the United States. Bethesda, MD: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2009. [Google Scholar]
- 5.Keith DS, Nichols GA, Gullion CM, Brown JB, Smith DH. Longitudinal follow-up and outcomes among a population with chronic kidney disease in a large managed care organization. Arch Intern Med. 2004 Mar 22;164(6):659–63. doi: 10.1001/archinte.164.6.659. [DOI] [PubMed] [Google Scholar]
- 6.Eddy AA, Neilson EG. Chronic kidney disease progression. J Am Soc Nephrol. 2006 Nov;17(11):2964–6. doi: 10.1681/ASN.2006070704. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Fang A, Riley CP, Wang M, Regnier FE, Buck C. Multi-dimensional liquid chromatography in proteomics--a review. Anal Chim Acta. 2010 Apr 7;664(2):101–13. doi: 10.1016/j.aca.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 9.Agron IA, Avtonomov DM, Kononikhin AS, Popov IA, Moshkovskii SA, Nikolaev EN. Accurate mass tag retention time database for urine proteome analysis by chromatography--mass spectrometry. Biochemistry (Mosc) 2010 May;75(5):636–41. doi: 10.1134/s0006297910050147. [DOI] [PubMed] [Google Scholar]
- 10.Brenton AG, Godfrey AR. Accurate Mass Measurement: Terminology and Treatment of Data. J Am Soc Mass Spectrom. 2010 Jun 19; doi: 10.1016/j.jasms.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, 2nd, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005 Jan–Feb;4(1):53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 12.Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics. 2009 Mar;8(3):443–50. doi: 10.1074/mcp.M800258-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002 May;1(5):376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Chalkley RJ, Hansen KC, Baldwin MA. Bioinformatic methods to exploit mass spectrometric data for proteomic applications. Methods Enzymol. 2005;402:289–312. doi: 10.1016/S0076-6879(05)02009-4. [DOI] [PubMed] [Google Scholar]
- 15.Grigoryev YA, Kurian SM, Nakorchevskiy AA, Burke JP, Campbell D, Head SR, Deng J, Kantor AB, Yates JR, 3rd, Salomon DR. Genome-wide analysis of immune activation in human T and B cells reveals distinct classes of alternatively spliced genes. PLoS One. 2009;4(11):e7906. doi: 10.1371/journal.pone.0007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurian SM, Heilman R, Mondala TS, Nakorchevsky A, Hewel JA, Campbell D, Robison EH, Wang L, Lin W, Gaber L, Solez K, Shidban H, Mendez R, Schaffer RL, Fisher JS, Flechner SM, Head SR, Horvath S, Yates JR, Marsh CL, Salomon DR. Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS One. 2009;4(7):e6212. doi: 10.1371/journal.pone.0006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakorchevsky A, Hewel JA, Kurian SM, Mondala TS, Campbell D, Head SR, Marsh CL, Yates JR, 3rd, Salomon DR. Molecular mechanisms of chronic kidney transplant rejection via large-scale proteogenomic analysis of tissue biopsies. J Am Soc Nephrol. 2010 Feb;21(2):362–73. doi: 10.1681/ASN.2009060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck LH, Jr, Bonegio RG, Lambeau G, Beck DM, Powell DW, Cummins TD, Klein JB, Salant DJ. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med. 2009 Jul 2;361(1):11–21. doi: 10.1056/NEJMoa0810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merchant ML, Perkins BA, Boratyn GM, Ficociello LH, Wilkey DW, Barati MT, Bertram CC, Page GP, Rovin BH, Warram JH, Krolewski AS, Klein JB. Urinary peptidome may predict renal function decline in type 1 diabetes and microalbuminuria. J Am Soc Nephrol. 2009 Sep;20(9):2065–74. doi: 10.1681/ASN.2008121233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Good DM, Zurbig P, Argiles A, Bauer HW, Behrens G, Coon JJ, Dakna M, Decramer S, Delles C, Dominiczak AF, Ehrich JH, Eitner F, Fliser D, Frommberger M, Ganser A, Girolami MA, Golovko I, Gwinner W, Haubitz M, Herget-Rosenthal S, Jankowski J, Jahn H, Jerums G, Julian BA, Kellmann M, Kliem V, Kolch W, Krolewski AS, Luppi M, Massy Z, Melter M, Neususs C, Novak J, Peter K, Rossing K, Rupprecht H, Schanstra JP, Schiffer E, Stolzenburg JU, Tarnow L, Theodorescu D, Thongboonkerd V, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics. 2010 Jul 8; doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossing K, Mischak H, Dakna M, Zurbig P, Novak J, Julian BA, Good DM, Coon JJ, Tarnow L, Rossing P. Urinary proteomics in diabetes and CKD. J Am Soc Nephrol. 2008 Jul;19(7):1283–90. doi: 10.1681/ASN.2007091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coon JJ, Zurbig P, Dakna M, Dominiczak AF, Decramer S, Fliser D, Frommberger M, Golovko I, Good DM, Herget-Rosenthal S, Jankowski J, Julian BA, Kellmann M, Kolch W, Massy Z, Novak J, Rossing K, Schanstra JP, Schiffer E, Theodorescu D, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clin Appl. 2008 Jul 10;2(7–8):964. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siuzdak G. The emergence of mass spectrometry in biochemical research. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11290–7. doi: 10.1073/pnas.91.24.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarov A, Scigelova M. Coupling liquid chromatography to Orbitrap mass spectrometry. J Chromatogr A. 2010 Jun 18;1217(25):3938–45. doi: 10.1016/j.chroma.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Heilier JF, Madalinski G, Genin E, Ezan E, Tabet JC, Junot C. Evaluation of accurate mass and relative isotopic abundance measurements in the LTQ-orbitrap mass spectrometer for further metabolomics database building. Anal Chem. 2010 Jul 1;82(13):5490–501. doi: 10.1021/ac100271j. [DOI] [PubMed] [Google Scholar]
- 26.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989 Oct 6;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 27.Hillenkamp F, Karas M. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 1990;193:280–95. doi: 10.1016/0076-6879(90)93420-p. [DOI] [PubMed] [Google Scholar]
- 28.Mann M, Kelleher NL. Precision proteomics: the case for high resolution and high mass accuracy. Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18132–8. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creese AJ, Cooper HJ. Liquid chromatography electron capture dissociation tandem mass spectrometry (LC-ECD-MS/MS) versus liquid chromatography collision-induced dissociation tandem mass spectrometry (LC-CID-MS/MS) for the identification of proteins. J Am Soc Mass Spectrom. 2007 May;18(5):891–7. doi: 10.1016/j.jasms.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper HJ, Hakansson K, Marshall AG. The role of electron capture dissociation in biomolecular analysis. Mass Spectrom Rev. 2005 Mar–Apr;24(2):201–22. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 31.Zubarev RA. Electron-capture dissociation tandem mass spectrometry. Curr Opin Biotechnol. 2004 Feb;15(1):12–6. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Palmblad M, Tsybin YO, Ramstrom M, Bergquist J, Hakansson P. Liquid chromatography and electron-capture dissociation in Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2002;16(10):988–92. doi: 10.1002/rcm.667. [DOI] [PubMed] [Google Scholar]
- 33.Hakansson K, Emmett MR, Hendrickson CL, Marshall AG. High-sensitivity electron capture dissociation tandem FTICR mass spectrometry of microelectrosprayed peptides. Anal Chem. 2001 Aug 1;73(15):3605–10. doi: 10.1021/ac010141z. [DOI] [PubMed] [Google Scholar]
- 34.Wenger CD, McAlister GC, Xia Q, Coon JJ. Sub-part-per-million precursor and product mass accuracy for high-throughput proteomics on an electron transfer dissociation-enabled orbitrap mass spectrometer. Mol Cell Proteomics. 2010 May;9(5):754–63. doi: 10.1074/mcp.M900541-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAlister GC, Berggren WT, Griep-Raming J, Horning S, Makarov A, Phanstiel D, Stafford G, Swaney DL, Syka JE, Zabrouskov V, Coon JJ. A proteomics grade electron transfer dissociation- enabled hybrid linear ion trap-orbitrap mass spectrometer. J Proteome Res. 2008 Aug;7(8):3127–36. doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams DK, Jr, McAlister GC, Good DM, Coon JJ, Muddiman DC. Dual electrospray ion source for electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007 Oct 15;79(20):7916–9. doi: 10.1021/ac071444h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Implementation of electron- transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007 May 15;79(10):3525–34. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007 Feb;7(3):340–50. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 39.Old WM, Meyer-Arendt K, veline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. MolCell Proteomics. 2005;4(10):1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., III Direct analysis of protein complexes using mass spectrometry. NatBiotechnol. 1999;17(7):676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 41.Yates JR, III, Link AJ, Schieltz D. Direct analysis of proteins in mixtures. Application to protein complexes. Methods MolBiol. 2000;146:17–26. doi: 10.1385/1-59259-045-4:17. [DOI] [PubMed] [Google Scholar]
- 42.Lopez MF, Rezai T, Sarracino DA, Prakash A, Krastins B, Athanas M, Singh RJ, Barnidge DR, Oran P, Borges C, Nelson RW. Selected reaction monitoring-mass spectrometric immunoassay responsive to parathyroid hormone and related variants. Clin Chem. 2010 Feb;56(2):281–90. doi: 10.1373/clinchem.2009.137323. [DOI] [PubMed] [Google Scholar]
- 43.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009 Aug 21;138(4):795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009 Jul;27(7):633–41. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meier-Kriesche HU, Ojo AO, Port FK, Arndorfer JA, Cibrik DM, Kaplan B. Survival improvement among patients with end-stage renal disease: trends over time for transplant recipients and wait-listed patients. J Am Soc Nephrol. 2001 Jun;12(6):1293–6. doi: 10.1681/ASN.V1261293. [DOI] [PubMed] [Google Scholar]
- 46.Paul LC. Chronic allograft nephropathy: An update. Kidney Int. 1999 Sep;56(3):783–93. doi: 10.1046/j.1523-1755.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- 47.Mengel M, Sis B, Halloran PF. SWOT analysis of Banff: strengths, weaknesses, opportunities and threats of the international Banff consensus process and classification system for renal allograft pathology. Am J Transplant. 2007 Oct;7(10):2221–6. doi: 10.1111/j.1600-6143.2007.01924.x. [DOI] [PubMed] [Google Scholar]
- 48.Cosio FG, Grande JP, Larson TS, Gloor JM, Velosa JA, Textor SC, Griffin MD, Stegall MD. Kidney allograft fibrosis and atrophy early after living donor transplantation. Am J Transplant. 2005 May;5(5):1130–6. doi: 10.1111/j.1600-6143.2005.00811.x. [DOI] [PubMed] [Google Scholar]
- 49.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001 Dec 1;73(23):5683–90. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 50.Systems URD. USRDS 2007 Annual Data Report: Atlas of Chronic Kidney Disease and End- Stage Renal Disease in the United States. National Institutes of Health NIoDaDaKD; Bethesda, MD: 2007. editor.2007. [Google Scholar]
- 51.Bauer C, Melamed ML, Hostetter TH. Staging of chronic kidney disease: time for a course correction. J Am Soc Nephrol. 2008 May;19(5):844–6. doi: 10.1681/ASN.2008010110. [DOI] [PubMed] [Google Scholar]
- 52.Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001 Mar;69(3):89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 53.Kolch W, Neususs C, Pelzing M, Mischak H. Capillary electrophoresis-mass spectrometry as a powerful tool in clinical diagnosis and biomarker discovery. Mass Spectrom Rev. 2005 doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- 54.Zurbig P, Decramer S, Dakna M, Jantos J, Good DM, Coon JJ, Bandin F, Mischak H, Bascands JL, Schanstra JP. The human urinary proteome reveals high similarity between kidney aging and chronic kidney disease. Proteomics. 2009 Apr;9(8):2108–17. doi: 10.1002/pmic.200800560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008 Jan;7(1):51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]